Abstract
Long non-coding RNAs (lncRNAs) participate in transcription and in epigenetic or post-transcriptional regulation of gene expression. They also have roles in epithelial to mesenchymal transition and in carcinogenesis. Because lncRNAs may also have a role in thyroid cancer progression, we examined a group of thyroid tumors which included papillary thyroid carcinomas and anaplastic thyroid carcinomas to determine the specific lncRNAs that were upregulated during thyroid tumor progression. An RT2 Profiler PCR Array Human Cancer Pathway Finder consisting of 84 lncRNAs (Qiagen) and fresh tissues of normal thyroid, PTCs, and ATCs with gene expression profiling was used to determine genes upregulated and downregulated in ATCs. Two of the most highly upregulated genes, prostate cancer antigen 3 (PCA3) and HOX antisense intergenic RNA myeloid 1 (HOTAIRM1 or HAM-1), were selected for further studies using a thyroid tissue microarray(TMA) with formalin-fixed paraffin-embedded tissues of normal thyroid (NT, n = 10), nodular goiters (NG, n = 10), follicular adenoma (FA, n = 32), follicular carcinoma (FCA, n = 28), papillary thyroid carcinoma (PTC, n = 28), follicular variant of papillary thyroid carcinoma (FVPTC, n = 28), and anaplastic thyroid carcinoma (ATC, n = 10). TMA sections were analyzed by in situ hybridization (ISH) using RNAscope technology. The results of ISH analyses were imaged with Vectra imaging technology and quantified with Nuance® and inForm® software. The TMA analysis was validated by qRT-PCR using FFPE tissues for RNA preparation. Cultured thyroid carcinoma cell lines (n = 7) were also used to analyze for lncRNAs by qRT-PCR. The results showed 11 lncRNAs upregulated and 7 downregulated lncRNAs more than twofold in the ATCS compared with PTCs. Two of the upregulated lncRNAs, PCA3 and HAM-1, were analyzed on a thyroid carcinoma TMA. There was increased expression of both lncRNAs in ATCs and PTCs compared with NT after TMA analysis. qRT-PCR analyses showed increased expression of both lncRNAs in ATCs compared with NT and PTCs. Analyses of these lncRNAs from cultured thyroid carcinoma cell lines by qRT-PCR showed the highest levels of lncRNA expression in ATCs. TGF-β treatment of cultured PTC and ATC cells for 21 days led to increased expression of PCA3 lncRNA in both cell lines by day 14. These results show that the lncRNAs PCA3 and HAM-1 are upregulated during thyroid tumor development and progression and may function as oncogenes during tumor progression.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Thyroid cancer is the most common malignancy of endocrine organs in humans [1]. Papillary thyroid carcinomas (PTCs), which are usually well-differentiated carcinomas, constitute approximately 80% of thyroid cancers. Most of these carcinomas are low-grade lesions with only about 10 to 15% constituting more aggressive carcinomas [1]. The rare poorly differentiated thyroid carcinomas (PDCs) are very uncommon in the USA and make up less than 2% of thyroid cancers, while the also rare undifferentiated or anaplastic thyroid carcinomas (ATCs) are the most aggressive thyroid carcinoma that frequently results in the death of patients in less than a year after diagnosis [1]. Extensive research on morphological classification and immunohistochemical analyses has contributed to the precise diagnosis and to predicting the biological behavior of these carcinomas [1], while molecular analyses have advanced our knowledge about the biological behavior and provided new diagnostic and predictive information about these malignancies [1,2,3].
Non-coding RNAs, which are RNA species that do not code for specific proteins, have been studied in thyroid neoplasms in recent years [4,5,6]. Most of the studies have been with microRNAs (composed of approximately 22 nucleotides). More recently, a growing number of studies have examined long non-coding RNAs (lncRNAs), which are non-coding RNAs greater than 200 nucleotides, in thyroid neoplasms [7,8,9,10]. LncRNAs may function as oncogenes or tumor suppressor genes and have been shown to regulate thyroid tumor progression. Most of the studies of lncRNAs in thyroid cancer have been done on PTCs [5, 10] and there are only a few studies of lncRNAs in ATCs [11,12,13]. Most of the studies of lncRNAs in ATCs have been done in cell lines rather than primary tumors [11,12,13]. To determine specific lncRNAs that are highly expressed in ATC, which may be important for thyroid cancer progression, we used an RT2 Profiler PCR Array Human Cancer Pathway Finder consisting of 84 lncRNAs and fresh tissues of normal thyroid, PTCs, and ATCs with gene expression profiling to determine some of the lncRNA genes upregulated and downregulated in ATCs compared with PTCs. This report analyzes various lncRNAs that are highly expressed in ATCs using formalin-fixed paraffin-embedded specimens from surgically resected materials and various thyroid cell lines.
