Abstract
An arabinanase gene was cloned by overlap-PCR from Penicillium sp. Y702 and expressed in Pichia pastoris. The recombinant enzyme was named AbnC702 with 20 U/mg of endo-arabinanase activity toward linear α-1,5-l-arabinan. The optimal pH and temperature of AbnC702 were 5.0 and 50 °C, respectively. The recombinant AbnC702 was highly stable at pH 5.0–7.0 and 50 °C. It could retain about 72.3 % of maximum specific activity at pH 5.0 after incubation for 2.5 h, which indicated AbnC702 was an acid-adapted enzyme. The K m and V max values were 24.8 ± 4.7 mg/ml and 88.5 ± 5.6 U/mg, respectively. A three-dimensional structure of AbnC702 was made by homology modeling, and the counting of acidic/basic amino residues within the region of 10 Å around the active site, as well the hydrogen bonds within the area of 5 Å around the active site, might theoretically interpret the acid adaptability of AbnC702. Analysis of hydrolysis products by thin layer chromatography (TLC) combined with high-performance liquid chromatography (HPLC) verified that the recombinant AbnC702 was an endo-1,5-α-l-arabinanase, which yielded arabinobiose and arabinotriose as major products. AbnC702 was applied in pectin extraction from apple pomace with synergistic action of α-L-arabinofuranosidase.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Arabinan is a homoglycan linked to the rhamnopyranosyl residues of pectins that is present in plant tissues [1]. The arabinan backbone is composed of l-arabinofuranosyl units bound by α-1,5-linkage, with arabinosyl side chains linked by α-1,2 and α-1,3 bonds [2]. Based on substrate specificity, arabinan-degrading enzymes are classified into two major groups: α-l-arabinofuranosidases (Abf, EC 3.2.1.55) and α-l-arabinanases (Abn, EC 3.2.1.99) including endo- and exo-types. These enzymes are classified as members of glycoside hydrolase families (GH) 3, 43, 51, 54, 62, and 93 [3, 4], based on their amino acid sequence similarities according to the CAZy database (http://www.cazy.org/fam/acc_GH.html). The arabinofuranosidases hydrolyze non-reducing terminal α-l-arabinofuranosyl residues and generate l-arabinose from arabino-oligosaccharides, which allow the arabinanases to attack the arabinan backbone [5, 6]. The Abfs and 1,5-α-l-arabinanases play a synergistic role in the production of arabino-oligosaccharides and liberation of l-arabinose from arabinan [7].
Due to the potential application in various industrial processes, there have been many researches focusing on arabinan-degrading enzymes in recent years [8]. In addition, oligosaccharides containing arabinose produced from arabinoxylans and arabinans have been identified as potential prebiotics [9]. Furthermore, l-arabinose may be used as substrates for pentose-fermented organisms to produce bioethanol and biohydrogen [10–12]. On the other hand, arabinanases play an important role in extraction of pectin as they may be classified as B-type protopectinases (PPases), such as PPase-C [13, 14], which seems to attack the hairy region of protopectin and splits the arabinan of the polysaccharides connecting the rhamnogalacturonan to the other constituents of the plant cell wall, releasing macromolecular pectin.
The great functional and nutritional characteristics of pectin make it quite useful in food, cosmetics, and medicine manufacture [15]. To date, pectin extraction by acid to break the bonds between protopectin and cellulose is the main method in industrial scale; however, this process brings about some disadvantages such as large amounts of acidic effluents produced, corrosion of equipment, and so on [16]. Due to the demand for “green” products and active demands of “green processing” for environment protection, environmentally friendly biotechnological approaches such as enzymatic conversion of some low value resource to value-added products are desirable. For instance, apple pomace is the solid residue produced in a large number during apple juice extraction in the industry, and more than 100 million tons of this byproduct resource is produced in China every year [17, 18]. Therefore, enzymes that result in pectin releasing from such material are significantly needed.
Microorganisms are the best sources for commercial production of arabinan-degrading enzymes. Endo-arabinanase is mostly from bacteria, such as Bacillus [19–22], hyperthermophilic bacteria Thermotoga thermarum [23] and Thermotoga petrophila [24]. Some fungi such as Penicillium sp. or Rhizomucor miehei also produce endo-1,5-α-l-arabinanase [25, 26]. Previous studies revealed that Penicillium strains could secrete an exo-arabinanase, Abnx; three arabinofuranosidases, AFQ1, AFS1, and AXS5; and two endo-arabinanases, AbnC and AbnS1 into the culture medium [27–30]. However, the obstacle being faced of endo-1,5-α-l-arabinanase being applied in food industrial processing such as juice clarification and pectin extraction is that materials of the above industrial processing are fruits with acidic pH. This will hinder the application of most endo-1,5-α-l-arabinanases that are stable in neutral condition. Molecular modification of enzymes is available for resolving such problem [31], but it costs many trials and labors. Therefore, an original robust endo-1,5-α-l-arabinanase with good acid applicability is preferred.
In this study, we found an endo-1,5-α-l-arabinanase, AbnC702, from Penicillium sp. Y702, which is a protopectinase (PPase)-producing strain. After the AbnC702 was expressed in Pichia pastoris, the biochemical properties of purified recombinant AbnC702 was investigated, and it was revealed to be acid-adapted. In order to take advantage of this enzyme in pectin extraction, it was applied in pectin extraction from apple pomace with a recombinant arabinofuranosidase.
