Abstract
Poly-3-hydroxybutyrate (P3HB) is a biopolymer, which presents characteristics similar to those of plastics derived from the petrochemical industry. The thermomechanical properties and biodegradability of P3HB are influenced by its molecular weight (MW). The aim of the present study was to evaluate the changes of the molecular weight of P3HB as a function of oxygen transfer rate (OTR) in the cultures using two strains of Azotobacter vinelandii, a wild-type strain OP, and PhbZ1 mutant with a P3HB depolymerase inactivated. Both strains were grown in a bioreactor under different OTR conditions. An inverse relationship was found between the average molecular weight of P3HB and the OTRmax, obtaining a polymer with a maximal MW (8000–10,000 kDa) from the cultures developed at OTRmax of 5 mmol L−1 h−1 using both strains, with respect to the cultures conducted at 8 and 11 mmol L−1 h−1, which produced a P3HB between 4000 and 5000 kDa. The increase in MW of P3HB was related to the activity of enzymes involved in the synthesis and depolymerization. Overall, our results show that it is possible to modulate the average molecular weight of P3HB by manipulating oxygen transfer conditions with both strains (OP and PhbZ1 mutant) of A. vinelandii.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Poly-3-hydroxybutyrate (P3HB) is a homopolymer of the polyhydroxyalkanoates (PHAs) family; this bioplastic is produced and accumulated in the form of intracellular inclusions as a carbon and energy reserve by the bacterium Azotobacter vinelandii and other bacteria and archaea [1,2,3]. P3HB has thermomechanical characteristics similar to those of conventional petroleum-based plastics, such as polypropylene and polyethylene, with the advantage that can be synthesized from renewable carbon sources [4]. This polymer is biodegradable and it is completely biocompatible, and therefore is not toxic to higher organisms, making it an ideal product for use in the biomedical and pharmaceutical areas [5]. The first step in P3HB production in the Azotobacter vinelandii is catalyzed by β-ketothiolase (PhaA) condensing two acetyl-CoA molecules to give acetoacetyl-CoA. The second step is reduced by the NADPH-dependent acetoacetyl-CoA reductase (PhbB) of the acetoacetyl-CoA to 3-hydroxybutyryl-CoA and finally through a P3HB synthase (PhbC) to form P3HB [6, 7]. The physicochemical and thermomechanical properties, such as elastic behavior, mechanical strength, and the degree of crystallinity of P3HB, are influenced by its molecular weight [8, 9]. Biopolymers with a high molecular weight also have a higher elongation to break and tensile strength. For example, P3HB fibers with an average mean molecular weight (MMW) of 300 kDa have a tensile strength of 190 MPa and an elongation at break of 5%, whereas when the MW is 5300 kDa, the fibers increase their tensile strength to 1320 MPa and their elongation at break to 58% [10, 11]. In addition, the biodegradability rate of P3HB is affected by the size of the molecules, observing that when the molecular weight of P3HB is higher, the degradation rate decreases [12].
P3HB is used as biomedical materials, as well as for bones and meniscal regeneration [13]. In recent years, several fermentation strategies using microbial models, such as A. vinelandii, have been developed in order to improve P3HB production and also the molecular mass composition of the polymer [2, 3, 14, 15]. One of the most important culture parameters that influence production and chemical characteristics of P3HB is dissolved oxygen tension (DOT) and the oxygen transfer rate (OTR) [3, 16,17,18].
About the influence of OTR on the chemical characteristics of P3HB, Myshkina et al. [19] observed, in cultures conducted in shake flasks with Azotobacter chroococcum 7B, that the molecular weight increased from 1480 to 1670 kDa, when the agitation rate decreased from 250 to 190 rpm and therefore the OTR. A previous study in our group has revealed that the MMW of P3HB is strongly influenced by both the aeration conditions and the strain used [3]. The OPN strain used in that study has an inactivation of ptsN, the gene coding for the IIANtr protein of the nitrogen-phosphoenolpyruvate-dependent phosphotransferase system (PTS-Ntr); the protein IIANtr has been shown to negatively regulate expression of the P3HB biosynthetic genes phbBAC and of phbR, the genes that encode for their transcriptional activation. In the cultures in shake flasks at high aeration, the molecular weight of P3HB was 1000 and 550 kDa for the OPN and parental strain (OP), respectively. An increase to a MMW of 2000 kDa was observed for the P3HB isolated from the cultures of OPN mutant under low aeration conditions at 60 h of cultivation. A similar behavior was observed in the polymer produced by the OP strain, obtaining P3HB with a MMW of 1650 kDa at the same culture time.
More recently, Millán et al. [20] reported that in cultures of A. vinelandii strain OP, under two fixed oxygen conditions, the DOT (1 and 15%) did not affect the mean molecular mass of P3HB. However, the MW changed through the culture stages (growth and stationary phase). This study showed that P3HB of high molecular mass was accumulated during the growth phase in the cultures grown 1% of DOT, and this was associated with a high activity of P3HB synthase; however, a decrease in the MMW of the polymer was observed at the end of the stationary phase, and this correlated with a higher P3HB depolymerase activity. In addition, the P3HB biosynthetic activity β-ketothiolase was high when the accumulation of P3HB increased, whereas the activity of this enzyme decreased when the accumulation of the polymer stopped.
Later, Adaya et al. [21] showed that the a PhbZ1 mutant of A. vinelandii, which has inactivated one of the P3HB depolymerases involved in the hydrolysis of P3HB, produced a polymer with a high molecular weight from the exponential growth phase and remained constant throughout the culture, under the same conditions of the parental strain OP. In addition, a higher P3HB accumulation was observed in the cultures of the PhbZ1 mutant. These results suggested that PhbZ1 mutant P3HB depolymerase has a role in the degradation of P3HB in cultures in bioreactors, and its inactivation allows the production of a polymer with a uniform high molecular weight over time.
