Abstract
Inflammasomes are intracellular macromolecular complexes assembled to activate inflammatory caspases such as caspase-1 and caspase-5, which perform critical roles during innate immune response. The NALP3 inflammasome comprises three protein components, NALP3, ASC, and caspase-1. ASC, which contains both a pyrin domain (PYD) and a caspase recruitment domain (CARD), acts as a bridge to recruit NALP3 using the PYD/PYD interaction and to recruit caspase-1 via the CARD/CARD interaction. In this study, we successfully purified and characterized ASC CARD and caspase-1 CARD. The results showed that ASC CARD was unable to interact with caspase-1 CARD in vitro; therefore, we proposed an interaction mode between ASC CARD and caspase-1 CARD from a structural based modeling study.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Inflammasomes are cytoplasmic multiprotein complexes (~700 kDa) that promote inflammation and inflammatory cell death (pyroptosis) through the activation of inflammatory caspase-1 and caspase-5, and resulting in the processing of their substrate proinflammatory cytokines, pro-interleukin-1 beta (pro-IL-1β), and pro-interleukin-18 (pro-IL-18) into active interleukin (IL)-1β and IL-18, which promotes inflammation and regulates immune responses [1]. It is formed upon recognition of pathogen-associated molecular patterns (PAMPs) or damage-associated molecular patterns (DAMPs), which participate in the inflammatory response for innate immunity [2, 3]. Inflammatory caspases such as caspase-1 and caspase-5 are recruited in the inflammasome and obtain proximity to be self-activated. Activated inflammatory caspases can process inflammatory cytokines such as IL-1β and IL-18, which can evoke numerous cytokine cascades [4–6]. These pro-inflammatory cytokines, IL-1β and IL-18, have been postulated to be pro-tumorigenic in inflammation-induced cancers. Moreover, cancer-associated inflammatory responses are associated with tumor initiation, progression, metastasis, and treatment. Targeting inflammasomes using compounds such as IL-1β neutralizing antibodies, recombinant IL-1RN (anakinra), IL-18 binding protein, and caspase-1 inhibitors are used to treat cancer [7]. The inhibition of pro-inflammatory caspases by the successful deployment of anti-IL-1 therapies are used to control autoinflammatory diseases caused by aberrant inflammasome signaling. Inhibition of IL-1 signaling has been proven beneficial in cryopyrin-associated periodic fever syndromes (CAPS) patients. Blocking of IL-1β signaling inhibits the tumor progression and metastasis in breast cancer cells and melanoma cells. Patients of CAPS and other disorders might benefit from inhibitors of caspase-1, VX-765, which prevented the secretion of IL-1β suggesting therapeutic potential in epilepsy [8]. Apoptosis-associated speck-like protein containing caspase recruitment domain (CARD) (ASC) is triggered in apoptosis in certain tumors, and the knockdown of ASC inhibited inflammatory caspase-1 activity and IL-1β secretion [9]. Thus, inflammasomes are emerging as targets for the development of novel anti-inflammatory drugs for the treatment of chronic inflammatory disease [10, 11].
Inflammasomes are assembled around pyrin domain (PYD)- and CARD-containing proteins. NLRPs, ASC, and IFI protein family harbor PYD domains, while NOD1, NOD2, NLRP1, NLRC4, ASC, and inflammatory caspases harbor CARD domains. It contains three proteins, NOD-like receptor (NLR), ASC, and caspase-1 [12]. Four distinct inflammasomes (NALP1, NALP2, NALP3, and AIM2) have been identified based on which NLR is recruited [13]. Different inflammasomes are activated by different signals/ligands. Several bacterial ligands, nucleic acids, synthetic antiviral compounds, cellular toxins, and some toll-like receptor agonists are known to trigger the assembly of inflammasomes [14–16]. NALP3 inflammasome is linked to many human inflammatory diseases including Muckle-Wells syndrome, familial cold autoinflammatory syndrome, chronic infantile neurological cutaneous, and articular syndrome, which are linked to mutations of NALP3 [17, 18]. NALP3 inflammasome, which is composed of NALP3, ASC, and caspase-1, is the most studied inflammasome [19]. NALP3 is a member of the NLR family that senses multiple danger signals [20, 21]. Adaptor protein ASC contains two protein interaction modules: PYD at the N-teminus and CARD at the C-terminus [22]. Caspase-1 has a CARD domain at the N-terminus. The ASC functions as a bridge between caspase-1 and NALP3 by offering the CARD for CARD/CARD interactions and PYD for PYD/PYD interactions [10, 12].
