Abstract
Background
Colorectal cancer is the third most common cancer and requires more prognostic biomarkers for precise treatment. GPR39 is a GPCR which can interact with Zn and modulate the colonocytes’ survival. The clinical significance of GPR39 in colon cancer has never been reported.
Materials
In our study, we compared GPR39 expression between colon cancers and tumor-adjacent tissues by retrieving TCGA data and detected the expression of GPR39 in colon cancers with qPCR and immunohistochemistry. The clinical significance of GPR39 was evaluated by analyzing the correlations with clinicopathological factors with the chi-square test. The prognostic significance of GPR39 was estimated with univariate and multivariate analyses. The expression of several other biomarkers including PPARG, EPCAM, and PD-L1 was investigated by re-analyzing TCGA data, qPCR, and IHC. The prognostic value of PPARG, EPCAM, and PD-L1 was also estimated with univariate analysis.
Results
In both TCGA database and our 15 colon cancer pairs, GPR39 expression was significantly upregulated in colon cancer tissues. GPR39 was an independent prognostic biomarker in colon cancer for poor prognosis. With TCGA data re-analysis, qPCR, and IHC, we showed that GPR39 expression was significantly correlated with the expression of EPCAM and PD-L1, but not PPARG. EPCAM and PD-L1 were also unfavorable prognostic biomarkers of colon cancer.
Conclusions
GPR39 was upregulated in colon cancer tissues compared with tumor-adjacent tissues. GPR39 was an independent prognostic biomarker in colon cancer for poor prognosis. EPCAM and PD-L1 were substantially associated with GPR39 expression, and they were also identified as prognostic biomarkers in colon cancers.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Colorectal cancer (CRC), including colon cancer and rectal cancer, is the third most common cancer and the second leading cause of cancer-related death worldwide [1, 2]. Although the standard chemotherapy and new targeted therapy have been improved in recent years, problems such as multidrug resistance remain unresolved and the prognosis of CRC is still unsatisfactory. In China, the 5-year survival rate for CRC is approximately 30% [3,4,5]. The high heterogeneity of CRC is a main reason of its poor response to some treatments and dismal prognosis [4, 5]. In this era of precise treatment, many breakthroughs have been made to classify CRC based on the molecular signatures, but few results of these studies were used in clinical practice to predict patients’ prognoses. The identification of new biomarkers is the basis of new treatment options and target therapies, so more effective biomarkers are required and more individualized treatment strategies of CRC are needed.
G protein-coupled receptors (GPCRs), also known as 7TM receptors, are a large group of cell surface receptors which respond to a variety of external signals and play essential roles in numerous cellular processes [6]. There are more than 600 members of GPCR, and GPR39 is a distinguished GPCR which can sense changes in extracellular Zinc (Zn2+) [7]. Zn2+ is an essential micronutrient participating in numerous vital physiological processes as a structural cofactor of numerous zinc finger transcription factors or enzymes [8]. In epithelial cells, Zn2+ can promote epithelial cell proliferation and survival and facilitate tumor progression originated from epithelial cells [9]. In many tissue types including neurons, colon epithelial cells, skin epidermal cells, pancreatic cells, prostate cancer cells, salivary gland cells, and in bones, Zn2+ has been reported to regulate the activity of GPR39.
GPR39 mainly couples Gq and activates the IP3 pathway to release intracellular Ca2+ from the endoplasmic reticulum [10]. Other reports showed that GPR39 can activate MAPK and AKT and thus increase cell proliferation and migration [10], which suggested a possible role for GPR39 in cancer progression. GPR39 was reported to be overexpressed in breast cancer and to promote breast cancer cell migration and proliferation [11]. The overexpression of GPR39 was also observed in several tumor types such as esophageal squamous cell carcinomas and gastric cancer [12, 13]. Moreover, the activation of GPR39 was also reported in androgen-independent prostate cancer cells [14]. GPR39 is a key receptor of Zn2+ which regulates Zn2+ signaling especially in epithelial cells including keratinocytes, colonocytes (colon epithelium cells), and salivary gland cells [15]. In colonocytes, GPR39 can reduce cell death by upregulating clusterin [15]. However, the expression and clinical significance of GPR39 in colon cancer are still unknown.