Materials and Methods
Tissue Microarrays
Tissue microarrays (TMAs) were constructed as described previously [14, 15] from formalin-fixed paraffin-embedded tissues (FFPE), including normal thyroid (NT, n = 10), nodular goiters (NG, n = 10), follicular adenoma (FA, n = 32), follicular carcinoma (FC, n = 28), conventional papillary thyroid carcinoma (PTC, n = 28), follicular variant of papillary thyroid carcinoma (FV, n = 29), and anaplastic thyroid carcinoma (ATC, n = 10). The TMA consisted of triplicate 0.6 mm cores made with a manual tissue microarrayer (Beecher Instruments, Sun Prairie, WI). The NT consisted of tissues from the opposite histologically normal thyroid lobe in patients with follicular or papillary carcinomas.
The study was approved by the Institutional Review Board at the University of Wisconsin–Madison.
In Situ Hybridization
TMAs were probed for prostate cancer antigen 3 (PCA3) (312201) and HOX antisense intergenic RNA myeloid 1 (HAM-1) (427421) expression using the RNAscope 2.5 HD-Brown Manual Assay (Advanced Cell Diagnostics, Newark, CA) as per manufacturer’s recommendations with the following modifications to manufacturer’s protocol, including a step in which antigen retrieval was performed in a Decloaker (Biocare Medical, Pacheco, CA) for 3 min and the probe was incubated overnight at 40 °C with a parafilm cover over sample in a humidified chamber. The probes used included positive control probes hs-PPIB (positive control, 313901), and negative control probe dapB (negative control, 310043) (Advanced Cell Diagnostics). Probe expression levels were visualized with diaminobenzidine (DAB).
Validation Cases
An independent set of cases consisting of whole tissue sections of 10 tall cell variant of PTCs and 7 cases of ATCs was used for ISH with lncRNA PCA3. These cases were quantified after ISH by visually counting the intensity of staining using a scale of 0 to 3+. Zero indicated no staining, 1+ was weak nuclear staining, 2+ was moderate nuclear staining, and 3+ was strong nuclear staining. Non-neoplastic tissues in the adjacent thyroid were also quantified with the same grading scale.
Automated Image Acquisition and Analysis
The stained TMA slides were visualized and analyzed with the Vectra slide scanner and associated software (PerkinElmer, Waltham, MA) as previously described [17]. Briefly, after image acquisition, any core with tissue folding or loss of tissue was excluded for analysis. The InForm 1.4.0 software was used to segment tissue compartments (epithelium vs. non-epithelium) and subcellular compartments (nucleus vs. cytoplasm). PCA3 and HAM-1 expression levels from each sample were quantitated as optical density (OD) per unit area (pixel). Only nuclear signal was used for analysis. Mean lncRNA expression level of each sample from the triplicated cores was used for further analysis.
Total RNA Extraction qPCR and PCR Array
PCR Array
Total RNA was isolated from fresh frozen tissue samples of PTC, ATC, and normal thyroid using TRIzol reagent (Thermo Fisher Scientific, Waltham, MA) according to the manufacturer’s instructions [18, 19], and RNA quality and concentrations were assessed with a NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA). One microgram of total RNA was reverse-transcribed using the RT2 First Strand Kit (330401, Qiagen) and assayed using the RT2 SYBR Green qPCR Mastermix (330504, Qiagen) on the BioRad CFX96 qPCR Thermocycler. The fresh frozen tissues were screened using the RT2 lncRNA PCR Array; Human Cancer Pathway Finder (LAHS-0027ZD-6, Qiagen). Each tissue was applied to an individual plate (one tissue per plate) which was pre-seeded with primers to 84 known cancer-related lncRNAs, 5 reference genes, and internal qPCR QC controls. Results were analyzed using the RT2 qPCR Array Analysis tool available through Qiagen and compared with the results of the control normal thyroid tissue. Results are expressed by the ΔΔ method and up/downregulation was determined as twofold above or below the expression of the normal thyroid tissue. The twofold change cutoff was recommended by the manufacturers and this is standard used in the literature for general gene expression analysis.