CARD is a member of the death domain superfamily, which is comprised four subfamilies: the death domain (DD), death effector domain (DED), CARD, and PYD [23, 24]. The death domain superfamily is one of the largest families of protein-interacting domains and highly prevalent in apoptotic and inflammatory signaling proteins [25]. CARD/CARD interaction in the inflammasome is critical to the formation of inflammasome. Given the fact that CARD-mediated inflammasome assembly followed by inflammation response is associated with many human diseases, including cancer, immune disorders, and aging, studies in these fields are of great clinical importance [26]. Despite the importance of CARD-mediated inflammasome formation, structural and biochemical studies have been limited due to insolubility of the CARD domain [27, 28]. In this study, we successfully purified and characterized ASC CARD and caspase-1 CARD. The results showed that ASC CARD is unable to interact with caspase-1 CARD in vitro. Finally, we proposed an interaction mode between ASC CARD and caspase-1 CARD based on a structural-based modeling study.
Materials and Methods
Bacterial Strains, Plasmid, and Antibiotics
Escherichia coli strain BL-21 (DE3) was used as a host, while the C-terminal His-tag vector pOKD (home-made vector) was used for the overexpression of human ASC CARD and caspase-1 CARD genes. PCR reagents, T4 DNA ligase, and restriction endonucleases were purchased from Solvent Co. Ltd. (Seoul, Korea Republic). Luria Bertani (LB) medium (Merck) with ampicillin antibiotic was used for aerobic culture of E. coli cells with recombinant plasmids.
Homology Modeling
Homology model of caspase-1 CARD was constructed using the SWISS-MODEL homology modeling server [29]. ICEBERG structure (PDBid: 1DGN) was used as the modeling template. The stereochemical quality of the constructed models was validated using a Ramachandran plot generated by PROCHECK [30]. Electrostatic surfaces and ribbon diagrams were generated using the PyMOL program (DeDeLano, W. L. (2002) The PyMOL Molecular Graphics System, DeLano Scientific, San Carlos, CA, USA).
Expression of ASC CARD and Caspase-1 CARD in E. coli
The cDNA of full-length human ASC and human caspase-1 were used as templates for PCR, while the amplicons were cloned into pOKD (home-made vector) [31]. The CARD domain of ASC encoding amino acid residues 108–195 (10.2 kDa) and the CARD domain of caspase-1 coding for amino acid residues 1–104 (12.6 kDa) were cloned into pOKD, resulting in recombinant plasmids pOKD-ASC CARD and pOKD-caspase-1 CARD, respectively. These recombinant plasmids were separately transformed into E. coli BL-21 (DE3)-competent cells and plated onto LB agar plates containing ampicillin (100 μg/ml), after which they were incubated at 37 °C overnight. A single colony was used to inoculate 5 ml of LB medium containing ampicillin, which was subsequently incubated at 37 °C in a shaking incubator overnight. Next, 5 ml of the overnight small-scale culture was used to inoculate 1 l of LB broth containing ampicillin antibiotic in a 2-l culture flask, and the culture growth was continued in a shaking incubator. When cells reached an optical density (O.D.) of 0.6–0.7 at 600 nm, gene expression was induced by overnight treatment with 0.5 mM isopropyl-β-d thiogalactopyranoside (IPTG) at 20 °C.
Purification of Target Proteins
Bacteria expressing each protein were pelleted by centrifugation, resuspended, and lysed by sonication in 80 ml of lysis buffer (20 mM Tris-HCl buffer pH 8.0, 500 mM NaCl, and 5 mM imidazole) supplied with a protease inhibitor cocktail (Roche, USA). The lysate was then centrifuged at 16,000 rpm for 60 min at 4 °C, after which the supernatant fractions were applied to a gravity column (Bio-Rad) packed with Ni-NTA affinity resin (Qiagen). Next, the unbound bacterial proteins were removed by washing with 100 ml of washing buffer (20 mM Tris buffer at pH 8.0, 500 mM NaCl, 30 mM imidazole), after which the target proteins were eluted from the column using elution buffer (20 mM Tris-HCl buffer pH 8.0, 500 mM NaCl, and 250 mM imidazole), with 0.8 ml elution fractions being collected over a total of 10 ml. Fractions containing greater than 80 % homogeneous target protein upon SDS-PAGE analysis were then selected and combined. In the final purification step, the sample was loaded onto a Superdex-200 gel-filtration column 10/30 (GE Healthcare, Sweden) equilibrated with buffer containing 20 mM Tris-HCl (pH 8.0) and 150 mM NaCl. Fractions containing the target protein were subsequently pooled and the purity of the protein was checked by SDS-PAGE.