In our study, we retrieved the data from TCGA database and detected the expression of GPR39 in colon cancers with quantitative real-time PCR (qPCR) and immunohistochemistry (IHC). Moreover, we compared GPR39 expression between colon cancers and tumor-adjacent tissues. The clinical significance of GPR39 was evaluated by analyzing the correlations with clinicopathological factors such as histological grade, tumor size, vascular invasion, tumor infiltration, lymphatic invasion, and metastasis. The prognostic significance of GPR39 was also estimated with univariate and multivariate analyses. By analyzing the TCGA database and literature review, we investigated several biomarkers which may be related with GPR39 expression including peroxisome proliferator-activated receptor gamma (PPARG), epithelial cell adhesion molecule (EPCAM), and programmed cell death 1 ligand 1 (PD-L1) and further investigated their expression and prognostic significance in colon cancer.
Materials and methods
Patients and follow-up
Our study was approved by the committee of Yidu Central Hospital and Qilu Hospital of Shandong University. A total of 273 patients were diagnosed with colon cancer and underwent radical surgery from 2010 to 2012 in Yidu Central Hospital and Qilu Hospital. A total of 161 patients were further selected into the final cohort if they had available follow-ups. All the specimens were obtained after the consents of patients. The whole study was approved and supervised by the Ethics Committee of Yidu Central Hospital and Qilu Hospital of Shandong University. The TNM stage was according to the 8th American Joint Committee on Cancer/Union for International Cancer Control (AJCC/UICC) staging system.
In silico analysis
The data from 275 colon cancers and 349 tumor-adjacent colon tissues were retrieved from the Cancer Genome Atlas (TCGA, http://cancergenome-nih.gov/) for data re-analysis. Transcripts Per Kilobase of exon model per Million mapped reads (TPMs) were applied to evaluate the expression of GPR39, PPARG, EPCAM, and PD-L1. The correlations between GPR39, PPARG, EPCAM, and PD-L1 were also assessed by TPMs. An online website GEPIA was used for data retrieval and analysis (http://gepia.cancer-pku.cn/detail.php).
RNA extraction and qPCR
The mRNA levels of GPR39, PPARG, EPCAM, and PD-L1 in the 15 pairs of colon cancers and the corresponding tumor-adjacent normal tissues were detected with qPCR. Firstly, TRIzol reagent (Thermo Fisher) and RNeasy protect mini kit (Qiagen, Hilden, Germany) were used to extract the total RNAs of these tissues. After that, Primescript RT reagent kit (Takara BIO INC.) was used for reverse transcription PCR. The quantification of qPCR was finally achieved by Thermo Fisher 7500 PCR System. The results were analyzed with the GAPDH as the internal control in a 2−ΔΔCt method. The qPCR primers were designed as follows: GPR39, forward: 5′-CAGGTCCCCGACAAGATCATA-3′, reverse: 5′-TGAGACCGTGTGGTACTTGAG-3′; PPARG, forward: 5′-GATGCCAGCGACTTTGACTC-3′, reverse: 5′-ACCCACGTCATCTTCAGGGA-3′; EPCAM, forward: 5′-AATCGTCAATGCCAGTGTACTT-3′, reverse: 5′-TCTCATCGCAGTCAGGATCATAA-3′; PD-L1, forward: 5′-TGGCATTTGCTGAACGCATTT-3′, reverse: 5′-TGCAGCCAGGTCTAATTGTTTT-3′; GADPH, forward: 5′-TGTGGGCATCAATGGATTTGG-3′, reverse: 5′-ACACCATGTATTCCGGGTCAAT-3′.