Most of the information generated so far comes from A. vinelandii grown in bioreactors, under controlled dissolved oxygen conditions (non-microaerophilic). However, under conditions without control of the dissolved oxygen, such as the shaken flaks, the cultures usually operate at DOT close to zero (> 1% of DOT, microaerophilic conditions). Under this condition, the oxygen transfer rate (OTR) profile shows a characteristic plateau region that indicates oxygen limitation [22]. Therefore, the objective of this study was to determine the influence of OTR on the molecular weight of P3HB produced by mutant strains of Azotobacter vinelandii cultured in 3 L bioreactor under microaerophilic conditions.
Material and Methods
Microbial Strain, Culture Medium, and Inoculum Preparation
Strains of A. vinelandii OP and its mutant derivative PhbZ1, containing a phbZ1::Gm gene inactivation, were used in the present study. Both strains were grown in PY medium, which contains sucrose (20 g L−1), yeast extract (3 g L−1), and peptone (5 g L−1). The strains were cryopreserved at − 70 °C in a 40% glycerol solution and maintained by monthly subculture on PY agar slopes and stored at 4 °C [3]. The pH of the medium was adjusted to 7.2 with addition of a 2N solution of NaOH, and the medium was sterilized at 121 °C for 20 min. The inoculum was incubated in Erlenmeyer flask on a rotatory shaker (New Brunswick Scientific Co., Model G 25, shaking radius 25 mm) at 200 rpm at 29 °C, to an absorbance (measured at 540 nm, after performing a dilution 1:50 dilution before the measurement) between 0.16 and 0.18 (corresponding to a cell dry weight between 0.08 and 0.1 g L−1). A total of 200 mL of the broth culture was centrifuged at 12,860g for 10 min at 4 °C. The supernatant was discarded, and the cells were suspended in 200 mL of fresh PY medium and transferred to the bioreactor containing 1.8 L of medium.
Culture Conditions in the Bioreactor
Cultures were carried out in a 3 L Applikon (Holland) bioreactor, with a work volume of 2 L, equipped with two Rushton turbines (impeller diameter/tank diameter = 0.35). The pH was measured with an Ingold probe (Applikon, ADI 1010) and controlled to 7.2 by automatically adding 2N NaOH during the growth phase or 2N HCl in the stationary phase of the culture. The DOT was measured by means of a polarographic oxygen electrode. The cultures were carried out at 29 °C using two agitation speeds (300 and 500 rpm); therefore, two different oxygen transfer rate (OTR) conditions were established under controlled aeration conditions (1 vvm). All experiments were conducted in triplicate, and the results presented are the average of independent runs.
Two gas analyzers (BCP-O2, BCP-CO2, BlueSens) were used to measure the O2 and CO2 in the gas flow from the bioreactor outlet. Estimation of the OTR was determined by online measurements of O2 in the exit gas and compared with measurements taken from the inlet gas flow rate, using the equation (Eq. 1) proposed by Zeng and Deckwer [23].
where VG is the gas flow at the inlet (L h−1), VL is the working volume (L), VN is the molar volume (L mol−1), and Xin and Xout are the mole fractions of oxygen at the inlet and outlet, respectively.
The equation (Eq. 2) used for determination of specific oxygen uptake rate (qO2) was as follows:
where OTRmax (mmol L−1 h−1) was the maximum oxygen uptake rate and Xmax (g L−1) was the maximum protein concentration.
Analytical Determinations
Biomass, Protein, Sucrose, and Poly-3-Hydroxybutyrate Determinations
Cell dry weight was determined gravimetrically using 6 mL of culture broth, which was centrifuged at 9660g during 10 min. The pellet was isolated, mixed in distilled water, and filtered through previously weighted Millipore filters (0.45 μm pore size). The filters were dried at 80 °C to constant weight. Protein concentration was determined by the Lowry method using bovine serum albumin as standard [24]. The specific growth rate (μ) was calculated based on the measured protein concentrations using the logistic model reported previously [25]. Sucrose was assayed for reducing power with the DNS reagent. Samples were previously hydrolyzed by using β-fructofuranosidase (from Bakers Yeast, Sigma-Aldrich, St. Louis, MO, USA) to generate glucose and fructose and then assayed for reducing power with the DNS reagent [26]. Quantification of P3HB was made by measuring crotonic acid concentration. P3HB inside the biomass was hydrolyzed with concentrated sulfuric acid at 90 °C for 1 h, causing the formation of crotonic acid [27], and measured using a high-performance liquid chromatography (HPLC) system with a UV detector (Waters 2996, USA) and an Aminex HPX-87H ion-exclusion organic acid column at 220 nm. Elution was performed with 0.014 N of H2SO4 at a flow rate of 0.65 mL min−1 at 50 °C, as described previously [20].
Analysis of Molecular Mass
Recovery of P3HB was performed as described previously [20]. The molecular mass analysis was performed by gel permeation chromatography (GPC) using a Shodex K-807 L column in an HPLC system (Waters 2695, USA) coupled with a refractive index detector (Waters 2414, USA). The mobile phase was chloroform at 30 °C at a flow rate of 1.0 mL min−1. A calibration curve was constructed with polystyrene standards (2.94 × 103 Da to 11.0 × 106 Da) as described [20]. Samples were dissolved in chloroform at a concentration of 2–3 mg mL−1 and were filtered through a 0.45-μm membrane before being injected into the HPLC.