Circular Dichroism Spectroscopy
The secondary structures were measured by circular dichroism (CD) spectroscopy using a J-715 spectropolarimeter at Korea Basic Science Institute in South Korea. The spectra were obtained from 200 to 250 nm at 25 °C in a 0.1-cm path-length quartz cuvette using a bandwidth of 1.0 nm, a speed of 50 mm/min, and a 5-s response time. The protein samples in buffer containing 20 mM Tris-HCl at pH 8.0 and 150 mM NaCl were diluted to 0.1 mg/ml prior to use. Four scans were accumulated and averaged, and the α-helical content was calculated from the molar ellipticity at 222 nm [32].
Complex Assay by Gel-Filtration Chromatography
Equal molars of purified ASC CARD was mixed with caspase-1 CARD and incubated for 1 h at 25 °C, after which the mixture was concentrated (at 4 °C) to 5–8 mg ml−1 using a Centricon centrifugal filter (Millipore, Bedford, MA) with a molecular mass cutoff of 3,000 Da. The concentrated protein mixture (~2 ml) was subsequently applied to a Superdex 200 gel-filtration column 10/30 (GE Healthcare) that had been pre-equilibrated with a solution of 20 mM Tris-HCl buffer at pH 8.0 and 150 mM NaCl. Complex formation was evaluated based on the positions of the eluted peaks at 280 nm followed by SDS-PAGE using Bio-rad Protean II Mini gel unit and casting stand. Initially, protein samples were diluted with 1:6 gel loading buffer (6×) and were boiled at 90 °C for 10 min before loading onto 15 % SDS-PAGE gel. This was electrophoresed at ~150 V for 90 min, and the resolved protein bands were visualized using Coomassie brilliant blue R-250 stain [33].
Results
Purification and Characterization of ASC CARD and Caspase-1 CARD
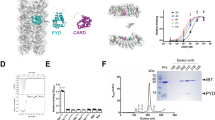
NALP3 inflammasome is the most well-known and studied inflammasome. NALP3, ASC, and caspase-1 are the main components of NALP3 inflammasome. Each inflammasome contains one member of the DD superfamily, which is the protein interaction module. Specifically, they contain CARD for ASC and caspase-1, and PYD for ASC and NALP3 to enable homotypic protein-protein interaction (Fig. 1a). Interaction between ASC and caspase-1 via CARD/CARD interaction is critical to assembly of inflammasome followed by recruitment and activation of caspase-1. Although the structure of ASC has been reported, the structure of caspase-1 CARD has not been elucidated to date. The structure of ASC CARD showed that it is composed of a six helix bundle, which is a general structural fold for the DD superfamily (Fig. 1b). As a first step to elucidation of the molecular basis of assembly of the NALP3 inflammasome via CARD/CARD interaction, we conducted homology modeling of caspase-1 CARD using the ICEBERG structure (PDBid: 1DGN) as the modeling template. ICEBERG shares 52 % amino acid sequence identity with caspase-1 CARD and is the most structurally similar CARD to caspase-1 CARD (Fig. 1c). The modeled structures of caspase-1 CARD were well constructed, possessing a six helixes bundle fold (Fig. 1d). The characteristics of the electrostatic surface of caspase-1 CARD are similar to those of other CARD domains, having two distinct oppositely charged regions (Fig. 1e).
Schematic representation of inflammasome and structure of CARDs. a Domain boundary of NALP3, ASC, and caspase-1. CARD, caspase recruitment domain; PYD, pyrin domain. PYD/PYD and CARD/CARD interactions for assembly of the inflammasome are indicated by dotted lines. b Ribbon diagram of ASC CARD structure. Six helixes are shown as H1–H6. c Sequence alignment between caspase-1 CARD and ICEBERG for sequence-based modeling. Identical sequences are indicated by bold characters. d Modeled structure of caspase-1 CARD. Six helixes are shown as H1–H6. e Molecular surface of caspase-1 CARD. The surface was calculated by PYMOL. Basic surfaces are blue, acidic surfaces are red
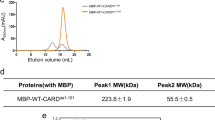
To reconstitute the ASC CARD (10.2 kDa) and caspase-1 CARD (12.6 kDa) complex in vitro, we initially attempted to purify each protein. Although purification of the CARD domain is difficult due to its insolubility, ASC CARD and caspase-1 CARD were expressed well in bacterial cells. Because the solubility of the death domain superfamily including CARD is sensitive to pH and salt concentration, we analyzed the characteristics of each CARD of ASC and caspase-1 under various buffer conditions [19, 27, 28, 34, 35]. Gel-filtration chromatography showed that ASC CARD behaved well in solution without any sensitivity against pH and salt concentrations (Fig. 2a, b). ASC CARD was eluted at around 18 ml, which indicates that it exists as a monomer in solution (Fig. 2a, b). In contrast, the solubility of caspase-1 CARD was found to be sensitive to pH. While caspase-1 CARD behaved well in pH 8.0 buffer, it was less soluble at pH 5.0 (Fig. 3a). Upon analysis of the effects of salt on the solubility of caspase-1 CARD, salt concentration did not have a great impact on the behavior of caspase-1 CARD in solution (Fig. 3b). Based on the characterization results of ASC CARD and caspase-1 CARD under various buffer conditions, we successfully purified ASC CARD and caspase-1 CARD in buffer containing 20 mM Tris buffer pH 8.0 and 150 mM NaCl. Quick two-step chromatography using affinity and gel-filtration chromatography produced >90 % pure proteins.