Tissue microarray (TMA) and immunohistochemistry
The 161 cases of formalin-fixed and paraffin-embedded colon cancer tissues were used to construct a TMA with 1 mm cylinders representing each sample. In the TMA, the expressions of GPR39, PPARG, EPCAM, and PD-L1 were detected with IHC. In brief, the TMAs were first deparaffinized with xylene and rehydrated with graded ethanol. Three percent of hydrogen peroxide was applied to inactivate the endogenous peroxidase activity. Slides were boiled in citrate buffer (pH = 6.0) for 10 min for optimal antigen retrieval, and then in 5% bovine serum albumin for 30 min to eliminate unspecific antigen binding. The primary antibodies of GPR39 (Novus Biologicals, catalog: NLS139), PPARG (Santa Cruz Biotechnology, catalog: sc-7273), EPCAM (BioLegend, catalog: 324,202), and PD-L1 (Invitrogen, catalog: 14–5983-80) were used to incubate the specimen at 4 °C overnight. The biotin-labeled secondary antibodies (Sangon, Shanghai, China) and streptavidin-peroxidase (Sangon) were used to incubate the slides. Finally, the visualization of slides was achieved by incubation 3, 3′-diaminobenzidine substrate for 10 min.
Evaluation of IHC results
The results of IHC were semi-quantified by evaluation with two senior pathologists who were unaware of the clinical information. The IHC score was defined as the score of staining intensity multiplied by the score of IHC positive cell percentage. The scores of staining intensity were set as score 0 representing negative staining, 1 representing weak staining, 2 representing moderate staining, and 3 representing strong staining. The scores of IHC positive cell percentage were defined as score 1 representing less than 25% of positive cells, score 2 representing 25–50% of positive cells, score 3 representing 50–75% of positive cells, and score 4 representing more than 75% of positive cells. So the final IHC scores ranged from 0 to 12. The patients were then divided into subgroups with low and high IHC scores of these biomarkers by the cut-off, which was identified by the receiver operating characteristic (ROC) curve.
Statistical analysis
SPSS 22.0 (IBM cooperation, Chicago, USA) was used to analyze the data. One-way ANOVA was used to compare TPMs of GPR39 in colon cancers and adjacent tissues. Paired t test was used to compare GPR39 expression in 15 pairs of colon cancer tissues. The correlations between the expression of GPR39 and the clinicopathological variables were analyzed with the chi-square test. Kaplan–Meier method was used to plot the overall survival (OS) curves, and the log-rank test was used to calculate the statistical difference between different groups. The Cox-regression hazard model was applied to identify the independent prognostic factors. P < 0.05 was considered as statistically significant.
Results
GPR39 was overexpressed in colon cancer compared with adjacent tissues
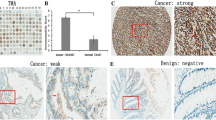
The expression of GPR39 was first evaluated by retrieved the data from TCGA database. TPMs were used to evaluate GPR39 mRNA level in 275 colon tumors and 349 tumor-adjacent colon tissues (Fig. 1A). The TPMs of GPR39 in tumor tissues were significantly higher than TPMs in adjacent tissues. Moreover, we collected 15 pairs of colon tumors and tumor-adjacent colon tissues and detected the GPR39 mRNA levels with qPCR. The qPCR results were in consistent with the mRNA sequencing results, which indicated that GPR39 was overexpressed in colon cancer tissues (Fig. 1B). Furthermore, we detected the expression of GPR39 with IHC in 161 colon cancer tissues (Fig. 1C). As a GPCR, GPR39 was expressed in the cell membrane in our study as expected (Fig. 1D). The 161 colon cancer tissues were further divided into subgroups which had low or high GPR39 expression, accounting for 55.28% (89/161) and 44.72% (72/161), respectively.
GPR39 expression in colon cancers. A The TPMs of GPR39 in 275 cases of colon cancers and 349 cases of adjacent tissues were compared with one-way ANOVA test. B GPR39 mRNA levels in 15 pairs of colon cancers and tumor-adjacent tissues were detected with qPCR. Statistical significance was evaluated with paired t test, and data were from 3 independent experiments. C IHC was used to detect GPR39 expression in 161 cases of colon cancer. D The magnified images of the boxes in C
GPR39 was associated with patients’ sex, T, and TNM stage
In the 161 patients, we performed the chi-square test to screen the potential tumor-associated progression which GPR39 may be involved in. In our study, GPR39 was significantly associated with T stage (P = 0.007) and TNM stage (P = 0.034). These results suggested that high expression of GPR39 in colon tumors indicated the advanced T stage and TNM stage (Table 1). Interestingly, we also observed that GPR39 correlated with patients’ sex in colon cancer. Female patients tended to have high GPR39 expression (P = 0.048).