Materials and Methods
Strains, Plasmid, Chemical Reagent, and Medium
Penicillium sp. Y702 was isolated from soil samples in a persimmon orchard (Shanghai, China). Escherichia coli strain DH5α (Tiangen Biotech, Beijing, China) and pMD19-T vector (TaKaRa Biotechnology Co. Ltd, Tokyo, Japan) were used for gene cloning. The plasmid pPIC9K and P. pastoris GS115 (his4) purchased from Invitrogen (San Diego, USA) were used as vector and host respectively for expression of the recombinant enzyme. Linear 1,5-α-l-arabinan, red debranched arabinan and arabinoxylan were purchased from Megazyme (Wicklow, Ireland). Pectin (apple), gum arabic, starch, and 4-nitrophenyl-α-arabinofuranoside was ordered from Sigma (St. Louis, MO, USA). T4 DNA ligase and DNA polymerase (Taq DNA polymerase) were ordered from TaKaRa (Tokyo, Japan). PCR purification kit and plasmid kit were purchased from Axygen Biotechnology Company (Hangzhou, China). All other molecular chemical reagents in analytical grade were from TaKaRa (Tokyo, Japan).
The medium used in the present work are as follows. The MD medium contains 13.4 g/l YNB, 20 g/l glucose, and 0.0004 g/l biotin; YPD contains 20 g/l peptone, 20 g/l glucose, 10 g/l yeast extract and geneticin with different concentrations; BMGY contains 13.4 g/l YNB, 20 g/l peptone, 10 g/l yeast extract, 10 g/l glycerol, and 0.0004 g/l biotin, and they were dissolved in 100 mM pH 6.0 PBS; BMGY contains 13.4 g/l YNB, 20 g/l peptone, 10 g/l yeast extract, 5 g/l methyl alcohol, and 0.0004 g/l biotin, and they were dissolved 100 mM pH 6.0 PBS.
Plasmid Construction, Transformation of Abnc702 Gene
The internal transcribed spacer (ITS) region, the small subunit (ITS1-5.8S-ITS2) of the rDNA genes, was amplified using forward primer ITS1F (5′-CTTGGTCATTTAGACG AAGTAA-3′) and reverse ITS4 (5′-TCCTCCGCTTATTGATATGC-3′) according to Gardes and Bruns [32]. The analytic ITS1-5.8S-ITS2 region sequences were checked by using the Blast Network Service with the sequences in the NCBI GenBank database as references. According to the Blast result, the present strain was 99 % homologous of P. chrysogenum. Then, the overlap-PCR primers (Table 1) were designed according to the cDNA of AbnC (GenBank accession no. XM_00Y702052) from P. chrysogenum, and the cDNA of AbnC702 was amplified through overlap PCR. The overlap-PCR protocol contained eight primers and was conducted in two steps. In the first step, four modified exon1, exon2, exon3, and exon4 of the AbnC702 gene were prepared. In the second step, the obtained four modified exons were mixed as templates and linked up by using the outermost primer pair (C1-1 and C4-2); PCRs were performed according to touchdown PCR method before primer pair (C1-1 and C4-2) was added. The overlap-PCR product was cloned into the pPIC9K vector and then transformed into P. pastoris GS115 cells by using an Eppendorf Eporator electroporation apparatus (Eppendorf, Germany) [33], and the transformants grew on solid MD medium at 30 °C for 2 days. Then, the transformants collected from solid MD medium were plated on solid YPD medium containing 0.5, 1, 2, 3, and 4 mg/ml geneticin. After 4 days of incubation, the single colony that appeared on the medium of the highest geneticin concentration was selected for AbnC702 expression.
Expression and Purification of AbnC702
A single colony was firstly grown in the YPD medium at 30 °C, 250 rpm, for 18–24 h. These cells were further inoculated in the liquid BMGY medium until the culture reached an OD600 = 2–6 (log-phase growth). To induce protein expression, the cells were resuspended in the liquid BMMY medium and grew at 28 °C, and methanol was added to a final concentration of 0.5 % for 120 h of induction.
The target protein was concentrated by ultrafiltration and dialyzed by using 50 mM Tris–HCl buffer (pH 7.3), then the enzyme solution was injected to a 5 ml Mono QTM (GE Healthcare, Waukesha, WI, USA) in an AKTA fast protein liquid chromatography purification system (GE Healthcare, Waukesha, WI, USA). The column was equilibrated with the same buffer mentioned above. The signal of protein was detected at 280 nm. Finally, the fractions were collected and stored in 100 mM of phosphate buffer (pH 7.0) mixed with 20 % glycerol, and then kept at −20 °C for further investigation.
The theoretical molecular mass and the isoelectric point (pI) of AbnC702 protein were calculated by the ProtParam tool ( http://us.expasy.org/tools/protparam.html ).
Determination of Enzyme Activity and Kinetic Parameters
Measurement of the amount of reducing sugars according to the dinitrosalicylic acid (DNS) method was used as the enzyme activity assay. The reaction mixture comprised of 50 μl of diluted enzyme solution and 200 μl of 0.25 % (w/v) various substrates in 100 mM acetate buffer (pH 5.0). One unit of arabinanase activity was defined as the release of 1 μmol of arabinose per minute [34].