Enzyme Activity Assays
Bacteria were harvested in 50-mL conical centrifuge tubes containing 45 mL culture by centrifugation at 9660g during 15 min at 4 °C and stored frozen until needed. Whole cells were suspended in 10 mM MgCl2, then the cells were disrupted by ultrasonic treatment at 4 °C (four pulses for 30 s at 6 W, Virsonic 60, Virtis) and the cell debris was removed by centrifugation at 9660g during 15 min at 4 °C. The total protein in the cell extracts was measured with a Lowry assay using and bovine serum albumin as standard. All enzymatic assays were measured using 96-well plates in Synergy H1 multi-mode microplate reader (BioTek) at 200 μL volume each well.
β-Ketothiolase Activity
β-Ketothiolase activity was measured spectrophotometrically by the method described by Segura et al. [7] in the thiolysis reaction with slight modifications. The reaction mixture contained 25 mM potassium phosphate buffer pH 7.8 with 40 mM MgCl2·6 H2O and 1 mM DL-dithiothreitol (DTT). After 2 min, 43.5 μM coenzyme A (20 μL) and 50 μM acetoacetyl-CoA (20 μL) were added. The reaction was started with the enzyme in 10 μL of crude cell extract with 2.6 μg protein concentration. The reaction progress was measured for 10 min to 29 °C and 303 nm, assuming that for the spectrophotometric assays, the extinction coefficient for our chemically mix acetoacetyl-CoA was 17,260 M−1 cm−1. The mean value was estimated from three independent experiments. The specific activity was measurement of the consumption of 1 μmol of Mg2+-chelated enolate of acetoacetyl-CoA corresponding to 1 μmol of product produced per minute.
Acetoacetyl-CoA Reductase Activity
Acetoacetyl-CoA reductase kinetics was determined using 40 μM acetoacetyl-CoA as substrate, 0.24 mM NADPH as cofactor, 100 mM potassium phosphate buffer pH 5.5 with 12 μM MgCl2·6H2O, and 0.5 mM DL-dithiothreitol (DTT) as reducing agent [28,29,30]. The reaction was determined at 29 ± 0.1 °C and started by adding 10 μL of crude cell extract (3.2 μg total protein) in the well. The activity was measured for 30 min following the decrease in absorbance resulting from NADPH (extinction coefficient at 6220 M−1 cm−1) consumption at a wavelength of 340 nm. The initial rates were measured as the activity for consuming 1 μmol of NADPH per minute.
P3HB Synthase Activity
Assay of PHA synthase activity was carried out according to the method described in the literature by Miyake et al. [31], which is based on the determination of thionitrobenzoic acid (TNB, extinction coefficient at 13,600 M−1 cm−1) formed that corresponded to the CoA released as a result of the condensation of monomers. The reactions were measured at 29 °C at a wavelength of 412 nm. The reaction was started by addition 10 μL of the insoluble fraction (granule-bound synthase) that contained enzyme (total protein concentration 4.4 μg for insoluble fraction). The mixture contained 25 mM potassium phosphate buffer pH 7.0, 0.1 mM of 5,5-dithiobis-2-nitrobenzoic acid (DTNB), and 55 μM of the substrate β-hydroxybutyryl-CoA. One unit was defined as the activity for producing 1 μmol of TNB anion per minute.
P3HB Granule Purification and Depolymerase Activity
Native P3HB (nPHB) granules were purified as reported by Gebauer and Jendrossek [32]. First, the cells were suspended in a lysis buffer pH 8.0 (50 mM of NaH2PO4, 300 mM NaCl, 10 mM imidazole, and protease inhibitor cocktail) and were disrupted in a French press (three passes at 900 psi) and centrifuged in a discontinuous glycerol gradient (87–40%) at 71,000g for 40 min at 4 °C. Later, the granule layer remaining between 50 and 60% glycerol fractions was recovered and the glycerol was removed by dialysis against 50 mM Tris-HCl pH 8.0 overnight at 4 °C. The granule suspension was adjusted to an OD600 to ~ 1.0. The P3HB depolymerase-specific activity was measured as granule degradation through changes in absorbance of the granule suspension incubated at 37 °C in shaker during 12, 24, 36, and 48 h, using a protein concentration of the cell extract of 7 μg mL−1 assay of activity. The values of absorbance measured in the assay were converted to P3HB concentration values by extrapolating in a calibration curve (absorbance values vs concentration of P3HB measured by HPLC). One unit of activity P3HB depolymerase was defined as the amount of polymer degraded per minute (mg P3HB min−1).
Results and Discussion
Evolution of the Oxygen Transfer Rate and Growth Kinetics in the A. vinelandii OP and PhbZ1 Cultures
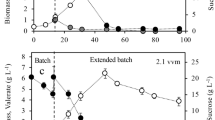
Considering the effect of oxygen transfer rate (OTR) on the production and composition of P3HB synthesized by the OP and PhbZ1 strains, in a first stage, the evolution of the OTR and OTRmax was determined in cultures with both strains grown in a 3-L stirred fermenter at two different agitation speeds (300 and 500 rpm). Figure 1 shows the typical OTR profiles and the growth kinetics measured as protein in the cultures using wild-type (OP) and modified strain (PhbZ1). Table 1 shows the influence of OTR defined by the agitation rate on OTRmax, qO2, specific growth rate, protein yield, P3HB accumulation, and production in cultures of A. vinelandii strains. In cultures conducted at 500 rpm, the highest sustained value of OTR during the culture (OTRmax) for the cultivations with the OP strain was 8.2 mmol L−1 h−1, which remained constant for approximately 20 h of cultivation, whereas using the PhbZ1 strain, an OTRmax of 11.2 mmol L−1 h−1 was achieved after 8 h of cultivation; this value remained constant for approximately 27 h. For cultivations conducted at 300 rpm, the OTRmax was 5.3 mmol L−1 h−1 for the two A. vinelandii strains evaluated. As expected, an increase in the agitation rate from 300 to 500 rpm increased the OTR. Regardless of the growth condition in both strains, the OTR profiles showed that the cultures were oxygen limited, with a plateau region characteristic of this kind of limitation, as previously described by Díaz-Barrera et al. [33] and Castillo et al. [34].