Purification and characterization of ASC CARD. a Effect of pH on the solubility of ASC CARD (10.2 kDa) as shown by gel-filtration chromatograms and fractions. The black and gray lines indicate the gel-filtration profile obtained at pH 8.0 and pH 5.0, respectively. SDS-PAGE of the peak fractions of gel-filtration chromatography from pH 8.0 stained by Coomassie blue are shown on the upper left. b Effect of salt on the solubility of ASC CARD as shown by gel-filtration chromatography. The black line shows the profile obtained under 0 mM NaCl using Tris-HCl buffer at pH 8.0. The gray line shows the profile obtained under 150 mM. The black-dotted line shows the profile obtained under 500 mM NaCl. SDS-PAGE of the fractions of gel-filtration chromatography from 150 mM NaCl stained with Coomassie blue are shown on the upper left side
Purification and characterization of caspase-1 CARD. a Effect of pH on the solubility of caspase-1 CARD (12.6 kDa) shown by gel-filtration chromatograms and fractions. The black and gray lines reflect the gel-filtration profiles obtained at pH 8.0 and pH 5.0, respectively. SDS-PAGE of the peak fractions of gel-filtration chromatography from pH 8.0 are shown. b Effect of salt on the solubility of caspase-1 CARD shown by gel-filtration chromatography. The black line shows the profile obtained under 0 mM NaCl using Tris-HCl buffer at pH 8.0. The gray line shows the profile obtained under 150 mM NaCl. The black-dotted line shows the profile obtained under 500 mM NaCl. SDS-PAGE of the fractions of gel-filtration chromatography from 150 mM NaCl are shown
Secondary Structures of Purified ASC CARD and Caspase-1 CARD Are Typical α-Helix Bundle Folds
To confirm the correct folding of purified ASC CARD and caspase-1 CARD, we evaluated the far UV circular dichroic spectra. Both proteins showed CD spectrum patterns typical of α-helical proteins, exhibiting two pronounced minima at 208 and 222 nm and a maxima at 195 nm, which matched the molecular structure of other members of the death domain superfamily well (Fig. 4a, b) [35]. The measured and predicted percentages of helix contents also agreed well, as indicated by 55 % predicted vs. 52 % measured for ASC CARD and 54 % predicted vs. 52 % measured for caspase-1 CARD.
Circular dichroic spectra of purified ASC CARD (a) and caspase-1 CARD (b). The spectra were recorded at 25 °C, and four scans were conducted and averaged using a J-715 spectropolarimeter (Jasco, Japan). The predicted percentage of helix contents was calculated using the GOR IV secondary structure prediction server
ASC CARD Did Not Interact with Caspase-1 CARD In Vitro
We next analyzed whether purified CARDs could interact with each other in vitro as shown in vivo in a previous study [12, 36]. To accomplish this, purified ASC CARD was mixed and incubated with purified caspase-1 CARD for 1 h at room temperature, after which the mixture was applied to a Superdex 200 gel-filtration column 10/30 (GE Healthcare) that had been pre-equilibrated with buffer containing 20 mM Tris buffer pH 8.0 and 150 mM NaCl. Complex formation was evaluated based on the positions of the eluted peak monitored at 280 nm followed by SDS-PAGE. ASC CARD and caspase-1 CARD were eluted at 16 and 18 ml, respectively, on the Superdex 200 gel-filtration column. Because the protein mixture was eluted at around 16 ml, where caspase-1 eluted, and 18 ml, where ASC CARD eluted, without any possible new complex peaks, we concluded that ASC CARD did not interact with caspase-1 CARD in vitro (Fig. 5). SDS-PAGE showed that ASC CARD and caspase-1 CARD did not co-migrate, indicating that they had not formed a complex (Fig. 5).