The determination of kinetic parameters was referenced as enzyme assay above. Linear 1,5-α-l-arabinan at concentrations from 1.0 to 10 mg/ml was performed using the above buffer in enzyme activity assay as the substrate for kinetics studies. The formation of reducing sugars was detected as products by DNS every 10 min during the reaction. Michaelis-Menten kinetics and Lineweaver-Burk plots were used to calculate the kinetic parameters.
Determination of pH and Temperature Features
The optimum pH was measured in the range of pH 4.0–9.0 in 100 mM different buffers: sodium acetate (pH 4.0–6.0), sodium phosphate (pH 6.0–7.0), and Tris–HCl (pH 7.0–9.0) at 37 °C. The enzyme was incubated at 30 °C in different pH buffers as mentioned above for 2.5 h for the pH stability profile. At regular incubation intervals, the residual enzyme activities were determined. The optimum temperature was investigated in a temperature range from 25 to 55 °C under the optimal pH for 30 min. The enzyme was assayed after incubation at different temperatures (35–55 °C) for the indicated time periods for temperature stability profile, and the residual enzyme activities were determined.
Effect of Metal Ions and Chelating Reagent on AbnC702
The effects of different metal ions and chelating reagent on the activity of recombinant AbnC702 were investigated by adding appropriate salts (1 mM) and chelating reagent (1 mM) to the enzyme reaction mixture as illustrated above, and the activity was assayed with enzyme activity in non-metal reaction as control.
Three-Dimensional Structure Model Analysis
The three-dimensional structure model of AbnC702 was built by Swiss Model server (http://swissmodel.expasy.org/), using the crystal structure of 1,5-α-l-arabinanase (PDB ID: 3CU9) as template and processed by Discovery Studio 4.0. The sequence similarity between target and template was 32.53 %, and the absolute model quality (QMEAN Z-score) of the modeling structure of AbnC702 was estimated as −3.34. The theoretical structure of the AbnZ1 construction was described by Wang et al. [31]. Furthermore, the number of acidic/basic amino residues within the region of 10 Å around the active site (with the center of the active site as core and 10 Å as radius) and hydrogen bonds in radius of 5 Å to the active site (with the center of the active site as core and 5 Å as radius) of both enzyme were estimated.
Substrate Specific and Degradation Pattern of AbnC702
Linear 1,5-α-l-arabinan, debranched arabinan, pectin (apple), arabic gum, starch, and arabinoxylan all in the concentration of 1 % (w/v), as well as 1 mM 4-nitrophenyl-α-arabinofuranoside were used in the substrate specificity evaluation.
For the degradation pattern, hydrolysis products of linear 1,5-α-l-arabinan at different reaction times were analyzed by both methods: thin layer chromatography (TLC) and high-performance liquid chromatography (HPLC). In TLC assay, hydrolysis products were spotted on the high silica gel plate GF254 (Jiangyou, Yantai, China) then developed in a solvent system (mixture of n-butanol-glacial/acetic acid/water = 4:3:1). The further analysis of hydrolysis products was carried out by HPLC (Agilent, USA) with an Inertsil HPLC NH2 column (Inertsil, Japan). The mobile phase contained water and acetonitrile in the ratio of 20:80 and the flow rate was 1 ml/min. The signal was monitored by an RI detector (Agilent Technologies 1200 series).
Hydrolysis of Apple Pomace
The apple pomace was washed to remove water-soluble sugars, pigments, and some other impurities, and then freeze-dried and smashed. One hundred microgram of such pretreated apple pomace were incubated with 1 U of AbnC702 and 19 U of arabinofuranosidase (AbfRumal) from Ruminococcus albus 7 (optimum pH and temperature were 6.0 and 50 °C, respectively), and 1 ml of 0.1 M HCl was used as control [15]. The reaction was carried out in 3 ml containing 50 mM sodium acetate, at pH 6 and 40 °C for 16 h; the reaction was terminated by 5 min of boiling. The liberated pectin was measured by the carbazole-H2SO4 method [35].
Result and Discussion
Analysis of Nucleotide and Amino Acid Sequence
The nucleotide sequence of cDNA has 963 nucleotides encoding 320 amino acids. A potential signal peptide was predicted with SignalP. Thus, the mature protein contained 299 amino acids, and the calculated molecular weight and pI are 33 kDa and 4.28, respectively.
AbnC702 was classified as a member of GH 43 by comparing the primary structure of AbnC702 against the Pfam database (http://pfam.sanger.ac.uk/search). Multiple sequence alignment (Fig. 1) indicates that three catalytic residues (Asp34, Asp148, and Glu199) are highly conserved in AbnC702. The Asp33 and Glu199 of AbnS1 from P. chrysogenum Wisconsin was reported acting as the catalytic base and acid, respectively [29]. Therefore, the Asp34 and Glu199 are inferred to serve the same functions in the mature AbnC702.
Alignment of the amino acid sequence of AbnC702 with other endo-arabinanases in the GenBank database revealed 67.6 % identity to P. chrysogenum Wisconsin AbnS1 (no. CAP92661), 68.5 % to AbnA from Aspergillus niveus (no. AEV23010), 31.8 % to Arb43a from Bacillus subtilis (no. AAV87172), and 26.0 % to Arb43A from Cellvibrio japonicus Ueda107 (no. ACE84667) using DNAman tool. Identical amino acids are written in black letters. The Asp34, Asp148, and Glu199 conserved catalytic residues of arabinanases are showed as closed inverted triangles. The predicted glycosylation sites (Ser75, Ser181, Ser183) are marked by closed circles
The amino acid sequence of AbnC702 shows relatively high similarity with those from fungi, showing 67.6 and 68.5 % similarity with the sequences of arabinanase from P. chrysogenum Wisconsin and Aspergillus niveus, respectively. However, the amino acid sequence of AbnC702 shows low identity as 31.8 and 26.0 % with arabinanase from bacteria B. subtilis and Cellvibrio japonicus Ueda107, respectively (Fig. 1).