Evolution of OTR and growth kinetics of strain OP (a), measured as protein of strains OP (b), and OTR growth kinetics of strain PhbZ1 (c), measured as protein of strain PhbZ1 (d), both in cultures developed at 300 and 500 rpm. Determinations were carried out in triplicate, and the average and standard deviation for each point is shown. (▲) OP at 300 rpm, (△) OP at 500 rpm, (•) PhbZ1 at 300 rpm, and (Ο) PhbZ1 at 500 rpm
On the other hand, an increase in the agitation rate from 300 to 500 rpm caused an increase in the specific growth rate and the maximum protein concentration. As shown in Fig. 1 (c, d) at 500 rpm, a maximum cellular protein concentration of 0.7 g L−1, with a μ of 0.08 h−1, was obtained for the cultures of strain OP, and 0.6 g L−1 with a μ of 0.11 h−1 for cultures of the PhbZ1 strain, whereas in the cultures at 300 rpm, the maximum protein concentration was close to 0.3 g L−1 and the values of μ were 32% lower than the values achieved at high OTR for both strains (Table 1).
As shown in Table 1, the highest value of specific oxygen consumption rate (qO2max) was achieved with the PhbZ1 mutant strain. In the cultures at high OTRmax (500 rpm), this value was 36% higher than the qO2max reached by cultivations of OP strain at the same conditions. However, for the cultures at low OTRmax (300 rpm), the qO2max was up to 33% higher in the cultures with the OP strain than with the PhbZ1 strain. These results suggest that at the highest OTRmax, the inactivation of phbZ1 can have an effect on the respiratory activity, and therefore, this strain could be channeling a higher amount of acetyl-CoA through the TCA cycle, but further experiments are needed to prove this hypothesis.
P3HB Production at Different Oxygen Transfer Rates
It is important to point out that in A. vinelandii, P3HB biosynthesis is a response to an imbalance of the NAD(P)H/NAD(P) ratio that modifies the carbon flux through the tricarboxylic acid (TCA) cycle. In this line, Díaz-Barrera et al. [35] and García et al. [36] reported that a decrease in the OTRmax improved the P3HB accumulation and production. Figure 2 shows the P3HB production and P3HB accumulation of strains OP and PhbZ1 mutant, in cultures developed at 300 and 500 rpm. In contrast to what was expected, in the present work, for both strains evaluated, the maximal P3HB accumulation (80–90%) and production (4.5 ± 0.5 g L−1) was reached in the cultures grown at 500 rpm (high OTRmax), whereas the lowest P3HB production (approximately 3.0 g L−1) was obtained at 300 rpm (low OTRmax).
P3HB production and P3HB accumulation of strains OP (a, b) and PhbZ1 (b, c), in cultures developed at 300 and 500 rpm. Determinations were carried out in triplicate, and the average and standard deviation for each point is shown. (▲) OP at 300 rpm, (△) OP at 500 rpm, (•) PhbZ1 at 300 rpm, and (Ο) PhbZ1 at 500 rpm
These results contrast with those reported by other authors, suggesting that under a higher agitation rate, and therefore, a higher qO2max, it is possible that the acetyl-CoA is channeled to the respiratory metabolism through the TCA cycle, and much of the carbon source is directed to P3HB production instead growth. This behavior could be related to the lack of the sigma factor AlgU, which is mutated in strain OP and its mutant derivatives [37], because this protein in turn positively regulates the transcriptional regulator CydR [38], which is a negative regulator of the phbBAC operon and some respiratory proteins [39, 40]. On the other hand, as shown in Table 1, the values of volumetric production of P3HB (qP3HB) were very similar in both strains under the different OTRmax conditions (around 0.116 ± 0.011 g L−1 at the highest OTRmax and 0.022 ± 0.004 at the lowest OTRmax).
Previous studies by Díaz-Barrera et al. [41] have shown that in cultures with the OP strain at different OTR, an increase of P3HB accumulation from 46 to 65% was observed, when the agitation rate of the culture and as consequence the OTRmax of the culture increased from 6.5 to 9.5 mmol L−1 h−1. These authors explain this behavior considering that under a higher agitation rate (600 rpm to 9.5 mmol L−1 h−1), physical forces in the fluid could damage the cells (sub-lethal damage), and it is possible that the higher P3HB accumulation could be a bacterial response to adverse conditions (hydrodynamic stress), such as has been postulated for alginate produced by A. vinelandii ATCC 9046.