Complex formation assay by gel-filtration chromatography. Separately purified ASC CARD (10.2 kDa) and caspase-1 CARD (12.6 kDa) were mixed and incubated at room temperature for 1 h. The incubated protein sample was then loaded onto a gel filtration column that had been pre-equilibrated with buffer containing 20 mM Tris pH 8.0 buffer and 150 mM NaCl. SDS-PAGE of gel filtration fractions obtained from the mixture is shown on the right side of the peak
A Predicted Model of the Interaction Between ASC CARD and Caspase-1 CARD for Assembly of the Inflammasome
The only structural information available for the CARD/CARD complex is for the Apaf-1 CARD/caspase-9 CARD complex, which is a core part of the apoptosome. Based on the known CARD complex structure, we predicted a model of the interaction between ASC CARD and caspase-1 CARD. To investigate the interaction, the sequences of ASC CARD and caspase-1 CARD were aligned with those of Apaf-1 and caspase-9, respectively (Fig. 6a). The structure of the Apaf-1CARD/caspase-9 CARD complex showed that the interaction is mainly mediated by charged interactions. Two negatively charged residues in Apaf-1 CARD, D27 and E40, and three positively charged residues in caspase-9 CARD, R13, R52, and R56, were identified as critical residues for the interaction (Fig. 6b). Sequence alignment indicated that important charged residues critical to the interaction between Apaf-1 CARD and caspase-9 CARD are conserved at ASC CARD (D28) and caspase-1 CARD (R55) (Fig. 6a). To analyze the interaction interface formed by ASC CARD and caspase-1 CARD, ASC CARD and modeled caspase-1 CARD were superposed with the previously solved Apaf-1 CARD/caspase-9 CARD complex structure (Fig. 6c). Both ASC CARD and caspase-1 CARD were well superimposed with Apaf-1 CARD and caspase-9 CARD, respectively. The gross features of each electrostatic surface of ASC CARD and caspase-1 CARD were very similar to those of Apaf-1 CARD and caspase-9 CARD in that the surface of CARD can be divided according to whether they are acidic or basic (Fig. 6c). Based on the modeling study, the acidic surface of ASC CARD might be involved in the interaction with the basic surface of caspase-1 CARD as indicated in Fig. 6c.
Model of the interaction between ASC CARD and caspase-1 CARD. a Sequence alignment between Apaf-1 CARD and ASC CARD, and between caspase-9 CARD and caspase-1 CARD. Residues involved in the interaction are indicated by red and blue color. b Previously reported structure of Afap-1 CARD/caspase-9 CARD complex. Residues involved in the interaction are shown in the stick model. c Superposition of ASC CARD and caspase-1 CARD to the Afap-1 CARD/caspase-9 CARD complex structure. A surface model was generated to show the surface charge
Discussion
NALP3 inflammasomes are molecular platforms activated upon stress and cellular infections that trigger the maturation and secretion of pro-inflammatory cytokines such as IL-1β to engage in innate immune defenses [12]. The assembly of NALP3 inflammasomes constitutes the interactions between NALP3, ASC, and caspase-1. NALP interacts with bipartite adaptor protein ASC through PYD/PYD domain interactions, while ASC interacts with caspase-1 through CARD/CARD domain interactions for downstream signaling. Many pro-apoptotic and pro-inflammatory proteins contain the CARD domain. The recruitment and activation of caspase-1 by ASC through the homotypic CARD/CARD interaction is critical to activation of caspase-1 by assembly of inflammasome. Basically, CARDs are members of the DD superfamily of six antiparallel α-helix bundle proteins that interact to form activation platforms in which interacting DD partners form short helical stacks or lock washer like disks [23]. Further knowledge regarding the higher resolution disk assembly of the CARD/CARD interaction clarifies the mechanism of activation of caspase-1 and therefore subsequent NF-κB activation.
Normally, the in vitro overexpression of ASC/caspase-1 CARD domains in bacterial systems are cumbersome, and most of the protein becomes insoluble, limiting structural studies of the CARD/CARD complex. However, the successful expression of recombinant CARD domains of ASC and caspase-1 in soluble form is also very unstable and oligomerizes easily. CARD interactions in ASC CARD/caspase-1 CARD have been reported in vivo in inflammasome assemblies [37]. However, the in vitro experiments conducted herein did not reveal any CARD/CARD interactions. Using the structure of ASC CARD and a model of caspase-1 CARD (based on the Iceberg NMR structure), Proell et al. [38] conducted several mutational analyses of caspase-1 CARD surfaces with respect to foci formation with ASC-CARD. The results revealed that mutants R10E, D27R, E41R, R55E, and D59R of caspase-1 CARD abolished loci formation by aborting interaction with ASC CARD. Kersse et al. [39] reported that D27R mutation of the caspase-1 CARD interrupts ASC/caspase-1 CARD signaling. They concluded that all mutants show loss of foci formation and interaction with ASC CARD residue in two main surfaces: one created by helices α1, α3, and α4 (Arg10, Lys42, and Asp59) and another located in helices α2 and α3 (Asp27 and Lys42) of caspase-1 CARD [38].