Expression and Purification of AbnC702
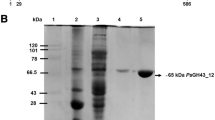
In this study, AbnC702 expression was induced by 1.5 % methanol and AbnC702 activity reached about 0.413 U/ml toward red debranched arabinan. After the ultrafiltration, the protein in the fermentation supernatant was separated and collected. Analysis by SDS-PAGE shows that the enzyme has a single band at around 35 kDa (Fig. 2), which is slightly higher than that theoretically expected (33 kDa). This difference may be due to the glycosylation by P. pastoris [36]. The final preparation of AbnC702 is 0.612 mg, which results in 7.94-fold purification with recovery rate of 30.0 %. The specific activity is 20 U/mg, which is in a relatively higher magnitude compared with some reported endo-1,5-α-l-arabinanase [16, 19–25].
Enzymatic Characteristics of Purified AbnC702
The maximal enzyme activity of AbnC702 appeared at pH 5.0 (Fig. 3a) and retained about 72.3 % after being incubated at such pH for 2.5 h (Fig. 3b). Furthermore, AbnC702 could maintain almost full activity at pH 6.0 and 7.0 for 2.5 h of incubation (Fig. 3b). These results verify that AbnC702 is an acid-adapted arabinanase. Most reported endo-1,5-α-l-arabinanases exhibited optimum activity in neutral pH range from 6.0 to 7.0 except RmArase from R. miehei with an optimal pH as 5.5 (Table 2). Concerning the pH stability, most arabinanases from the bacteria referred in Table 2 were stable under more neutral pH range except a thermostable one, AbnA from T. petrophila, which could be stable at lower pH. However, the incubation time for this one was 0.5 h, which is not comparable to AbnC702. It was notable that two arabinanases from fungi showed better pH stability than those of bacteria. One was a cold-adapted enzyme from P. chrysogenum and could even keep most activity in a broad pH range when incubated at 20 °C for 16 h. The only disadvantage of this enzyme was the relative lower specific activity which might hamper its application in large scale.
Biochemical properties of purified AbnC702. a Effect of pH on enzyme activity. b Effect of pH on enzyme stability. c Effect of temperature on enzyme activity. d Effect of temperature on enzyme stability. The activity was determined in 0.1 M sodium acetate (pH 5.0) at 37 °C for 30 min. All experiments were performed in duplicate. e Effect of metal ions and chelating reagent on recombinant AbnC702 activity. The activity was determined in 0.1 M sodium acetate (pH 5.0) which was incubated at 50 °C for 30 min. All experiments were performed in duplicate
AbnZ1 was a cold-adapted endo-arabinanase cloned from Paenibacillus polymyxa which retained only 5.78 % of maximal enzyme activity when incubated at pH 5.0 for 0.5 h [31]. Pectin extraction from fruit pomace usually performed at pH about 4.0–4.5. Thus, AbnC702 can be applied directly in such process without modifying the pH. This would significantly lower cost and pollution.
The optimal temperature of the recombinant AbnC702 is 50 °C (Fig. 3c). Over 90 % and nearly 70 % of its specific activity could remain after AbnC702 was incubated at 35 and 50 °C for 2.5 h, respectively (Fig. 3d). And even over 65 % of specific activity was kept at 25 °C incubation for a period, which indicated that AbnC702 had a wide range of temperature, especially lower temperature for application. In summary, AbnC702 can catalyze the reaction under low pH and temperature, suggesting that it has a good prospect for application in the food processing under such conditions.
AbnC702 exhibited 90 % enzyme activity after the metal ions were removed by EDTA (Fig. 3e), which made the recombinant AbnC702 turn out to be a metal-independent enzyme. Results also show that most of the metal ions tested have an activating effect on AbnC702 activity, except that Cu2+ strongly inhibits AbnC702 activity. Ca2+ enhances enzyme activity to 122 % compared with control. It was reported and confirmed that the presence of a Ca2+ ion in the catalytic domain of BsArb43B structure played an important role in the enzymatic mechanism of BsArb43B [37]. Kinetic studies were conducted by using linear α-1,5-l-arabinan as substrate at optimal pH and temperature. The K m and V max values for AbnC702 are 24.8 ± 4.7 mg/ml and 88.5 ± 5.6 U/mg, respectively.