GPR39 expression was correlated with the OS in colon cancers
In addition, we analyzed the correlation between the clinicopathological factors, GPR39 expression, and the OS with univariate analysis (Table 2). In our cohort, clinicopathological factors including histological grade (P < 0.001), lymphatic invasion (P < 0.001), metastasis (P = 0.002), vascular invasion (P = 0.016), and TNM stage (P = 0.001) were all prognostic factors. High histological grade, positive lymphatic invasion, metastasis and vascular invasion, and advanced TNM stage were all significantly associated with low OS rates in colon cancer (Fig. 2A–E). Moreover, GPR39 was identified as a significant biomarker predicting the poor prognosis of colon cancer (P = 0.002). The 5-year OS rates of patients with low and high expression of GPR39 were 67.9% and 49.7%, respectively (Fig. 2F). Other clinicopathological factors including T stage (P = 0.389) and tumor size (P = 0.671) exhibited no significant statistical difference towards the OS rate in colon cancer(Fig. 2G, H).
Survival significance of clinicopathological factors and GPR39 expression. A–F Histological grade, lymph node invasion, metastasis, vascular invasion, TNM stage, and GPR39 expression were all prognostic factors in colon cancers. Kaplan–Meier method was used to plot the OS curves and the log-rank test was used to calculate the statistical differences. G and H The correlations between T stage, tumor size, and the OS rate had no significant difference
GPR39 as an independent prognostic factor in colon cancer
All the prognostic factors in univariate analysis were enrolled into the multivariate analysis for the identification of independent prognostic factors of colon cancer in our study. The enrolled factors included histological grade, lymphatic invasion, metastasis, vascular invasion, and GPR39 expression (Table 3). TNM stage was a consequent factor constituted by T, N, and M stage, so it was excluded from the multivariate analysis. In the Cox-regression model for multivariate analysis, GPR39 expression was identified as an independent risk for unfavorable prognosis (P = 0.002). The cancer-related death odds of patients with high GPR39 expression was 2.17-fold higher than the patients with low GPR39 expression (HR = 2.17). In addition, high histological grade (P < 0.001), positive lymph node invasion (P = 0.002), and positive metastasis (P < 0.001) all indicated the poor prognosis independently in colon cancer (Table 3).
GPR39 expression was significantly associated with EPCAM and PDL1 expression
From the TCGA database, we screened the potential genes which may be associated with GPR39. PPARG and EPCAM were selected because they had possible positive correlations with GPR39 in TCGA database (Fig. 3A), and they were reported to be involved in tumor progression and prognosis in colon cancer [16,17,18]. In our study, PD-L1 was also detected because of its essential role in the response to immunotherapy. We detected their mRNA correlations with GPR39 mRNA in the 15 patients with colon cancer. Interestingly, there were no substantial correlations between GPR39 and PPARG, though PPARG was shown to be associated with GPR39 with mRNA sequencing in TCGA database. EPCAM and PD-L1 mRNA expression had significant correlations with GPR39 in the consecutive cohort consisting of 15 colon cancers (Fig. 3B). Moreover, we detected PPARG, EPCAM, and PD-L1 in the 161 cases of colon cancer and divided them into subsets according to the different expressions of PPARG, EPCAM, and PD-L1 (Fig. 3C). In colon cancer, PPARG was mainly localized in cell nucleus, EPCAM was localized in both cell nucleus and cytoplasm, and PD-L1 was mainly expressed in membrane (Fig. 3D). In consistent with the qPCR results in Fig. 3B, patients with high GPR39 expression had a high expression level of EPCAM and PD-L1, but not PPARG (Fig. 3E).