Influence of the OTRmax Conditions on the Molecular Weight of P3HB
Figure 3 and Table 2 show that the molecular weight is strongly influenced by the OTRmax of the culture and the strain tested. A maximal molecular weight of 9400 ± 250 kDa was reached for the P3HB isolated from the cultures of the P3HB depolymerase mutant PhbZ1 under low OTRmax condition, at 24 h of cultivation. This value was 16% higher with respect to that of the P3HB produced by the OP strain that showed molecular weight of 8100 ± 200 kDa at 48 h of cultivation (Fig. 3a). In contrast, in the cultures at high OTRmax, the molecular weight of the P3HB accumulated was considerably lower for both strains with values of 4800 ± 450 kDa and 3500 ± 100 kDa, for the PhbZ1 and OP strain respectively (Fig. 3a). Again, the PhbZ1 strain showed a slightly higher molecular weight than that of the OP strain, which would be expected for a mutant that has a P3HB depolymerase gene inactivated. The effect of oxygen observed had been previously reported by Peña et al. [3] using A. vinelandii OP and OPN (carrying a mutation on ptsN, the gene encoding enzyme IIANtr of the PTSNtr system) cultured in shaken flasks. These authors observed that a polymer having a high molecular weight (3670 kDa) was produced under low aeration conditions with the OPN strain, a value of 40% higher (2600 kDa) than the polymer obtained from the cultures at high aeration conditions. However, in those experiments in shaken flasks, the pH was not controlled, and the range of OTRmax evaluated in both culture systems was different. For example, in shaken flasks, the maximum OTRmax was between 2.8 and 5.5 mmol L−1 h−1, whereas in the bioreactor, the range of OTRmax was between 5 and 11 mmol L−1 h−1. To our knowledge, this is the first report about the synthesis of P3HB of ultra-high molecular weight (more than 8000 kDa) using mutant strains of A. vinelandii.
To better understand the changes observed in the molecular weight of P3HB, Fig. 3 b shows the weight-average molecular weight distribution of the P3HB accumulated at 24 h. In the case of the polymer produced by PhbZ1 and OP strains at high OTRmax, the P3HB was constituted by fractions with molecular weights from 100 to 10,000 kDa. These findings were reflected on the molecular weight, which was almost three-fold lower in those cultures conducted with an OTR of 11.5 mmol L−1 h−1, compared with those developed at the lowest OTRmax, where the percentage of P3HB molecules was from 1000 to 50,000 kDa. Interestingly, under the same culture condition, the molecular weight distribution of the polymer produced by the P3HB depolymerase mutant PhbZ1, although similar to that of the OP mutant, showed higher molecular weights (Fig. 3b).
Biochemical Analysis of the P3HB Synthesis Cycle
In the present study, a biochemical monitoring through measurement of the enzymatic activities involved in the synthesis and degradation of P3HB was carried out. It is known that simultaneous synthesis and degradation of P3HB occur in several bacteria [29, 42,43,44], so the activity of P3HB synthase and P3BH depolymerase was measured in total extracts 24 h after the start of cultivation. In an effort to follow-up the synthesis of P3HB upstream, the enzymes β-ketothiolase and acetoacetyl-CoA reductase were also analyzed, thus having an overview on the whole cycle of P3HB metabolism.
As shown in Table 2, P3HB synthase activity showed significant differences between the two OTRmax conditions. With the parental strain (OP), there was an almost 2-fold higher activity (0.058 ± 0.023 U/mg prot) at 300 rpm in comparison with the 500 rpm condition, where the synthase activity was 0.031 ± 0.017 U/mg prot (see Table 2). The same behavior was observed with the mutant strain PhbZ1, where the synthase activity was 2-fold higher at 300 rpm (0.035 ± 0.011 U/mg prot) than at 500 rpm (0.016 ± 0.007 U/mg prot).
On the other hand, the P3HB depolymerase activity was much lower in the cultures with the PhbZ1 mutant compared with the parental strain, due to the lack of depolymerase. In this study, we found differences between strains, when compared at the same OTRmax condition. As expected, the parental strain had a higher P3HB depolymerase activity than the PhbZ1 mutant strain in both OTRmax conditions (Table 2). Depolymerase activity was 2.5-fold higher in the OP strain at 300 rpm and 4-fold higher at 500 rpm than what is observed with the PhbZ1 strain.
The significant changes observed for the activity of P3HB depolymerase between the parental strain OP and PhbZ1 in the cultures conducted at 300 and 500 rpm are mainly the reflection of the high efficiency of this PhbZ1 enzyme (Avin_03910) to depolymerize the product and are related to that observed by Adaya et al. (2018). It is interesting to note that residual depolymerase activity in the PhbZ1 mutant strain could be due to the other 6 possible P3HB depolymerase enzymes that are found in its genome [44].
The depolymerase activity did not show significant difference between the OTR evaluated; it reached 0.029 ± 0.020 U/mg prot for cultures developed at 500 rpm and 0.035 ± 0.012 U/mg prot at 300 rpm with the PhbZ1 mutant strain. In this same manner, no significant difference was found for depolymerase activity of the parental strain (OP) between conditions (Table 2).
Overall, the differences in the molecular weight found in the P3HB produced at high and low OTR might be related to the activity of P3HB synthase between 300 and 500 rpm for both strains, being higher for the low OTRmax condition of 5 mmol L−1 h−1 (300 rpm) (Table 2). On the other hand, an increase of 2.5-fold in the P3HB depolymerase activity in cultures developed at 300 rpm in A. vinelandii OP compared with PhbZ1 mutant strain could explain the drop observed in the MW of the isolated polymer from 9400 ± 250 for the PhbZ1 mutant strain to 8100 ± 200 kDa with the OP strain. It is interesting to point out that 1.6 g L−1 of P3HB was accumulated in the OP strain, against 3.0 g L−1 in the PhbZ1 mutant strain, demonstrating an accumulation of the product up to 2-fold higher compared with its parent (Table 1).