Three-Dimensional Model of AbnC702
A three-dimensional structure of AbnC702 was made by the Swiss Model with 1,5-α-l-arabinanase (PDB ID: 3CU9) as template (Fig. 4a). The three-dimensional structure of AbnZ1 of our previous work was used as a control (Fig. 4b) [31]. As shown in Fig. 4a, the AbnC702 has the typical β-propeller fold structure of GH 43 family, which contains a five-bladed β-propeller around a pseudo axis [30, 38]. The catalytic domain of AbnC702 includes three active site residues: Asp34, Asp148, and Glu199. According to the crystal structure of arabinanase BsArb43B and multiple sequence alignment (Fig. 1), the function of these three key active site residues can be identified [37]. Asp34 in AbnC702 is a general catalytic nucleophile and Glu199 is a catalytic hydrogen donor [38]. Asp148 is believed to elevate the pKa of Glu199 and maintain the correct bonding between the general acid residue and the substrate [39]. Such three essential amino acids of AbnZ1 are Asp49, Asp165, and Glu215 [31]. The catalytic core of enzymes in the GH 43 family is highly conserved; however, the alignment of residues around the active site is different among them, which results in the diversity of enzymatic characteristics of each.
3D model structure of AbnC702 (a) and AbnZ1 (b) as control created by Swiss model and processed by Discovery Studio 4.0. Both enzymes contain a five-blade β-propeller, which is a characteristic structure in GH43. The α-helices and β-sheets are shown in red and cyan, respectively. Loops and other secondary structure are shown in green. The catalytic residues Asp34, Asp148, and Glu199 of AbnC702, and Asp49, Asp165, and Glu215 of AbnZ1 are shown in purple. Within the radius of 10 Å to the active site, the basic (blue) and acidic residues (orange) were also marked, which for AbnC702 (a), the basic amino acid residues were Arg47, Lys221, Lys234, Lys236, His291, and Lys304 and acidic residues were Asp86, Asp91, Glu109, Asp 191, Glu231, Glu232; and for AbnZ1 (b), the basic amino acid residues were His 48, Arg81, Arg129, Arg154, Lys188, Arg208, Arg209, His218, Lys237, Lys244, His297, and Lys308 and acidic residues were Asp44, Asp104, Asp109, Asp233, Asp240. Internal hydrogen bonds of AbnC702 (c) and AbnZ1 (d) as control created by Swiss model and processed by Discovery Studio 4.0. Hydrogen bonds are shown in green dashed line. In the radius of 5 Å to the active site, the number of hydrogen bonds for AbnC702 (c) are 48, and the number of hydrogen bonds for AbnZ1 (d) are 31
Some work has been performed to improve the activity at low pH by site-directed mutagenesis with basic amino acids being replaced by acidic residues. The optimal pH of wild-type AbnZ1 was 6.0, while the optimal pH of mutant H218D was 5.5. Besides, the mutant H218D was more stable at pH 5.5, and the stability was improved by 20 % when compared to its wild counterpart [31]. The specific activity of a wild-type α-amylase from B. subtilis is 108.2 U/mg at pH 4.5. After the His was substituted by Asp in some sites, the specific activity of mutants was improved from 20 to 100 %. Therefore, such site-directed mutagenesis strategy efficiently resulted in more stable α-amylase at low pH [40]. The results from such work indicate that acidic amino acids around the active site may be responsible for the acid adaptability of enzymes. Accordingly, the acidic and basic amino acids within the sphere of 10 Å (with the center of the active site as core and 10 Å as radius) were marked, and the distance of these residues to the center of the active site was determined in the structure of AbnC702, by using the AbnZ1 as the control (Table 3). As the result, AbnC702 has more acidic amino acids and much less basic residues than AbnZ1 does in this region, which may be a reason of its acid-adapted feature.
Protein stability may depend on the hydrogen bonding network around the active site. For instance, the mutant H218D shows three more hydrogen bonds compared with the wild AbnZ1 which has eight hydrogen bonds in the same area, which may explain the stability enhancement of H218D [31]. Likewise, after single mutation or combinational mutation of His was replaced by Asp of the α-amylase, the hydrogen bonds of mutants increased from 500 to 507 [40], which resulted in more stable structure of enzyme molecule. Hence, the hydrogen bonds in the area around the catalytic domain within 5 Å of AbnC702 were counted. Figure 4c shows that AbnC702 has 48 hydrogen bonds in the chosen area, while Fig. 4d shows that AbnZ1 had 31 hydrogen bonds in the same area. More hydrogen bonds around the catalytic domain of AbnC702 might lead to a more stable structure of AbnC702 than that of AbnZ1 and account for its acid-adapted property.
Substrate specificity and mode of action of AbnC702
Substrate specificity was determined using different materials. The AbnC702 shows activity to linear α-1,5-l-arabinan, red debranched arabinan, and pectin but did not exhibit activity to starch, arabinoxylan, and 4-nitrophenyl-α-arabinofuranoside (Table 4). Additionally, the degradation of red debranched arabinan showed that the enzyme acted in an endo-mode. These results suggest that AbnC702 may be an endo-α-1,5-l-arabinanase and the AbnC702 approaches to α-1,5-arabinan backbone would be interfered by side chains of α-l-arabinofuranosyl.
The products of linear α-1,5-l-arabinan degradation were monitored by TLC and HPLC. At the beginning of the reaction, arabino-oligosaccharides were mainly produced (Fig. 5a). As the reaction proceeded, the long-chain arabino-oligosaccharides were gradually degraded into smaller ones, and smaller ones could be detected after reaction for 2 h. Since the migrating rates of arabinose and arabinotriose are approximately the same on a TLC plate, arabinose cannot be distinguished from arabinotriose. Thus, the hydrolysis compounds are further identified by HPLC analysis. HPLC shows arabinobiose and arabinotriose are produced and accumulate while the reaction progressed, but arabinose can hardly be detected (Fig. 5b). Generally, arabinose or arabinobiose accumulates during the hydrolysis reaction of exo-acting enzymes, and arabino-oligosaccharides are formed in the case of endo-acting enzymes [41, 42]. Therefore, the end products generated by AbnC702 indicates that the enzyme is a typical endo-arabinanase that displays randomly internal cleavage of linear α-1,5-l-arabinan backbone.