GPR39 expression correlated with EPCAM and PD-L1 expression. A In TCGA database, the GPR39 expression exhibited significant correlation with PPARG and EPCAM in 275 colon cancers. R value was calculated by the Spearman analysis. B The correlations between PPARG, EPCAM, and PD-L1, and GPR39 expression were analyzed in the 15 colon cancer tissues with the Spearman analysis. C and D PPARG, EPCAM, and PD-L1 expressions in 161 colon cancer tissues were detected with IHC. E Patients with high GPR39 expression had more expression of EPCAM and PD-L1, but not PPARG. * represents P < 0.05, ** represents P < 0.01 with t test
EPCAM and PD-L1 were also prognostic biomarkers of colon cancer
After detection of EPCAM, PD-L1, and GPR39 expression with IHC, we analyzed the correlation between EPCAM, PD-L1, and GPR39 expression with the chi-square test. EPCAM (P = 0.001) and PD-L1 (P = 0.001) expression were all positively associated with GPR39 expression (Table 4). The prognostic value of PPARG, EPCAM, and PD-L1 was evaluated with univariate analysis. Intriguingly, PPARG exhibited no correlation with the OS in colon cancer (P = 0.350) (Fig. 4A), while both EPCAM (P = 0.019) and PD-L1 (P = 0.033) indicated the poor prognoses of colon cancer (Fig. 4B, C).
Discussion
Zn2+ is a critical metal elements in human and the deficiency of Zn2+ is linked to many diseases mainly in the digestive, immune, nervous, endocrine, and integumentary systems, such as impaired learning and memory, diarrhea and taste disorders [19, 20]. Zn2+ is an essential structural element and cofactor in enzymes and transcription factors, and there are approximate 3000 proteins which have Zn2+ binding sites and their activities were regulated by Zn2+ interaction [21,22,23]. Although it has been decades since the essential role of Zn2+ has been defined, the mechanisms underlying Zn2+-involved cellular processes and tumor progression are still poorly understood [24]. GPR39 is known as Zn2+ sensing receptor can interact with extracellular Zn2+ at physiological concentrations, couples Gq-protein and therefore triggers the IP3 pathway to release intracellular Ca2+ [10]. In colonocytes, GPR39 was showed to activate downstream signaling pathways such as MAPK-ERK signaling and PI3K-AKT pathway [25,26,27]. In our study, we for the first time investigated the expression of GPR39 in colon cancer and the corresponding tumor-adjacent tissues. Furthermore, we identified GPR39 as an independent prognostic biomarker of colon cancers. This is an important supplement of the oncogenic role of GPR39 because there are quite few studies on the function of GPR39 in cancer. The expression of GPR39 was only studied in prostate cancer, breast cancer and gastric cancer till now.
The AJCC TNM system is the only well-accepted system to predict the prognosis and select patients for adjuvant therapy until now [28]. In recent years, several studies tried to classify CRC according to the molecular characteristics [29]. Personalized medicine is relied on the stratification of tumors into subtypes by molecular fingerprints associated with different prognoses or therapeutic responses. For example, in colorectal cancer, KRAS mutations correlates with poor prognosis and guide therapeutic decisions [30, 31]. However, the high-throughput methods such as mRNA sequencing need the validation of protein level detection, and the results need further verification of larger cohorts and multi-centers. In our study, we identified GPR39 as an independent prognostic biomarker in colon cancer for the first time, which suggested that detection of GPR39 expression could stratify the high-risk patients more precisely. This is an important supplement to the post-operational detection of colon cancer, and may be a new elicitation to the precise treatment of patients with colon cancer.
Till now, there is no well-accepted biomarker and criteria to predict the response to immunotherapy, mainly including the immune checkpoint inhibitor such as PD1 antagonist. It is still controversial whether PD-L1 expression is associated with the response to PD1 inhibitor [32]. Here we detected the expression of PD-L1 in colon tumor cells and interestingly demonstrated that PD-L1 was a prognostic biomarker of colon cancer. All the patients in our cohort received no immunotherapy because the establishment of cohort is more than 10 years ago, when the therapy of immune checkpoint inhibitor is not so common. In these patients, high expression of PD-L1 indicated the poor prognosis of colon cancer, which may be explained that high PD-L1 in tumor cells could inhibit the immune cells and promote the immune escape in the microenvironment of colon cancer. However, whether these patients have better response to immunotherapy requires further clinical study to demonstrate.