In addition, in cultures developed at 500 rpm, a 4-fold decrease was observed for activity of P3HB depolymerase with the mutant PhbZ1 strain in comparison with the parental OP strain, explaining the lower MMW observed in the parental strain OP (3500 ± 100 kDa) compared with the PhbZ1 strain (4800 ± 450 kDa). It is clear that with the OP strain, there is a significant depolymerization, due in part to the presence of depolymerase PhbZ1. In the case of the PhbZ1 mutant strain, it appears that the MMW (higher than that of the OP strain) results from the combined effect of very low depolymerase activity and a favored synthesis of high molecular weight molecules.
It is important to point out that no significant changes were found in the activity of β-ketothiolase (phbA); however, our findings on acetoacetyl-CoA reductase (phbB) show that there is a higher activity (0.5-fold) in the cultures conducted at higher transfer conditions (500 rpm) in both strains, favoring monomer synthesis 3-hydroxybutyryl-CoA, compared with 300 rpm (5 mmol L−1 h−1), thus evidencing that there is more substrate to accumulate P3HB under this condition at high OTRmax (8–11 mmol L−1 h−1). This behavior contrast with the P3HB synthase activity, which is lower at high OTRmax, supporting the fact that P3HB de novo is being synthesized under this condition.
On this regard, it was observed that at low OTRmax (5 mmol L−1 h−1), there were higher MW P3HB molecules but lower accumulation of the polymer, whereas at high oxygen transfer condition (8–11 mmol L−1 h−1), P3HB accumulation increased but its MW decreased. These data contrast with that found by Millán et al. [20], who observed that the higher the OTR, the lower the P3HB accumulated; possibly, this behavior is due to the total transfer that is being supplied in each system, as in that particular study DOT was controlled at < 1%. Regarding accumulation, in our study, it was observed that for the condition of 500 rpm, an 88% accumulation of P3HB was reached with the OP strain, against 94% for the PhbZ1 mutant. For cultures developed at 300 rpm, 56% was of P3HB with OP strain and 82% for the PhbZ1 strain.
One strategy that we have developed, in order to obtain a high PHB production with a high molecular weight, is the use of two feeding-pulses fed-batch, employing the OPNA mutant strain of A. vinelandii, where the highest polymer concentration (27.6 g/L) was obtained, with a polymer having an ultra-high molecular mass (close to 6 MDa) (Castillo et al. [14]. In the case of batch culture, a strategy would use an intermediate OTR between 5 and 11 mmol L-1 h-1.
The behavior found under microaerophilic conditions (DOT < 1%) seems to indicate a dichotomy or trade-off between the percentage of P3HB accumulation and the molecular weight; that is, the decrease in the final concentration of the polymer is correlated with a significant increase in its molecular weight. It is possible that for PHA, synthase is favorable to increase the length of the molecule that is already in the active site of the enzyme, instead of starting de novo synthesis of new P3HB molecules, but further analysis is needed to prove this hypothesis.
In a previous study on P3HB synthases, it is suggested that class I synthase, such as the one present in A. vinelandii, generates longer molecules without restarting the process to generate new P3HB molecules, compared with class II or class III synthases [45]. When the ratio of substrate to enzyme (S/E) is low, synthase I can initiate polymerization on its own hydroxybutyrate-CoA-loaded. Furthermore, it is suggested that class I synthase has hydrolase activity, which would allow self-regulation and modulate the size of the polymer and its re-initiation [45]. Given the above, one possibility would be that under microaerophilic conditions, and under the specific conditions of this study (DOT < 1%), P3HB synthase is regulating itself in such a way that it does not make use of its hydrolase activity, as it would happen when it is under higher oxygen transfer conditions. This could also be related to the increased activity that was observed in the enzymatic analyses at low oxygen transfer with both strains.
Conclusions
Overall, our results have demonstrated for the first time that under microaerophilic conditions, the molecular mass of P3HB produced by OP and PhbZ1 mutant strains is affected positively by the OTRmax of the culture, finding a maximal molecular mass in the range of 8000 to 9000 kDa in cultures developed at OTRmax of 5 mmol L−1 h−1. This phenomenon was due to the simultaneous activities of enzymes involved in the polymerization of P3HB, mainly synthase activity and, to a lesser extent, the depolymerase activity. The results of this study reveal the feasibility to control the average molecular weight of P3HB by manipulating the conditions of oxygen transfer in the cultures of OP and PhbZ1 mutant strains of A. vinelandii.
References
Chen, G., & Page, W. (1997). Production of poly-b-hydroxybutyrate by Azotobacter vinelandii in a two-stage fermentation process. Biotechnology Techniques., 11(5), 347–350.
Garcia, A., Segura, D., Espin, G., Galindo, E., Castillo, T., & Pena, C. (2014). High production of poly-beta-hydroxybutyrate (PHB) by an Azotobacter vinelandii mutant altered in PHB regulation using a fed-batch fermentation process. Biochemical Engineering Journal, 82, 117–123.
Peña, C., Castillo, T., García, A., Millán, M., & Segura, D. (2014). Biotechnological strategies to improve production of microbial poly-(3-hydroxybutyrate): a review of recent research work. Microbial Biotechnology, 7(4), 278–293.
Laycock, B., Halley, P., Pratt, S., Werker, A., & Lant, P. (2014). The chemomechanical properties of microbial polyhydroxyalkanoates. Progress in Polymer Science, 38, 536–583.
Chen, G. (2009). A microbial polyhydroxyalkanoates (PHA) based bio- and materials industry. Chemical Society Reviews, 38(8), 2434–2446.
Manchak, J., & Page, W. (1994). Control of polyhydroxyalkanoate synthesis in Azotobacter vinelandii strain UWD. Microbiology, 140(4), 953–963.
Segura, D., Vargas, E., & Espín, G. (2000). β-Ketothiolase genes in Azotobacter vinelandii. Gene, 260(1–2), 113–120.