Hydrolysis products of AbnC702 acting on linear arabinan. a TLC analysis of reaction products. Lane 1: arabinose; Lane 2: arabinobiose; Lane 3: arabinotriose; Lane 4: arabinotetraose; Lane 5–8: hydrolysis products of linear 1,5-α-l-arabinan after incubation for 0.5, 1, 2, and 16 h, respectively. (b) HPLC chromatograms of enzymatic products of linear 1,5-α-l-arabinan with the recombinant AbnC702 after different reaction times. Ara2, arabinobiose; Ara3, arabinotriose
Hydrolysis of Apple Pomace
When 0.1 M HCl, AbfRumal, or AbnC702 was used individually to hydrolyze apple pomace, 0.75, 0.23, and 0.65 mg of pectin was released, respectively (Fig. 6). AbnC702 had pectin releasing activity with pretreated apple pomace because it could split the α-1,5-l-arabinan chain in rhamnogalacturonan, indicating that AbnC702 belongs to B-type protopectinase [12, 13]. In the presence of both AbnC702 and AbfRumal, the pectin yield increased to 1.9 mg, which was 240 % of the pectin amount released by HCl. The structure of pectic polysaccharides is composed of homogalacturonan (HG), xylogalacturonan (XGA), apiogalacturonan (AGA), rhamnogalacturonan II (RG-II), and rhamnogalacturonan I (RG-I) [43]. The RG-I backbone, RG-I arabinan, and galactan side chains were four-linked with α-(1,5)-linked-l-Araf and β-(1,4)-linked-d-Galp chains in the cell walls of apple [44]. And, long branch chains of mono- or dimeric l-arabinan were three-linked to α-1,5-l-arabinan chains that originate from the RG-I backbone [45]. Abf can hydrolyze (1,3)- and (1,5)-arabinosyl linkages of arabinan from the non-reducing terminal of arabinose-containing polysaccharides in exo-type. Thus, when the arabinofuranoside branch chains were preferentially removed by AbfRumal, it would facilitate the binding of AbnC702 to arabinan side chains in RG I to cleave α-(1,5) glycosidic linkages, which promoted the release of pectin from the apple pomace.
Conclusions
In this study, the heterogeneous expression of an endo-α-1,5-l-arabinanase (AbnC702) from Penicillium sp. Y702 in P. pastoris GS115 was reported. AbnC702 exhibited relative high activity, wide temperature range, and good acidic stability, indicating that it had prospects for application in the food industry. Especially, when AbnC702 cooperated with arabinofuranosidase for apple pomace hydrolysis, higher pectin yield was achieved, which was nearly 2.5 times of that released by HCl. So, these results indicated that AbnC702 could also play a significant role in pectin extraction. Then, the coming work will be fermentation optimization for the production of AbnC702 and the scale-up application of such enzyme.
References
Janaswamy, S., & Chandrasekaran, R. (2005). Polysaccharide structures from powder diffraction data: molecular models of arabinan. Carbohydrate Research, 340, 835–839.
Shulami, S., Raz-Pasteur, A., Tabachnikov, O., Gilead-Gropper, S., Shner, I., & Shoham, Y. (2011). The L-arabinan utilization system of Geobacillus stearothermophilus. Journal of Bacteriology, 193, 2838–2850.
Lim, Y. R., Yoon, R. Y., Seo, E. S., Kim, Y. S., Park, C. S., & Oh, D. K. (2010). Hydrolytic properties of a thermostable α-L-arabinofuranosidase from Caldicellulosiruptor saccharolyticus. Journal of Applied Microbiology, 109, 1188–1197.
Numan, M. T., & Bhosle, N. B. (2006). Alpha-L-arabinofuranosidases: the potential applications in biotechnology. Journal of Industrial Microbiology & Biotechnology, 332, 247–260.
Hong, Y., Hitomi, I., Mitsutoshi, N., Hideyuki, K., & Satoshi, K. (2006). Synergy between an α-L-arabinofuranosidase from Aspergillus oryzae and an endo-arabinanase from Streptomyces coelicolor for degradation of arabinan. Food Science and Technology Research, 12, 43–49.
Damasio, A. R. D., Pessela, B. C., Segato, F., Prade, R. A., Guisan, J. M., & Polizeli, M. D. T. M. (2012). Improvement of fungal arabinofuranosidase thermal stability by reversible immobilization. Process Biochemistry, 47, 2411–2417.
Yang, X. Z., Shi, P. J., Ma, R., Luo, H. Y., Huang, H. Q., Yang, P. L., & Yao, B. (2015). A new GH43 α-arabinofuranosidase from Humicola insolens Y1: biochemical characterization and synergistic action with a xylanase on xylan degradation. Applied Biochemistry and Biotechnology, 175, 1960–1970.
Seiboth, B., & Metz, B. (2011). Fungal arabinan and L-arabinose metabolism. Applied Microbiology and Biotechnology, 89, 1665–1673.