Cell Lines
Total RNA was extracted from the ATC cell lines 8505C and THJ-16T (16T) [18, 19] with TRIzol reagent (Thermo Fisher Scientific, Waltham, MA) according to the manufacturer’s instructions, and RNA quality and concentrations were assessed with a NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA). One microgram of total RNA was reverse-transcribed using the All-in-One First Strand cDNA Synthesis Kit (GeneCopoeia, Rockville, MD). RT-qPCR was performed on a CFX96 PCR detection system (Bio-Rad Laboratories, Hercules, CA) using Bullseye EvaGreen qPCR master mix (MIDSCI, St. Louis, MO), normalized to 18S rRNA; relative fold change was determined by the ΔΔ CT method as previously reported [18, 19]. The qPCR primers for 18S included Forward 5′-GTAACCCGTTGAACCCCATT-3′ and Reverse 5′-CCATCCAATCGGTAGTAGCG-3′. The primers used for PCA3 (LPH05631A-200) and HAM-1 (LPH10483A-200) analysis were purchased from Qiagen (Germantown, MD.).
FFPE Tissues
During the construction of the TMA, additional cores of normal thyroid, nodular goiter, PTC, and ATC tissues were taken for qPCR analysis. Total RNA was extracted from the cores using the NucleoSpin Total RNA FFPE Kit purchased from Thermo Fisher Scientific (Madison, WI) according to manufacturer’s instructions. cDNA synthesis and qPCR were performed using the methods described in the cell line section above.
Cell Culture and EMT Induction with TGF-β
The papillary thyroid carcinoma cell line TPC1 and the anaplastic thyroid carcinoma cell line THJ-16T were cultured and treated with TGF-β to induce EMT as previously described [18, 19]. Cells were treated with serum-free media with and without TGF-β (2 ng/ml) for 7, 14, and 21 days [18, 19].
Statistics
Student’s t test was used to analyze lncRNA expression data collected from RNA ISH and RT-qPCR. Two-tailed P values of < 0.05 were considered to be statistically significant. Data are expressed as means ± standard error of the mean (SEM).
Results
Clinicopathologic Findings
The clinicopathologic findings of the cases on the TMA were previously reported [14]. Patients with ATCs had the most aggressive malignancies resulting in the death of several patients. Follow-up of patients from the validation set of cases showed more extensive and lethal disease in patients with ATCS, since all patients with ATCs with available follow-up died of their disease and only one patient with tall cell variant of PTC with available follow-up died of the disease (Table 1).
RT2 Profiler PCR Array Human Cancer Pathway Finder
The RT2 Profiler PCR Array Human Cancer Pathway Finder consisting of 84 lncRNAs with fresh tissues of two PTCs and two ATCs used gene expression profiling to determine genes upregulated and downregulated in ATCs. There were 11 upregulated and 7 downregulated genes by more than twofold for ATCs compared with PTCs (Table 2). Two of these genes, PCA3 and HOTAIRM-1 or HAM-1, were selected for further studies by ISH.
In Situ Hybridization
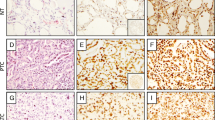
In situ hybridization (ISH) results using the thyroid TMA showed nuclear localization of both PCA3 and HAM-1 (Fig. 1). The positive control probe PPIB showed predominantly nuclear localization with weaker cytoplasmic localization (Fig. 1). The negative control probe resulted in no staining of the tissues (data not shown). Staining among the three replicate cores was fairly consistent. In seven cases on the TMA, one core was lost and the average staining was evaluated with two cores. Quantitative image analysis with Vectra and InForm software showed significantly higher levels of PCA3 in ATC, PTC, and FVPTC compared with NT and significantly higher levels of HAM-1 in ATCs and PTCs compared with NT (Fig. 2). An independent set of samples consisting of whole sections of tall cell variant of PTCs (n = 10) and ATCs (n = 7) was analyzed by ISH and manual counting. The adjacent normal thyroid was also evaluated for PCA3 expression. Comparison of this analysis is shown in Fig. 3 indicating that the ATC cases showed the highest levels of PCA3 expression. Both ATCs and PTCs were significantly higher than the normal thyroid samples.