Since the last decade, several models have been proposed to solve the structure of CARD/CARD interaction. Zhou et al. [40] reported solution of the structure of the Apaf-1 CARD domain and its surface of interaction with caspase-9 CARD. By using chemical shift perturbation data, a model was proposed in which α1 and α4 of caspase-1 CARD bound to α2, α3, and α5 of Apaf-1 CARD. The acidic α2 and α3 of Apaf-1 CARD form a typical antiparallel four-helix bundle with the basic α1 and α4 of caspase-1 CARD. Later, Qin et al. [41] reported the crystal structure of the CARD/CARD complex between Apaf-1 and procaspase-9, which is the only CARD/CARD complex that has been structurally elucidated to date. The CARD/CARD interface primarily consists of electrostatic interactions between an acidic and convex surface patch of helices α1 and α4 within the CARD domain of Apaf-1 and a basic and concave surface path of helices α2 and α3 within the CARD of procaspase-9. The crucial residues involved in interactions (D27, E39, E40, and E41) are localized within the acidic region of Apaf-1. Multiple sequence alignment of Apaf-1/caspase-9 and ASC/caspase-1 CARD domains showed conserved residues in an acidic patch (Apaf-1: D27, S38, E39, E40, E41; ASC: D134, E144, Q145, Y146, Q147; caspase-1: D27, E38, E39, M40, E41; and caspase-9: D27, H38, M39, I40, E41) and a basic patch (Apaf-1: R12, E46, Q49, A54; ASC: R15, E152, N155, R56; caspase-1: R10, N47, V50, R55; and caspase-9: R13, G47, R51, R56) [42]. Although the primary sequence conservation between CARD domains is very low, there is a high degree of structural homology and the interface residues of CARD/CARD of Apaf-1/C9 are highly conserved among NLR effector domains, caspases, and adaptor proteins. In particular, the first and last residues in the acidic patch and the basic patch are highly conserved among CARD domains. These findings suggest that the CARD/CARD pattern of interaction between ASC/C1 would be similar to that described for Apaf-1/C9. However, recently, Lu et al. [43] suggested a unified model of ASC-dependent inflammasome assembly in two successive steps of nucleation-induced polymerization. AIM2/NLRP3 upon activation nucleates helical ASC clusters through PYD/PYD interactions. The oligomerized ASC CARDs then acts as a platform for caspase-1 CARD to nucleate into filaments, which brings caspase domains into proximity inducing dimerization, trans-autocleavage, and activation in situ. Thus, nucleation-induced filament formation mechanism is involved in ASC-dependent inflammasome formation. In vitro reconstitution of the ternary inflammasome suggests 3.5-fold overstoichiometry of caspase-1 to ASC. Santiveri et al. [44] proposed a seven-member ring-like model for ASC self-association via the CARD domain. Self-oligomerization of ASC PYD domain prevents complex formation in the NALP3 inflammasome in vitro [45]. This study also suggests that ASC CARD and caspase-1 CARD did not interact with each other because of self-oligomerization of caspase-1 CARD. CARD domain-mediated self-oligomerization of caspase-1 might be regulating process for the assembly of inflammasome.
References
Kolb, R., Liu, G. H., Janowski, A. M., Sutterwala, F. S., & Zhang, W. (2014). Inflammasomes in cancer: a double-edged sword. Protein & Cell, 5, 12–30.
Dostert, C., Petrilli, V., Van Bruggen, R., Steele, C., Mossman, B. T., & Tschopp, J. (2008). Innate immune activation through Nalp3 inflammasome sensing of asbestos and silica. Science, 320, 674–677.
Martinon, F., Burns, K., & Tschopp, J. (2002). The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Molecular Cell, 10, 417–426.
Ghayur, T., Banerjee, S., Hugunin, M., Butler, D., Herzog, L., Carter, A., Quintal, L., Sekut, L., Talanian, R., Paskind, M., Wong, W., Kamen, R., Tracey, D., & Allen, H. (1997). Caspase-1 processes IFN-gamma-inducing factor and regulates LPS-induced IFN-gamma production. Nature, 386, 619–623.
Kanneganti, T. D., Ozoren, N., Body-Malapel, M., Amer, A., Park, J. H., Franchi, L., Whitfield, J., Barchet, W., Colonna, M., Vandenabeele, P., Bertin, J., Coyle, A., Grant, E. P., Akira, S., & Nunez, G. (2006). Bacterial RNA and small antiviral compounds activate caspase-1 through cryopyrin/Nalp3. Nature, 440, 233–236.
Sutterwala, F. S., Ogura, Y., Szczepanik, M., Lara-Tejero, M., Lichtenberger, G. S., Grant, E. P., Bertin, J., Coyle, A. J., Galan, J. E., Askenase, P. W., & Flavell, R. A. (2006). Critical role for NALP3/CIAS1/Cryopyrin in innate and adaptive immunity through its regulation of caspase-1. Immunity, 24, 317–327.