Aoyagi, Y., Yamashita, K., & Doi, Y. (2002). Thermal degradation of poly[(R)-3-hydroxybutyrate], poly[ε-caprolactone], and poly[(S)-lactide]. Polymer Degradradation and Stability, 76(1), 53–59.
Kusaka, S., Iwata, T., & Doi, Y. (1999). Properties and biodegradability of ultra-high-molecular-weight poly[(R)-hydroxybutyrate] produced by a recombinant Escherichia coli. International Journal of Biological Macromolecules, 25(1–3), 87–94.
Iwata, T. (2005). Strong fibers and films of microbial polyesters. Macromolecular Bioscience, 5(8), 689–701.
Ragan, B. (2008). Molecular weight modulation in polyhydroxybutyrate fermentations. Thesis (Ph.D.) Dept. of Chemical Engineering. Massachusetts Institute of Technology, Massachusetts, USA.
Bonartsev, A. P., Boskhomodgiev, A. P., Iordanskii, A. L., Bonartseva, G. A., Rebrov, A. V., Makhina, T. K., Myshkina, V. L., Yakovlev, S. A., Filatova, E. A., Ivanov, E. A., Bagrov, D. V., & Zaikov, G. E. (2012). Hydrolytic degradation of poly(3-hydroxybutyrate), polylactide and their derivatives: kinetics, crystallinity, and surface morphology. Molecular Crystals and Liquid Crystals, 556(1), 288–300.
Chen, G. Q., & Wang, Y. (2013). Medical applications of biopolyesters polyhydroxyalkanoates. Chinese Journal of Polymer Science (English Edition), 31(5), 719–736.
Castillo, T., Flores, C., Segura, D., Espín, G., Sanguino, J., Cabrera, E., Barreto, J., Díaz-Barrera, A., & Peña, C. (2017). Production of polyhydroxybutyrate (PHB) of high and ultra-high molecular weight by Azotobacter vinelandii in batch and fed-batch cultures. Journal of Chemical Technology & Biotechnology, 92(7), 1809–1816.
García, A., Pérez, D., Castro, M., Urtuvia, V., Castillo, T., Díaz-Barrera, A., Espín, G., & Peña, C. (2019). Production and recovery of poly‐3‐hydroxybutyrate [P(3HB)] of ultra‐high molecular weight using fed‐batch cultures of Azotobacter vinelandii OPNA strain. Journal of Chemical Technology & Biotechnology, 94(6), 1853–1860.
Galindo, E., Peña, C., Núñez, C., Segura, D., & Espin, G. (2007). Molecular and bioengineering strategies to improve alginate and polyhydroxyalkanoate production by Azotobacter vinelandii. Microbial Cell Factories, 6, 1–16.
Verlinden, R., Hill, D., Kenward, M., Wiliams, C., & Radecka, I. (2007). Bacterial synthesis of biodegradable polyhydroxyalkanoates. Journal of Applied Microbiology, 102(6), 1437–1449.
Peña, C., Galindo, E., & Büchs, J. (2011). The viscosifying power, degree of acetylation and molecular mass of the alginate produced by Azotobacter vinelandii in shake flasks are determined by the oxygen transfer rate. Process Biochemistry, 46(1), 290–297.
Myshkina, V., Nikolaeva, D., Makhina, T., Bonartsev, A., & Bonartseva, G. (2008). Effect of growth conditions on the molecular weight of polyhydroxybutyrate produced by Azotobacter chroococcum 7B. Applied Biochemistry and Microbiology, 44(5), 482–486.
Millán, M., Segura, D., Galindo, E., & Peña, C. (2016). Molecular mass of poly-3-hydroxybutyrate (P3HB) produced by Azotobacter vinelandii is determined by the ratio of synthesis and degradation under fixed dissolved oxygen tension. Process Biochemistry, 51(8), 950–958.
Adaya, L., Millán, M., Peña, C., Jendrossek, D., Espín, G., Tinoco-Valencia, R., Guzmán, J., Pfeiffer, D., & Segura, D. (2018). Inactivation of an intracellular poly-3-hydroxybutyrate depolymerase of Azotobacter vinelandii allows to obtain a polymer of uniform high molecular mass. Applied Microbiology and Biotechnology, 102(6), 2693–2707.
Anderlei, T., & Büchs, J. (2001). Device for sterile online measurement of the oxygen transfer rate in shaking flasks. Biochemical Engineering Journal, 7(2), 157–162.
Zeng, A. P., Byun, T., Posten, C., & Deckwer, W. D. (1994). Use of the respiratory quotient as a control parameter for optimum oxygen supply and scale-up of 2,3-butanediol production under microaerobic conditions. Biotechnology and Bioengineering, 44(9), 1107–1114.
Lowry, O. H., Rosebrough, N. J., Farr, A. L., & Randall, R. J. (1951). Protein measurement with the Folin phenol reagent. The Journal of Biological Chemistry, 193(1), 265–275.
Klimek, J., & Ollis, D. (1980). Extracellular microbial polysaccharides: kinetics of Pseudomonas sp., Azotobacter vinelandii and Aureobasidium pullulans batch fermentations. Biotechnology and Bioengineering, 22(11), 2321–2342.
Miller, G. L. (1959). Use of dinitrosalicylic acid reagent for determination of reducing sugar. Analytical Chemistry, 31(3), 426–428.
Law, H., & Slepecky, R. (1961). Assay of poly-β-hydroxybutyric acid. Journal of Bacteriology, 82(1), 33–36.
Senior, P. J., & Dawes, E. A. (1973). Poly--hydroxybutyrate biosynthesis and the regulation of glucose metabolism in Azotobacter beijerinckii. Biochemical Journal, 125(1), 55–66.