Representative H&E sections and in situ hybridization (ISH) localization of lncRNS1 HAM-1 and PCA3 in formalin-fixed paraffin-embedded TMA sections (NTs: a–d; PTC: e–h; ATC: i–l). These represent two of eleven upregulated lncRNAs in ATCs. The housekeeping gene peptidyl-prolyl cis-trans isomerase B (PPIB) was used as a RNA preservation marker. Insets show a complete section from a TMA core
Nuclear expression of PCA3 (a) and HAM-1 (b) in normal thyroid tissue and thyroid neoplasms. The highest levels of expression were observed in PTCs and ATCs for both lncRNAs, which is statically significantly higher than that of NTs. * P < 0.05, compared with normal thyroid tissue. Error bars expressed as SEM
Validation set of independent cases to show differences in PCA3 expression determined by RNA scope ISH assay in non-neoplastic thyroid (NT), tall cell variant of papillary thyroid carcinoma (T-PTC) (n = 10), and anaplastic thyroid carcinoma (ATC) (n = 7). The ATC group showed the highest level of expression. * P < 0.05, compared with NT and compared with T-PTC. Error bars expressed as SEM
qRT-PCR Analysis
qRT-PCR analysis of paraffin-embedded sections of thyroid tissues for PCA3 and HAM-1 expression showed the highest levels of both lncRNAs in ATCs, although only the differences in the HAM-1 probe were significantly different (Fig. 4).
Cell Culture Experiments
Various cell lines including PTCs (BcPAP and TPC1) and ATCs (16T, FRO, and cancer stem cell clones A3 and D3) [18] grown in 10% serum (BcPAP, TPC1, 16T, and FRO) or in modified serum-free media (A3 and D3) were analyzed for expression of PCA3 and HAM-1. Both lncRNAs were significantly higher in the ATCs compared with the PTC cell lines (Fig. 5). Treatment of the TPC1 and 16T cell lines with TGF-β led to increase expression of PCA3 lncRNA after days 7 and 14. There was a decrease in PCA3 expression on day 21 in both cell lines (Fig. 6).
Analysis of PCA3 (a) and HAM-1 (b) expression in cultured cell lines. The two papillary thyroid carcinoma cell lines (BCPAP and TPC1) expressed lower levels of PCA3 and HAM-1 compared with the anaplastic thyroid carcinoma cell lines (16T, FRO. A3 and D3). The A3 and D3 are two cancer stem cell lines derived from 16T. * P < 0.05, ** P < 0.01 compared with PTC (BCP and TPC)
lncRNA PCA3 expression during EMT induction. TPC-1 and THJ-16T cells were treated with 2 ng/ml TGF-β in serum-free media to induce EMT over a 21-day period. RT-qPCR results show a significant upregulation in PCA3 in both TPC-1 and THJ-16T cells at day 14 of treatment. However, TPC-1 cells only showed this upregulation at day 14 only, whereas the THJ-16T cells showed upregulation of PCA3 expression up to day 21 as well. * P < 0.05, ** P < 0.01
Discussion
Although there has been a rapidly growing number of studies on lncRNA expression in PTCs [5, 8, 10, 16, 20], there are only a few studies about lncRNA expression in ATCs [11,12,13]. This study shows for the first time that there are a significant number of lncRNAs that are expressed at higher levels in ATCs compared with PTCs. Our study with primary thyroid tissues and cell lines shows that PCA3 and HAM-1 are both highly expressed in ATCs as well as PTCs. The early studies with PCA3 indicated that this lncRNA was highly expressed in prostate cancer and some studies suggested that this lncRNA may have been largely restricted to prostate tissues [21,22,23]. However, recent studies have shown that other tissues express this lncRNA including normal ovarian tissues and ovarian cancer [24] and normal white blood cells and leukemic blood cells [25]. Since PCA3 is overexpressed in prostate and other malignancies, the data suggest that it may function as an oncogene in various cancers. HAM-1 has been shown to play a role in the progression of several types of cancers including pancreatic ductal adenocarcinomas [26], promyelocytic leukemia [27], and glioblastoma multiforme [28]. This lncRNA has also been shown to function as a tumor suppressor gene in colon cancer [29]. This latter finding is of interest, since several studies have shown that lncRNAs such as MALAT1 may function as oncogenes in some tumors and tumor suppressor genes in other tumors [30, 31]. These findings suggest that the interaction of lncRNAs with other molecules such as miRNAs and epigenetic regulatory proteins may determine the oncogenic role of these molecules in different tumor types.
