Abstract
This paper presents a novel approach for automatic segmentation of MS lesion including both of number and volume. The novelty includes the combination of the multiplicative intrinsic component optimization algorithm (Li et al. in Magn Reson Imaging 32:913–923, 2014) in bias field correction and normal tissue segmentation simultaneously, and the development of a multi-atlas and multi-channel (MAMC) segmentation approach. The first research focus is the classification of brain tissue into white matter, cerebrospinal fluid and gray matter in T1-w image and FLAIR image. The second research focus is the segmentation of MS lesion in white matter region using atlas. In label fusion, the coefficient as a specific weight is assigned to target label image based on the correlation function between atlases. This novel MAMC approach is evaluated by 20 training cases obtained from Medical Image Computing and Computer Aided Intervention Society 2008 MS Lesions Segmentation Challenge. The numerical results are presented in terms of accuracy, specificity and absolute volume difference. A comparison of MAMC approach and other conventional approaches is presented in terms of the true positive rate and the positive predictive value. Furthermore, the total lesion volume is calculated and compared with expert delineation. It can be seen that the MAMC approach is able to acquire a larger mean value of the Dice similarity coefficient than the other conventional approaches do. Therefore, this novel approach is an added value for the clinical evaluation of MS patients.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Multiple sclerosis (MS) is a frequent, chronic and degenerative disease in human central nervous system, especially it causes disabilities in young adults. There are 1.3–2.5 million MS patients worldwide and among them roughly three times more common in women than men [2]. Magnetic resonance imaging (MRI) technique is sensitive enough to detect MS plaque and quantify the number and volume of lesions. Therefore, MRI is the most important clinical approach for diagnosing MS due to its high image quality and less radiation. In general, MRI sequences are divided into different categories, including gadolinium-enhanced T1-weighted, T2-weighted, proton density (PD) and fluid-attenuated inversion recovery (FLAIR). A comparison of Fig. 1a, b shows that the tissue borders of white matter (WM), gray matter (GM) and cerebrospinal fluid (CSF) are more distinguishable in T1-w image than that in FLAIR image, but MS lesion is easier to be recognized in FLAIR image than that in T1-w image. Figure 1c shows a MS lesion segmentation image based on FLAIR, in which the red area except the area within the white circle is the MS lesion and the area within the white circle is the artifact caused by ventricle pulsation. Clearly, the artifacts make the accurate segmentation of MS lesion to be difficult. For example, as shown in Fig. 1c, the red area within white circle is considered as normal tissue. In this case, it is hard to distinguish lesion from artifact in FLAIR image since the artifact intensity is very similar to the lesion intensity.
The identification of number and volume of MS lesion is a crucial procedure in the diagnosis, which is characterized by the presence of WM lesion and is typically delineated manually by a radiologist. The manual delineation is time consumable and suffers from intra- and inter-observer variability. In contrast, the automatic segmentation of MS lesion is able to save time and decrease observer dependency. However, noise, artifacts and bias field in MR image [3,4,5] make the accurate and automatic segmentation to be a challenge.
The investigation on automatic segmentation in MR image started 10 years ago. Vovk [6] and Balafar [7] summarized the early works on the methodology for automatic segmentation in MR image, in which [6] focused on the correction of intensity inhomogeneity while [7] focused on the brain MR images. They indicated that the challenges for automatic segmentation in MR image would be the accuracy of feature extraction and relatively required large computing power. Furthermore, a number of automatic approaches for MS lesion segmentation have been investigated recently, including k nearest neighbors [8], artificial neural networks [9], expectation–maximization (EM) [10, 11], spatial decision forest [12, 13], Markov random field (MRF) [14], anatomical and topological atlas [15, 16], and energy minimization [17]. These lesion segmentation approaches can be categorized as the supervised learning approach and the unsupervised approach [18]. The supervised learning approach requires a highly efficient and accurate training database. In contrast, the unsupervised approach segments the MS lesion using priori information, which are not included in the training database. In spite of the segmentation approaches as mentioned above [7,8,9,10,11,12,13,14,15,16], there is no satisfactory solution can be found due to the poor contrast, which can exist between lesion and neighboring ROI. On the other hand, the performance with long processing time is one of the main weaknesses for them not be accepted by certain clinical applications.
Atlas-based approach employs an expert defined prior image, which is used to segment the target image. Furthermore, multi-atlas-based segmentation is able to compensate the potential errors comparing to a single atlas and improve segmentation accuracy [17]. In this approach, each atlas image is registered to the target image and the calculated deformation matrix is applied to warp the atlas label image into the target label image [19]. Hence, a specific approach is able to combine these target label images into a final target label image. This has been an active topic of research in recent years. In contrast, label fusion is another active research topic in this field, where the most of existing label fusion approaches are based on weighted voting [20, 21]. The weight of atlas is derived based on the similarity between the atlas and the target image. Then, it is preferable to assign high weight value to the accurate segmentation. Alternatively, Jacobian determinant values are used to measure the similarity between the atlas and the target image [21]. The Jacobian determinant gives a best linear approximation of a transformation, but the determinant Jacobian value of a rotation matrix is always equal to 1, which is a significant limitation of label fusion. In order to overcome this limitation, joint label fusion approach is proposed in [22]. In contrast, the normalized mutual information (NMI) for label fusion between the atlas and target image is presented in this paper.