Grootaert, C., Delcour, J. A., Courtin, C. M., Broekaert, W. F., Verstraete, W., & Van de Wiele, T. (2007). Microbial metabolism and prebiotic potency of arabinoxylan oligosaccharides in the human intestine. Trends in Food Science & Technology, 18, 64–71.
Li, J. Z., Ren, N. Q., Li, B. K., Qin, Z., & He, J. G. (2008). Anaerobic biohydrogen production from monosaccharides by a mixed microbial community culture. Bioresource Technology, 99, 6528–6537.
Becker, J., & Boles, E. (2003). A modified Saccharomyces cerevisiae strain that consumes L-arabinose and produces ethanol. Applied and Environmental Microbiology, 69, 4144–4150.
Bettiga, M., Bengtsson, O., Hahn-Hägerdal, B., & Gorwa-Grauslund, M. (2009). Arabinose and xylose fermentation by recombinant Saccharomyces cerevisiae expressing a fungal pentose utilization pathway. Microbial Cell Factories, 8, 40–52.
Sakamoto, T., Yoshinaga, J., Shogaki, T., & Sakai, T. (1993). Studies on protopectinase-C mode of action: analysis of the chemical structure of the specific substrate in sugar beet protopectin and characterization of the enzyme activity. Bioscience Biotechnology and Biochemistry, 57, 1832–1837.
Takao, M., Yamaguchi, A., Yoshikawa, K., Terashita, T., & Sakai, T. (2002). Molecular cloning of the gene encoding thermostable endo-1,5-α-L-arabinase of Bacillus thermodenitrificans TS-3 and its expression in Bacillus subtilis. Bioscience Biotechnology and Biochemistry, 66, 430–433.
Ptichkina, N. M., Markina, O. A., & Runlyantseva, G. N. (2008). Pectin extraction from pumpkin with the aid of microbial enzymes. Food Hydrocolloids, 22, 192–195.
Panouille, M., Thibault, J. F., & Bonnin, E. (2006). Cellulase and protease preparations can extract pectins from various plant byproducts. Journal of Agricultural and Food Chemistry, 54, 8926–8935.
Li, X. L., He, X. L., Lv, Y. P., & He, Q. (2014). Extraction and functional properties of water-soluble dietary fiber from apple pomace. Journal of Food Process Engineering, 37, 293–298.
Cheng, L., Sun, Z. T., Du, J. H., & Jian, W. (2008). Response surface optimization of fermentation conditions for producing xylanase by Aspergillus niger SL-05. Journal of Industrial Microbiology and Biotechnology, 35, 703–711.
Park, J. M., Jang, M. U., Kang, J. H., Kim, M. J., Lee, S. W., Song, Y. B., Shin, C. S., Han, N. S., & Kim, T. J. (2012). Detailed modes of action and biochemical characterization of endo-arabinanase from Bacillus licheniformis DSM13. Journal of Microbiology, 50, 1041–1046.
Seo, E. S., Lim, Y. R., Kim, Y. S., Park, C. S., & Oh, D. K. (2010). Characterization of a recombinant endo-1,5-alpha-L-arabinanase from the isolated Bacterium Bacillus. Biotechnology and Bioprocess Engineering, 15, 590–594.
Leal, T. F., & de Sa-Nogueira, I. (2004). Purification, characterization and functional analysis of an endo-arabinanase (AbnA) from Bacillus subtilis. FEMS Microbiology Letters, 241, 41–48.
Inácio, J. M., & de Sá-Nogueira, I. (2008). Characterization of abn2 (yxiA), encoding a Bacillus subtilis GH43 arabinanase, Abn2, and its role in arabino-polysaccharide degradation. Journal of Bacteriology, 190, 4272–4280.
Shi, H., Ding, H. H., Huang, Y. J., Wang, L. L., Zhang, Y., Li, X., & Wang, F. (2014). Expression and characterization of a GH43 endo-arabinanase from Thermotoga thermarum. BMC Biotechnology, 14, 1–9.
Squina, F. M., Santos, C. R., Ribeiro, D. A., Cota, J., de Oliveira, R. R., Ruller, R., Mort, A., Murakami, M. T., & Prade, R. A. (2010). Substrate cleavage pattern, biophysical characterization and low-resolution structure of a novel hyperthermostable arabinanase from Thermotoga petrophila. Biochemical and Biophysical Research Communications, 399, 505–511.
Sakamoto, T., Ihara, H., Kozaki, S., & Kawasaki, H. (2003). A cold-adapted endo-arabinanase from Penicillium chrysogenum. Biochimica et Biophysica Acta-General Subjects, 1624, 70–75.
Chen, Z., Liu, Y., Yan, Q. J., Yang, S. Q., & Jiang, Z. Q. (2015). Biochemical characterization of a novel endo-1,5-alpha-L-arabinanase from Rhizomucor miehei. Journal of Agricultural and Food Chemistry, 63, 1226–1233.
Sakamoto, T., & Thibault, J. F. (2001). Exo-arabinanase of Penicillium chrysogenum able to release arabinobiose from alpha-1,5-L-arabinan. Applied and Environmental Microbiology, 67, 3319–3321.
Sakamoto, T., & Kawasaki, H. (2003). Purification and properties of two type-B α-L-arabinofuranosidases produced by Penicillium chrysogenum. Biochimica Et Biophysica Acta, 1621, 204–210.