Analysis of the discovery set of thyroid cases from the TMA and the validation set of PTC and ATC cases using whole sections of FFPE tissues showed similar results with increased expression of PCA3 in PTCs and ATCs compared with the normal thyroid tissues. Although the results were quantified differently (Vectra and InForm software vs. manual counting), the trend towards higher expression in the ATC compared with PTC was observed by both methods. Tall cell variant of PTC is a more aggressive variant of well-differentiated thyroid carcinoma and recent studies of genome analysis of a large series of PTCs by the TGCA [32] (have shown that this subtype of PTC has more mutations than other variants of PTC such as classical and follicular variant of PTC, suggesting that they have undergone dedifferentiation and are closer to ATCs). Recent studies have shown that ATCs arise from well-differentiated thyroid carcinomas through the accumulation of key additional genetic abnormalities, many of which have prognostic and possible therapeutic relevance [33]. Nevertheless, there were detectable differences in the expression of PCA3 in tall cell variant of PTC and ATCs. These findings suggest that during epithelial to mesenchymal transition (EMT), there may be increased expression of PCA3 in thyroid carcinomas.
In a recent study of PVT1 expression in thyroid tumors by ISH and qRT-PCR from our group, there was a slightly higher expression of the lncRNA PVT1 in PTCs compared with ATCs by qRT-PCR. However, in our current RT2 Profiler PCR Array Human Cancer Pathway Finder, lncRNA PVT1 was more highly expressed in ATCs compared with the PTCs. This slight difference in expression may be due to the fact that the primers used in the RT2 Profiler PCR Array Human Cancer Pathway Finder were different from those used for our earlier qRT-PCR experiments. To resolve this slight discrepancy, we will need to use the same set of primers for both procedures as is planned for future experiments.
Analysis of various cell lines including those from PTC and ATC as well as cancer stem-like (CSC) lines (A3 and D3) showed the highest levels of PCA3 in the two CSC lines and higher levels in the ATC cell lines compared with the PTC cell lines. Since there have been reports of increased numbers of CSCs in ATCs compared with PTCs [34], our findings suggest that PCA3 may also be associated with increases in CSCs in thyroid cancer. Our results also showed that TGF-β stimulated EMT in PTC and ATC cell lines. Our previous study [18] showed that TGF-β increased the number of thyroid CSCs along with increases in EMT [18, 19]. These observations implicate PCA3 in both EMT and CSC regulation in thyroid carcinoma development and progression.
In summary, these results show that during progression from PTC to ATC, there are specific lncRNAs that are upregulated and a smaller number that are downregulated. Analysis of two of the upregulated lncRNAs, PCA3 and HAM-1, showed the highest expression levels in ATCs and PTCs with higher levels of both lncRNAs in cultured cell lines. These two lncRNAs appear to have regulatory roles in thyroid cancer progression.
References
Lloyd RV, Osamura RY, Kloppel G, Rosai J. WHO Classification of Tumours of Endocrine Organs, 4th Edition, International Agency for Research on Cancer (IARC) Lyon, France, 2017.
Nikiforov YE, Nikiforova MN. Molecular genetics and diagnosis of thyroid cancer. Nat Rev Endocrinol. 2011;7(10):569–580.
Livoisi VA. Papillary thyroid carcinoma: An update. Mod Pathol. 2011 ;24(Suppl 2):S1–S9.
Li X, Wu Z, Fu X, Han W. Long noncoding RNAs: Insights from biological features and functions to diseases. Med Res Rev. 2013;33(3):517–553.
Zhang R, Hardin H, Chen J, Guo Z, Lloyd RV. Non-coding RNAs in Thyroid Cancer Endocr Pathol. 2016;27 :12–20.
Sui F, Ji M, Hou P. Long non-coding RNAs in thyroid cancer: Biological functions and clinical significance. Molecular and Cellular Endocrine. 2018, 469:11–22..
He H, Nagy R, Liyanarachchi S, Jiao H, Li W, Suster S, Kere J, de la Chapelle A. A susceptibility locus for papillary thyroid carcinoma on chormosome 8q24. Cancer Res. 2009;69:625–631.