PPARG is a kind of nuclear receptor which interacts with peroxisome proliferators such as hypolipidemic drugs and fatty acids. PPARG rs3856806 C > T polymorphism increased the risk of colorectal cancer in Eastern Chinese Han population [33], but PPARG was also reported to be correlated with good prognosis in colorectal cancer in a cohort consisting of American patients [16], which was not validated in our cohort. In addition, plenty of evidence showed that EPCAM could promote the progression of colon cancer [17, 18], and indicated that EPCAM antibody can help treat colon cancer [34]. In our stydy, we screened the potential biomarkers which may be associated with GPR39 expression in colon cancer in TCGA database and selected PPARG and EPCAM by literature review. Interestingly, only EPCAM had positive correlation with GPR39 and OS rate, though PPARG and EPCAM exhibited strong correlations with GPR39 in mRNA sequencing results in TCGA. This result showed that the validation of protein level to high-throughput sequencing is necessary because that the probes of mRNA sequencing may be not that special and many post-translational changes after mRNA translation could not be ignored. For the first time, we showed that PD-L1 and EPCAM expressions were significantly associated with GPR39, which is an interesting result and needs further study to elucidate the mechanism. Our results indicated that PD-L1 or EPCAM may be downstream factors responsible for the GPR39-induced prognosis of colon cancer. This may elicit a new direction of GPR39 and colon cancer study.
The importance of GPR39 has been investigated in many diseases, including inflammatory bowel diseases, epilepsy, depression, and cancer [7]. Moreover, GPR39 knock-out mice have heavier body weight, more food intake, and energy expenditure, indicating that GPR39 may be also involved in metabolic processes [35]. Taken together, GPR39 is a promising drug target considering the essentiality of Zn2+ in numerous cellular processes and the clinical significance of GPR39. With a variety of high-throughput screenings, several agonists for GPR39 have been proposed but very few are successfully validated by in vivo experiments [36]. Recently, an oral active GPR39 agonist, called Cpd1324, was developed as a weight-lowering agent due to its stimulatory effect on GLP-1 secretion [37]. However, the specific inhibitor of GPR39 has not been identified. To screen and validate the effective inhibitor of GPR39 is a promising but challenging topic to elucidate the underlying mechanism of GPR39-involved cellular processes and help develop new therapeutic strategies to treat GPR30-related diseases.
In summary, we demonstrated that GPR39 was significantly upregulated in colon cancer tissues compared with tumor-adjacent tissues by TCGA database re-analysis and qPCR. Moreover, we detected GPR39 expression in 161 colon cancer tissues and showed that GPR39 was an independent prognostic biomarker. With TCGA data re-analysis, qPCR, and IHC, we showed that GPR39 expression was significantly correlated with the expression of EPCAM and PD-L1, but not PPARG. EPCAM and PD-L1 were also unfavorable prognostic biomarkers of colon cancer.
Abbreviations
- GPR39:
-
G-protein coupled receptor 39
- PPARG:
-
Peroxisome proliferator-activated receptor gamma
- EPCAM:
-
Epithelial cell adhesion molecule
- PD-L1:
-
Programmed cell death 1 ligand 1
- qPCR:
-
Quantitative real-time PCR
- IHC:
-
Immunohistochemistry
- TCGA:
-
The Cancer Genome Atlas
- CRC:
-
Colorectal cancer
- MAPK:
-
Mitogen-activated protein kinase
- AKT:
-
RAC-alpha serine/threonine-protein kinase
- TPM:
-
Transcripts per million
References
Siegel RL, Miller KD, Fedewa SA et al (2017) Colorectal cancer statistics, 2017. CA Cancer J Clin 67(3):177–193. https://doi.org/10.3322/caac.21395