Dinarello, C. A. (2011). Interleukin-1 in the pathogenesis and treatment of inflammatory diseases. Blood, 117, 3720–3732.
Lamkanfi, M., & Dixit, V. M. (2014). Mechanisms and functions of inflammasomes. Cell, 157, 1013–1022.
Liu, W., Luo, Y., Dunn, J. H., Norris, D. A., Dinarello, C. A., & Fujita, M. (2013). Dual role of apoptosis-associated speck-like protein containing a CARD (ASC) in tumorigenesis of human melanoma. Journal of Investigative Dermatology, 133, 518–527.
Davis, B. K., Wen, H., & Ting, J. P. (2011). The inflammasome NLRs in immunity, inflammation, and associated diseases. Annual Review of Immunology, 29, 707–735.
Agostini, L., Martinon, F., Burns, K., McDermott, M. F., Hawkins, P. N., & Tschopp, J. (2004). NALP3 forms an IL-1beta-processing inflammasome with increased activity in Muckle-Wells autoinflammatory disorder. Immunity, 20, 319–325.
Schroder, K., & Tschopp, J. (2010). The inflammasomes. Cell, 140, 821–832.
Fernandes-Alnemri, T., Yu, J. W., Datta, P., Wu, J., & Alnemri, E. S. (2009). AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature, 458, 509–513.
Kolly, L., Busso, N., Palmer, G., Talabot-Ayer, D., Chobaz, V., & So, A. (2010). Expression and function of the NALP3 inflammasome in rheumatoid synovium. Immunology, 129, 178–185.
Liu-Bryan, R. (2010). Intracellular innate immunity in gouty arthritis: role of NALP3 inflammasome. Immunology and Cell Biology, 88, 20–23.
Martinon, F., Petrilli, V., Mayor, A., Tardivel, A., & Tschopp, J. (2006). Gout-associated uric acid crystals activate the NALP3 inflammasome. Nature, 440, 237–241.
Feldmann, J., Prieur, A. M., Quartier, P., Berquin, P., Certain, S., Cortis, E., Teillac-Hamel, D., Fischer, A., & de Saint Basile, G. (2002). Chronic infantile neurological cutaneous and articular syndrome is caused by mutations in CIAS1, a gene highly expressed in polymorphonuclear cells and chondrocytes. The American Journal of Human Genetics, 71, 198–203.
Hoffman, H. M., Mueller, J. L., Broide, D. H., Wanderer, A. A., & Kolodner, R. D. (2001). Mutation of a new gene encoding a putative pyrin-like protein causes familial cold autoinflammatory syndrome and Muckle-Wells syndrome. Nature Genetics, 29, 301–305.
Bae, J. Y., & Park, H. H. (2011). Crystal structure of NALP3 protein pyrin domain (PYD) and its implications in inflammasome assembly. Journal of Biological Chemistry, 286, 39528–39536.
Martinon, F. (2008). Detection of immune danger signals by NALP3. Journal of Leukocyte Biology, 83, 507–511.
Duncan, J. A., Bergstralh, D. T., Wang, Y., Willingham, S. B., Ye, Z., Zimmermann, A. G., & Ting, J. P. (2007). Cryopyrin/NALP3 binds ATP/dATP, is an ATPase, and requires ATP binding to mediate inflammatory signaling. Proceedings of the National Academy of Sciences U.S.A, 104, 8041–8046.
Shiohara, M., Taniguchi, S., Masumoto, J., Yasui, K., Koike, K., Komiyama, A., & Sagara, J. (2002). ASC, which is composed of a PYD and a CARD, is up-regulated by inflammation and apoptosis in human neutrophils. Biochemical and Biophysical Research Communications, 293, 1314–1318.
Park, H. H., Lo, Y. C., Lin, S. C., Wang, L., Yang, J. K., & Wu, H. (2007). The death domain superfamily in intracellular signaling of apoptosis and inflammation. Annual Review of Immunology, 25, 561–586.
Park, H. H., Logette, E., Rauser, S., Cuenin, S., Walz, T., Tschopp, J., & Wu, H. (2007). Death domain assembly mechanism revealed by crystal structure of the oligomeric PIDDosome core complex. Cell, 128, 533–546.
Damiano, J. S., & Reed, J. C. (2004). CARD proteins as therapeutic targets in cancer. Current Drug Targets, 5, 367–374.
Franchi, L., Eigenbrod, T., Munoz-Planillo, R., & Nunez, G. (2009). The inflammasome: a caspase-1-activation platform that regulates immune responses and disease pathogenesis. Nature Immunology, 10, 241–247.
Jang, T. H., & Park, H. H. (2011). Generalized semi-refolding methods for purification of the functional death domain superfamily. Journal of Biotechnology, 151, 335–342.