Sakamoto, T., Ogura, A., Inui, M., Tokuda, S., Hosokawa, S., Ihara, H., & Kasai, N. (2011). Identification of a GH62 α-L-arabinofuranosidase specific for arabinoxylan produced by Penicillium chrysogenum. Applied Microbiology and Biotechnology, 90, 137–146.
Sakamoto, T., Inui, M., Yasui, K., Tokuda, S., Akiyoshi, M., Kobori, Y., Nakaniwa, T., & Tada, T. (2012). Biochemical characterization and gene expression of two endo-arabinanases from Penicillium chrysogenum 31B. Applied Microbiology and Biotechnology, 93, 1087–1096.
Wang, S. H., Yang, Y., Yang, R. J., Zhang, J., Chen, M., Matsukawa, S., Xie, J. L., & Wei, D. Z. (2014). Cloning and characterization of a cold-adapted endo-1,5-alpha-L-arabinanase from Paenibacillus polymyxa and rational design for acidic applicability. Journal of Agricultural and Food Chemistry, 62, 8460–8469.
Gardes, M., & Bruns, T. D. (1993). ITS primers with enhanced specificity for basidiomycetes-application to the identification of mycorrhizae and rusts. Molecular Ecology, 2, 113–118.
Wu, S. X., & Letchworth, G. J. (2004). High efficiency transformation by electroporation of Pichia pastoris pretreated with lithium acetate and dithiothreitol. Biotechniques, 36, 152–154.
Sogabe, Y., Kitatani, T., Yamaguchi, A., Kinoshita, T., Adachi, H., Takano, K., Inoue, T., Mori, Y., Matsumura, H., & Sakamoto, T. (2011). High-resolution structure of exo-arabinanase from Penicillium chrysogenum. Acta Crystallographica, Section D: Biological Crystallography, 67, 415–422.
Takao, M., Akiyama, K., & Sakai, T. (2002). Purification and characterization of thermostable endo-1,5-α-L-arabinase from a strain of Bacillus thermodenitrificans. Applied and Environmental Microbiology, 68, 1639–1646.
Kazenwadel, C., Klebensberger, J., Richter, S., Pfannstiel, J., Gerken, U., Pickel, B., Schaller, A., & Hauer, B. (2012). Optimized expression of the dirigent protein AtDIR6 in Pichia pastoris and impact of glycosylation on protein structure and function. Applied Microbiology and Biotechnology, 97, 7215–7227.
de Sanctis, D., Inacio, J. M., Lindley, P. F., de Sa-Nogueira, I., & Bento, I. (2010). New evidence for the role of calcium in the glycosidase reaction of GH43 arabinanases. The FEBS Journal, 277, 4562–4574.
Pons, T., Naumoff, D. G., Martínez-Fleites, C., & Hernández, L. (2004). Three acidic residues are at the active site of a β-propeller architecture in glycoside hydrolase families 32, 43, 62, and 68. Proteins, 54, 424–432.
Davies, G., & Henrissat, B. (1995). Structures and mechanisms of glycosyl hydrolases. Structure, 3, 853–859.
Yang, H. Q., Liu, L., Shin, H. D., Chen, R. R., Li, J. H., Du, G. C., & Chen, J. (2013). Structure-based engineering of histidine residues in the catalytic domain of α-amylase from Bacillus subtilis for improved protein stability and catalytic efficiency under acidic conditions. Journal of Biotechnology, 164, 59–66.
Sakamoto, T., Ihara, H., Kozaki, S., & Kawasaki, H. (2003). A cold-adapted endo-arabinanase from Penicillium chrysogenum. Biochimica et Biophysica Acta, 1624, 70–75.
Wong, D. W., Chan, V. J., & McCormack, A. A. (2009). Functional cloning and expression of a novel endo-α-1,5-L-arabinanase from a metagenomic library. Protein and Peptide Letters, 16, 1435–1441.
Ridley, B. L., O’Neill, M. A., & Mohnen, D. A. (2001). Pectins: structure, biosynthesis, and oligogalacturonide-related signaling. Phytochemistry, 57, 929–967.
Schols, H. A., Mutter, M., Voragen, A. G., Niessen, W. M., van der Hoeven, R. A., van der Greef, J., & Bruggink, C. (1994). The use of combined high-performance anion-exchange chromatography-thermospray mass spectrometry in the structural analysis of pectic oligosaccharides. Carbohydrate Research, 261, 335–342.
Glushka, J. N., Terrell, M., York, W. S., O’Neill, M. A., Gucwa, A., Darvill, A. G., Albersheim, P., & Prestegard, J. H. (2003). Primary structure of the 2-O-methyl-alpha-L-fucose-containing side chain of the pectic polysaccharide, rhamnogalacturonan II. Carbohydrate Research, 338, 341–352.
Acknowledgments
This work was supported by National Special Fund for State Key Laboratory of Bioreactor Engineering (2060204) and partially supported by the National Natural Science Foundation of China (Nos. 21506057 and 21506057), the National High Technology Research and Development Program of China (No. 2013AA102109), and the Natural Science Foundation of Shanghai (No. 2013ZR1412100).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Lang, C., Yang, R., Yang, Y. et al. An Acid-Adapted Endo-α-1,5-l-arabinanase for Pectin Releasing. Appl Biochem Biotechnol 180, 900–916 (2016). https://doi.org/10.1007/s12010-016-2141-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-016-2141-5