Song, H., Yingxin, Z., Weibao, K., & Chungu, X. (2012). Activities of key enzymes in the biosynthesis of poly-3-hydroxybutyrate by Methylosinus trichosporium IMV3011. Chinese Journal of Catalysis, 33(11), 1754–1761.
Volova, T. G., Kalacheva, G. S., Gorbunova, O. V., & Zhila, N. O. (2004). Dynamics of activity of the key enzymes of polyhydroxyalkanoate metabolism in Ralstonia eutropha B5786. Applied Biochemistry and Microbiology, 40(2), 170–177.
Miyake, M., Kataoka, K., Shirai, M., & Asada, Y. (1997). Control of poly-beta-hydroxybutyrate synthase mediated by acetyl phosphate in cyanobacteria. Journal of Bacteriology, 179(16), 5009–5013.
Gebauer, B., & Jendrossek, D. (2006). Assay of poly(3-hydroxybutyrate) depolymerase activity and product determination. Applied and Environmental Microbiology, 72(9), 6094–6100.
Díaz-Barrera, A., Andler, R., Martinez, I., & Peña, C. (2016). Poly-3-hydroxybutyrate production by Azotobacter vinelandii strains in batch cultures at different oxygen transfer rates. Journal of Chemical Technology and Biotechnology, 91(4), 1063–1071.
Castillo-Marenco, T., Lopez, I., Flores, C., Segura, D., García, A., Galindo, E., & Peña, C. (2018). Oxygen uptaken rate in alginate producer (algU+) and nonproducer (algU-) strains of Azotobacter vinelandii under nitrogen-fixation conditions. Journal of Applied Microbiology, 125(1), 181–189.
Díaz-Barrera, A., Urtuvia, V., Padilla-Córdova, C., & Peña, C. (2019). Poly(3-hydroxybutyrate) accumulation by Azotobacter vinelandii under different oxygen transfer strategies. Journal of Industrial Microbiology & Biotechnology, 46(1), 13–19.
García, A., Ferrer, P., Albiol, J., Castillo, T., Segura, D., & Peña, C. (2018). Metabolic flux analysis and the NAD(P)H/NAD(P)+ ratios in chemostat cultures of Azotobacter vinelandii. Microbial Cell Factories, 17(1), 10.
Segura, D., Vite, O., Romero, Y., Moreno, S., Castañeda, M., & Espín, G. (2009). Isolation and characterization of Azotobacter vinelandii mutants impaired in alkylresorcinol synthesis: alkylresorcinols are not essential for cyst desiccation resistance. Journal of Bacteriology, 191(9), 3142–3148.
León, R., & Espín, G. (2008). flhDC, but not fleQ, regulates flagella biogenesis in Azotobacter vinelandii, and is under AlgU and CydR negative control. Microbiology (Reading, England), 154(Pt 6), 1719–1728.
Wu, G., Cruz-Ramos, H., Hill, S., Green, J., Sawers, G., & Poole, R. K. (2000). Regulation of cytochrome bd expression in the obligate aerobe Azotobacter vinelandii by CydR (Fnr). Journal of Biological Chemistry, 275(7), 4679–4686.
Wu, G., Moir, A., Sawers, G., Hill, S., & Poole, R. (2001). Biosynthesis of poly-beta-hydroxybutyrate (PHB) is controlled by CydR (Fnr) in the obligate aerobe Azotobacter vinelandii. FEMS Microbiology Letters, 194(2), 215–220.
Doi, Y., Segawa, A., Kawaguchi, Y., & Kunioka, M. (1990). Cycle nature of poly(3-hydroxyalkanoate) metabolism in Alacaligenes eutrophus, FEMS Microbiology. Letters, 67(1-2), 165–170.
Ren, Q., de Roo, G., Ruth, K., Witholt, B., Zinn, M., & Thöny-Meyer, L. (2009). Simultaneous accumulation and degradation of polyhydroxyalkanoates: futile cycle or clever regulation? Biomacromolecules, 10(4), 916–922.
Volova, T. G., Zhila, N. O., Kalacheva, G. S., Brigham, C. J., & Sinskey, A. J. (2013). Effects of intracellular poly(3-hydroxybutyrate) reserves on physiological-biochemical properties and growth of Ralstonia eutropha. Research in Microbiology, 164(2), 164–171.
Setubal, J. C., Dos Santos, P., Goldman, B. S., Ertesvåg, H., Espin, G., Rubio, L. M., et al. (2009). Genome sequence of Azotobacter vinelandii, an obligate aerobe specialized to support diverse anaerobic metabolic processes. Journal of Bacteriology, 191(14), 4534–4545.
Tian, J., Sinskey, A. J., & Stubbe, J. (2005). Class III Polyhydroxybutyrate Synthase : Involvement in Chain Termination and Reinitiation. Biochemistry, 44(23), 8369–8377.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Highlights
• The MW of P3HB obtained under oxygen limitation was as high as 10,000 kDa.
• The MW of P3HB increased by decreasing the OTRmax of the culture.
• An A. vinelandii P3HB depolymerase mutant strain accumulated up to 94% of intracellular P3HB.
Rights and permissions
About this article
Cite this article
Gómez-Hernández, E., Salgado-Lugo, H., Segura, D. et al. Production of Poly-3-Hydroxybutyrate (P3HB) with Ultra-High Molecular Weight (UHMW) by Mutant Strains of Azotobacter vinelandii Under Microaerophilic Conditions. Appl Biochem Biotechnol 193, 79–95 (2021). https://doi.org/10.1007/s12010-020-03384-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-020-03384-w