Park, H. H. (2011). Structural analyses of death domains and their interactions. Apoptosis, 16, 209–220.
Schwede, T., Kopp, J., Guex, N., & Peitsch, M. C. (2003). SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Research, 31, 3381–3385.
Laskowski, R. A., Rullmannn, J. A., MacArthur, M. W., Kaptein, R., & Thornton, J. M. (1996). AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. Journal of Biomolecular NMR, 8, 477–486.
Dzivenu, O. K., Park, H. H., & Wu, H. (2004). General co-expression vectors for the overexpression of heterodimeric protein complexes in Escherichia coli. Protein Expression and Purification, 38, 1–8.
Chen, Y. H., Yang, J. T., & Martinez, H. M. (1972). Determination of the secondary structures of proteins by circular dichroism and optical rotatory dispersion. Biochemistry, 11, 4120–4131.
Laemmli, U. K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 227, 680–685.
Bae, J. Y., & Park, H. H. (2011). Crystallization and preliminary X-ray crystallographic studies of the PYD domain of human NALP3. Acta Crystallographica Section F Structural Biology and Crystallization Communications, 67, 1421–1424.
Jang, T. H., Bae, J. Y., Park, O. K., Kim, J. H., Cho, K. H., Jeon, J. H., & Park, H. H. (2010). Identification and analysis of dominant negative mutants of RAIDD and PIDD. Biochimica et Biophysica Acta, 1804, 1557–1563.
Srinivasula, S. M., Poyet, J. L., Razmara, M., Datta, P., Zhang, Z., & Alnemri, E. S. (2002). The PYRIN-CARD protein ASC is an activating adaptor for caspase-1. Journal of Biological Chemistry, 277, 21119–21122.
Stehlik, C., Lee, S. H., Dorfleutner, A., Stassinopoulos, A., Sagara, J., & Reed, J. C. (2003). Apoptosis-associated speck-like protein containing a caspase recruitment domain is a regulator of procaspase-1 activation. Journal of Immunology, 171, 6154–6163.
Proell, M., Gerlic, M., Mace, P. D., Reed, J. C., & Riedl, S. J. (2013). The CARD plays a critical role in ASC foci formation and inflammasome signalling. Biochemistry Journal, 449, 613–621.
Kersse, K., Lamkanfi, M., Bertrand, M. J., Vanden Berghe, T., & Vandenabeele, P. (2011). Interaction patches of procaspase-1 caspase recruitment domains (CARDs) are differently involved in procaspase-1 activation and receptor-interacting protein 2 (RIP2)-dependent nuclear factor kB signalling. Journal of Biological Chemistry, 286, 35874–35882.
Zhou, P., Chou, J., Olea, R. S., Yuan, J., & Wagner, G. (1999). Solution structure of Apaf-1 CARD and its interaction with caspase-9 CARD: a structural basis for specific adaptor/caspase interaction. Proceedings of the National Academy of Sciences U.S.A, 96, 11265–11270.
Qin, H., Srinivasula, S. M., Wu, G., Fernandes-Alnemri, T., Alnemri, E. S., & Shi, Y. (1999). Structural basis of procaspase-9 recruitment by the apoptotic protease-activating factor 1. Nature, 399, 549–557.
Proell, M., Riedl, S. J., Fritz, J. H., Rojas, A. M., & Schwarzenbacher, R. (2008). The Nod-like receptor (NLR) family: a tale of similarities and differences. PloS One, 3, e2119.
Lu, A., Magupalli, V. G., Ruan, J., Yin, Q., Atianand, M. K., Vos, M. R., Schroder, G. F., Fitzgerald, K. A., Wu, H., & Egelman, E. H. (2014). Unified polymerization mechanism for the assembly of ASC-dependent inflammasomes. Cell, 156, 1193–1206.
Santiveri, C. M., Oroz, J., & de Alba, E. (2014). A ring-like model for ASC self-association via the CARD domain. Inflammasome, 1, 44–54.
Narayanan, K. B., Jang, T. H., & Park, H. H. (2014). Self-oligomerization of ASC PYD domain prevents the assembly of inflammasome in vitro. Applied Biochemistry and Biotechnology, 172, 3902–3912.
Acknowledgments
This study was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) of the Ministry of Education, Science and Technology (NRF-2012R1A2A2A01010870) and a grant from the Korea Healthcare Technology R&D Project, Ministry of Health and Welfare, Republic of Korea (HI13C1449).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Narayanan, K.B., Park, H.H. Purification and Analysis of the Interactions of Caspase-1 and ASC for Assembly of the Inflammasome. Appl Biochem Biotechnol 175, 2883–2894 (2015). https://doi.org/10.1007/s12010-014-1471-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-014-1471-4