He H, Li W, Liyanarachchi S, Jendrzejewski J, Srinivas M, Davuluri RV, Nagy R, de la Chapelle A. Genetic predisposition to papillary thryoid carcinoma: Involvement of FOXE1, TSHR, and a Novel lincRNA Gene, PTCSC2. J Clin Endocrinol Metab. 2015;100:E164–E172.
Jendrzejewski J, He H, Randomska HS, Li W, Tomsic J, Liyanarachchi S, Davuluri RV, Nagy R, de la Chapelle A. The polymorphism rs944289 predisposes to papillary thryoid carcinoma through a large intergenic noncoding RNA gene of tumor suppressor type. Proc Natl Acad Sci USA. 2012;109:8646–8651.
Zhang R, Hardin H, Huang W, Chen J, Asioli S, Righi A, Maletta F, Sapino A, Lloyd RV. MALAT1 Long non-coding RNA expression in thyroid tissues: Analysis by in situ hybridizastion and real-time PCR. Endocr Pathology 2017 ;28:7–12,
Zhou Q, Chen J, Feng J, Wang J Long noncoding RNA PVT` modulates thyroid cancer cell proliferation by recruiting EZH2 and regulating thyroid stimulating hormone receptor (TSHR) Tumor Biol 2016;37:3105–3113.
Wang Y, Hou Z. Li D, Long noncoding RNA UCA1 promotes anaplastic thyroid cancer cell proliferation via miR-135a-mediated cmyc activation. Mol Med Rep 2018;18:3068–3076.
Samimi H, Haghpanah V, Irani S, Arefian E, Sohi AN, Fallah P, Soleiman M. Transcript-levelregulation of MALAT1-mediated cell cycle and apoptosis genes using dual MEK/Aurora kinase inhibitor “BI-847325” on anaplastic thyroid carcinoma. Daru M 2019 ,doi.https://doi.org/10.1007/s40199-018-0231-3, 27, 1, 7,.
Buehler D, Hardin H, Shan W, Montemayor-Garcia C, Rush PS, Asioli S, Chen H, Lloyd RV. Expression of epithelial-mesenchymal transition regulators SNAI2 and TWIST1 in thyroid carcinomas. Mod Pathol. 2012; 26:54–61.
Guo Z, Hardin H, Montemayor-Garcia C, Asioli S, Righi A, Maletta F, Sapino A, Lloyd RV. In situ hybridization analysis of miR-146b-5p and miR-21 in thyroid nodules: Diagnostic implications. Endocr Pathol. 2015; 26:157–163.
Zhang R, Hardin H, Huang W, Buehler D, Lloyd RV. Long Non-coding RNA Linc-ROR is upregulated in papillary thyroid carcinoma. Endocr Pathol 29:1–8, 2018.
Yu Q, Hardin H, Chu YH, Rehrauer W, Lloyd RV. Parathyroid neoplasms: Immunohistochemical characterization and long noncoding RNA (lncRNA) expression. Endocrine Path 2019; 30: 96–105.
Hardin H, Guo Z, Shan W, Montemayor-Garcia C, Asioli S, Yu XM, Harrison AD, Chen H, Lloyd RV. The roles of the epithelial-mesenchymal transition marker PRRX1 and miR-146b-5p in papillary thyroid carcinoma progression. Am J Pathol. 2014; 184:2342–2354.
Hardin H, Yu X-M, Harrison AD, Larrain C, Zhang R, Chen J, Chen H, Lloyd RV. Generation of novel thyroid cancer stem-like cell clones effects of resveratrol and valproic acid. Am J Pathol. 2016; 186:1662–1673.
Di W, Li Q, Shen W, Guo H, Zhao S. The long non-coding RNA HOTAIR promotes thyroid cancer cell growth, invasion and migration through the miR-1-CCND2 axis. Am J Cancer Res. 2017; 7: 1298–1309.
Walsh AL, Tuzova AV Bolton EM, Lynch TH, Perry AS. Long noncoding RNAs and prostate carcinogenesis: the missing ‘line’? Trends Mol Med. 2014; 20:428–436.
Wang Y, Liu XJ, Yao XD. Function of PCA3 in prostate tissue and clinical research progress on developing a PCA3 score. Chin J Cancer Res. 2014; 26:493–500.