Torre LA, Bray F, Siegel RL et al (2015) Global cancer statistics, 2012. CA Cancer J Clin 65(2):87–108. https://doi.org/10.3322/caac.21262
Lee YC, Lee YL, Chuang JP, Lee JC (2013) Differences in survival between colon and rectal cancer from SEER data. PLoS One 8(11):e78709. https://doi.org/10.1371/journal.pone.0078709
Liu H, Xu Y, Zhang Q et al (2017) Correlations between TBL1XR1 and recurrence of colorectal cancer. Sci Rep 7:44275. https://doi.org/10.1038/srep44275
Liu H, Xu Y, Zhang Q et al (2017) Prognostic significance of TBL1XR1 in predicting liver metastasis for early stage colorectal cancer. Surg Oncol 26(1):13–20. https://doi.org/10.1016/j.suronc.2016.12.003
Hilger D, Masureel M, Kobilka BK (2018) Structure and dynamics of GPCR signaling complexes. Nat Struct Mol Biol 25(1):4–12. https://doi.org/10.1038/s41594-017-0011-7
Hershfinkel M (2018) The zinc sensing receptor, ZnR/GPR39, in health and disease. Int J Mol Sci 19(2):439. https://doi.org/10.3390/ijms19020439
Levaot N, Hershfinkel M (2018) How cellular Zn(2+) signaling drives physiological functions. Cell Calcium 75:53–63. https://doi.org/10.1016/j.ceca.2018.08.004
McCormick NH, Hennigar SR, Kiselyov K, Kelleher SL (2014) The biology of zinc transport in mammary epithelial cells: implications for mammary gland development, lactation, and involution. J Mammary Gland Biol Neoplasia 19(1):59–71. https://doi.org/10.1007/s10911-013-9314-4
Hershfinkel M, Moran A, Grossman N, Sekler I (2001) A zinc-sensing receptor triggers the release of intracellular Ca2+ and regulates ion transport. Proc Natl Acad Sci U S A 98(20):11749–11754. https://doi.org/10.1073/pnas.201193398
Mero M, Asraf H, Sekler I et al (2019) ZnR/GPR39 upregulation of K(+)/Cl(-)-cotransporter 3 in tamoxifen resistant breast cancer cells. Cell Calcium 81:12–20. https://doi.org/10.1016/j.ceca.2019.05.005
Alen BO, Leal-Lopez S, Alen MO et al (2016) The role of the obestatin/GPR39 system in human gastric adenocarcinomas. Oncotarget 7(5):5957–5971. https://doi.org/10.18632/oncotarget.6718
Xie F, Liu H, Zhu YH et al (2011) Overexpression of GPR39 contributes to malignant development of human esophageal squamous cell carcinoma. BMC Cancer 11:86. https://doi.org/10.1186/1471-2407-11-86
Dubi N, Gheber L, Fishman D et al (2008) Extracellular zinc and zinc-citrate, acting through a putative zinc-sensing receptor, regulate growth and survival of prostate cancer cells. Carcinogenesis 29(9):1692–1700. https://doi.org/10.1093/carcin/bgn027
Cohen L, Azriel-Tamir H, Arotsker N et al (2012) Zinc sensing receptor signaling, mediated by GPR39, reduces butyrate-induced cell death in HT29 colonocytes via upregulation of clusterin. PLoS One 7(4):e35482. https://doi.org/10.1371/journal.pone.0035482
Ogino S, Shima K, Baba Y et al (2009) Colorectal cancer expression of peroxisome proliferator-activated receptor gamma (PPARG, PPARgamma) is associated with good prognosis. Gastroenterology 136(4):1242–1250. https://doi.org/10.1053/j.gastro.2008.12.048
Liang KH, Tso HC, Hung SH et al (2018) Extracellular domain of EpCAM enhances tumor progression through EGFR signaling in colon cancer cells. Cancer Lett 433:165–175. https://doi.org/10.1016/j.canlet.2018.06.040
Zhou FQ, Qi YM, Xu H et al (2015) Expression of EpCAM and Wnt/ beta-catenin in human colon cancer. Genet Mol Res 14(2):4485–4494. https://doi.org/10.4238/2015.May.4.6
Roohani N, Hurrell R, Kelishadi R, Schulin R (2013) Zinc and its importance for human health: an integrative review. J Res Med Sci 18(2):144–157
Sandstead HH (2012) Zinc nutrition from discovery to global health impact. Adv Nutr 3(5):718–719. https://doi.org/10.3945/an.112.002485
Doboszewska U, Mlyniec K, Wlaz A et al (2019) Zinc signaling and epilepsy. Pharmacol Ther 193:156–177. https://doi.org/10.1016/j.pharmthera.2018.08.013
Maret W (2013) Zinc biochemistry: from a single zinc enzyme to a key element of life. Adv Nutr 4(1):82–91. https://doi.org/10.3945/an.112.003038
Vallee BL, Falchuk KH (1993) The biochemical basis of zinc physiology. Physiol Rev 73(1):79–118. https://doi.org/10.1152/physrev.1993.73.1.79
Maret W (2017) Zinc in cellular regulation: the nature and significance of “Zinc Signals.” Int J Mol Sci. https://doi.org/10.3390/ijms18112285
Fassnacht M, Weismann D, Ebert S et al (2005) AKT is highly phosphorylated in pheochromocytomas but not in benign adrenocortical tumors. J Clin Endocrinol Metab 90(7):4366–4370. https://doi.org/10.1210/jc.2004-2198
Hasson SP, Rubinek T, Ryvo L, Wolf I (2013) Endocrine resistance in breast cancer: focus on the phosphatidylinositol 3-kinase/akt/mammalian target of rapamycin signaling pathway. Breast Care (Basel) 8(4):248–255. https://doi.org/10.1159/000354757
Miller TW, Rexer BN, Garrett JT, Arteaga CL (2011) Mutations in the phosphatidylinositol 3-kinase pathway: role in tumor progression and therapeutic implications in breast cancer. Breast Cancer Res 13(6):224. https://doi.org/10.1186/bcr3039
Ma J, Mai HQ, Hong MH et al (2001) Is the 1997 AJCC staging system for nasopharyngeal carcinoma prognostically useful for Chinese patient populations? Int J Radiat Oncol Biol Phys 50(5):1181–1189. https://doi.org/10.1016/s0360-3016(01)01537-1
Sadanandam A, Lyssiotis CA, Homicsko K et al (2013) A colorectal cancer classification system that associates cellular phenotype and responses to therapy. Nat Med 19(5):619–625. https://doi.org/10.1038/nm.3175
Allegra CJ, Jessup JM, Somerfield MR et al (2009) American Society of Clinical Oncology provisional clinical opinion: testing for KRAS gene mutations in patients with metastatic colorectal carcinoma to predict response to anti-epidermal growth factor receptor monoclonal antibody therapy. J Clin Oncol 27(12):2091–2096. https://doi.org/10.1200/JCO.2009.21.9170
Pricolo VE, Finkelstein SD, Wu TT et al (1996) Prognostic value of TP53 and K-ras-2 mutational analysis in stage III carcinoma of the colon. Am J Surg 171(1):41–46. https://doi.org/10.1016/S0002-9610(99)80071-3
Kuol N, Stojanovska L, Nurgali K, Apostolopoulos V (2018) PD-1/PD-L1 in disease. Immunotherapy 10(2):149–160. https://doi.org/10.2217/imt-2017-0120
Lin J, Chen Y, Tang WF et al (2019) PPARG rs3856806 C>T polymorphism increased the risk of colorectal cancer: a case-control study in Eastern Chinese Han Population. Front Oncol 9:63. https://doi.org/10.3389/fonc.2019.00063
Liao MY, Lai JK, Kuo MY et al (2015) An anti-EpCAM antibody EpAb2-6 for the treatment of colon cancer. Oncotarget 6(28):24947–24968. https://doi.org/10.18632/oncotarget.4453
Moechars D, Depoortere I, Moreaux B et al (2006) Altered gastrointestinal and metabolic function in the GPR39-obestatin receptor-knockout mouse. Gastroenterology 131(4):1131–1141. https://doi.org/10.1053/j.gastro.2006.07.009
Frimurer TM, Mende F, Graae AS et al (2017) Model-based discovery of synthetic agonists for the Zn(2+)-sensing G-protein-coupled receptor 39 (GPR39) reveals novel biological functions. J Med Chem 60(3):886–898. https://doi.org/10.1021/acs.jmedchem.6b00648
Grunddal KV, Diep TA, Petersen N et al (2021) Selective release of gastrointestinal hormones induced by an orally active GPR39 agonist. Mol Metab 49:101207. https://doi.org/10.1016/j.molmet.2021.101207
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Wu, X., Dou, Y., Xu, H. et al. The expression and clinical significance of GPR39 in colon cancer. Ir J Med Sci 191, 1577–1585 (2022). https://doi.org/10.1007/s11845-021-02792-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11845-021-02792-z