Clinical prospects of long noncoding RNAs as novel biomarkers and therapeutic targets in prostate cancer. Prostate Cancer Prostatic Dis. 2016; 19:14–20.
Liu Y Zong ZH, Guan X, Wang LL, Zhao Y. The role of long non-coding RNA PCA3 in epithelial ovarian carcinoma tumorigenesis and progression. Gene. 2017; 63342–47.
Saijadi E, Atashi A, Taijrishi MAMH, Saei Z. Gene expression analysis of noncoding PCA3 gene in patients with chronic myeloid leukemia. J Cancer Res Ther. 2018; 14:1079–1082.
Luo Y, He Y, Ye X, Song J, Wang Q, Li Y, Xie X. High expression of long noncoding RNA HOTAIRM1 is associated with the proliferation and migration in pancreatic ductal adenocarcinoma. Pathol Oncol Res. 2019. doi:https://doi.org/10.1007/s12253-018-00570-4
Zhang X, Weissman SM, Newburger PE. Long intergeneic non-coding RNA HOTAIRM1 regulates cell cycle progression during myeloid maturation in NB4 human promyelocytic leukemia cells. RNA Biol. 2014; 11:777–787.
Li Q, Dong C, Cui J, Wang Y, Hong X. Over-expressed IncRNA HOTAIRM1 promotes tumor growth and invasion through up-regulating HOXA1 and sequestering G9a/EZH2/Dnmts away from the HOXA1 gene in gioblastoma multiforme. J Exp Clin Cancer Res. 2018; 37:265.
Wan L, Kong J, Tang J, Wu Y, Xu E, Lai M, Zhang H. HOTAIRM1 as a potential biomarker for diagnosis of colorectal cancer functions the role in the tumour suppressor. J Cell Mol Med. 2016; 20:L2036–L2044.
Pang EJ, Yang R, Fu XB, Liu YF. 2015. Overexpression of long non-coding RNA MALAT1 is correlated with clinical progression and unfavorable prognosis in pancreatic cancer. Tumour Biol 36:2403–2407.
Kim J, Piao HL, Kim BJ, Yao F, Han Z, Wang Y, Xiao Z, Siverly AN, Lawhon SE, Ton BN, Lee H, Zhou Z, Gan B, Nakagawa S, Ellis MJ, Liang H, Hung MC, You MJ, Sun Y, Ma L 2018. Long noncoding RNA MALAT1 suppresses breast cancer metastasis. Nat Genet 50:1705–1715.
Cancer Genome Atlas Research Network. Integrated genomic characterizatin of papillary thyroid carcinoma. Cell. 2014;159:676–690.
.Landa I., Ibrahimpasic T., Boucai L., Sinha R., Knauf J.A., Shah R.H., Dogan S., Ricarte-Filho J.C., Krishnamoorthy G.P., Xu B., Schultz N., Berger M.F., Sander C., Taylor B.S., Ghossein R., Ganly I., Fagin J.A., 2016. Genomic and transcriptomic hallmarks of poorly differentiated and anaplastic thryoid cancers. J Clin. Invest. 2016,126, 1052–1066.
Todaro M. Lovino F, Eterno V, Cammaren P, Gambara G, Espinma V, Gu, Lotta G, Dieli F. S. Giordano, De Maria R, Stassi G. Tumorigenic and metastatic activity of human thyroid cancer stem cells. Cancer Res. 2010;70:8874–8885.
Acknowledgements
We kindly thank Dr. John A. Copland III (Mayo Clinic, Jacksonville, FL) for the THJ-16T cell line, Dr. Rebecca E. Schweppe (University of Colorado, Denver, CO) for the BCPAP cell line, Dr. Daniel T. Ruan (Brigham and Women’s Hospital, Boston, MA) for the TPC-1 cell line, and the staff of the Translational Research Initiatives in Pathology (TRIP) Laboratory for technical assistance.
Funding
Dr. Y. Wang received a research grant from the Department of Pathology and Laboratory Medicine at the University of Wisconsin School of Medicine and Public Health.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The study received ethical approval from the local Institutional Review Board.
Conflict of Interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Wang, Y., Hardin, H., Chu, YH. et al. Long Non-coding RNA Expression in Anaplastic Thyroid Carcinomas. Endocr Pathol 30, 262–269 (2019). https://doi.org/10.1007/s12022-019-09589-y
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12022-019-09589-y