A significant research work in this field is called multiplicative intrinsic component optimization (MICO), which is an energy minimization algorithm developed by Li [1], where the MICO algorithm has been applied to bias field estimation and tissue segmentation. The experimental results presented in [1] show that the MICO is able to effectively correct the errors in bias field estimation and simultaneously to segment GM, WM and CSF tissue using MR image of normal brains. To the best of our knowledge, the research work presented in this paper is the first instance, in which the MICO is specifically applied to segment MS lesion by taking the advantage in the decomposition of MR image. Since intensity values in MS lesions are very similar to the intensity values of gray matter (GM) [2]. From this point of view, the MICO cannot be directly applied to the segmentation of MS lesion. In order to overcome this limitation, a novel multi-atlas and multi-channel (MAMC) approach is developed to associate with the MICO simultaneously.
Furthermore, this paper presents a multiple-atlas scheme to implement the multiple WM probability atlas images, in which the probability of occurrence is much lower for artifact than that for lesion. This scheme is able to take the advantage of occurrence probability during lesion segmentation.
Finally, in order to reduce the matching ambiguities and to increase segmentation accuracy, a multiple channel mechanism combining of T1-w and FLAIR image is proposed. This mechanism is able to dynamically assign weight coefficient to the corresponding target label image based on the correlation between atlases. The performance evaluation is compared with the approaches proposed by Souplet et al. [23] and by Weiss et al. [24]. In [23] a hyperintense MS lesion segmentation method combining with threshold and EM was used for T2-FLAIR sequence image. The principal idea in [24] was to segment lesion as outlier from image patch, which was reconstructed using dictionary learning and sparse coding.
The rest of this paper is organized as follows: The MAMC algorithm is discussed in Sect. 2, the numerical results obtained from experimental tests are presented in Sect. 3, a discussion on the experimental results is given in Sect. 3, and the conclusion is in Sect. 4.
2 Methods
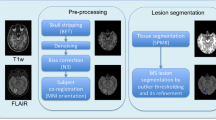
Figure 2 shows a diagram of the proposed MAMC approach for MS lesion segmentation. First, the FMRIB’s Software Library (FSL) is used to preprocess the both of target image and atlas image. Since the images acquired by different MRI scanners or from different sources may be in different formats, the original T1-w image and FLAIR image must be uniformly transformed into a standard format: (1) LAS orientation (from left to right, from anterior to posterior, from superior to inferior), (2) image size in terms of voxels is presented as 256 × 256 × 170, and (3) skull stripping.
Second, T1-w atlas and FLAIR atlas, named as probability-based atlas, are registered as the T1-w target image and the FLAIR target image, respectively. From Fig. 1, it can be seen that the intensity level for WM, GM and CSF tissues in T1-w image is different from that in FLAIR image. For example, WM tissue is the brightest tissue in T1-w image, but it is a tissue with intermediate gray color in FLAIR image. In contrast, GM tissue appears as an intermediate gray in T1-w image, but it is the brightest tissue in FLAIR image. Moreover, CSF tissue is the darkest tissue in both T1-w image and FLAIR image. Note that such characteristics may cause the cross-registration errors between T1-w image and FLAIR image, so that the cross-registration between the T1-w atlas and FLAIR target image is prohibited.
In preprocessing, it needs to pay a special attention to the lesion with small voxel size. For example, the MNI152 is a standardized image in format of voxel size of 91 × 109 × 91. When a FLAIR image of 256 × 256 × 170 voxel is transformed to the MNI152 image format, then it can be seen that small lesions in the original FLAIR image may become invisible due to the downsize of voxel during the transform. Likewise, the same situation can be seen in the transferring of T1-w image into the MNI152 standard. As a result, the lesion detection error for MR image with MNI152 format is larger than that in the original image. To avoid such error, the multiple atlases need to be transformed into target image.
Third, the lesion is segmented using the MAMC approach, simultaneously the FLAIR image and T1-w image are segmented into PVE label images using the MICO algorithm, respectively. The reason is that the T1-w and FLAIR PVE label image can be used to determine WM lesions (WMLs), which can be used to classify the corresponding GM tissue because of WML intensity level is similar to GM intensity level [25]. The final goal is to generate a new probability-based atlas, in which WMLs have an obviously larger probability than that of GM tissue. Hence, WMLs can be identified by a threshold value. Furthermore, label image of WMLs can be acquired from each atlas in T1-w and FLAIR image, respectively. By using label fusion, the final label image can be acquired for WMLs.
2.1 Preprocessing
FSL software is sued to preprocess the image, including uniform transformation of the original T1-w, FLAIR and atlas images into the LAS orientation, respectively.
Step 1. the images from different sources have different size of voxels, for example, the GG-853-T1-2.0 mm image is 91 × 109 × 91 voxels and the training images provided by MICCAI 2008 are 512 × 512 × 512 voxels. In order to equally process these images in the following stages, all images are uniformly resampled into the same size of voxels as 256 × 256 × 170.
Step 2. the T1-w image with MNI152 atlases is cropped and automatically registered using linear 12-parameter affine and nonlinear transformations, respectively.
Step 3. the skull-stripping aims to remove the skull and extra-cerebral tissue such as scalp and Dura, which is a very important preprocessing step in most automatic brain MRI applications [26]. Otherwise, these skull and extra-cerebral tissues are able to seriously affect segmentation accuracy. In this research, the skull-stripping process uses the brain extraction tool (BET) to extract brain tissue. BET is a widely available boundary-based method [27].
2.2 MICO algorithm
Intensity inhomogeneity can blur image due to machine error and patient head moving, which is a major factor affecting the segmentation accuracy. Therefore, intensity inhomogeneity is generally considered as the multiplicative or additive function in originally homogeneous regions. On the other hand, noise is usually considered to be independent of inhomogeneity. For a given MR image I, it can be described as follows:
where x represents the voxel, \( I(x) \) is the intensity of voxel x in the MR image, \( b(x) \) is the inhomogeneity function, \( J(x) \) is the intensity in the originally homogeneous image, and \( n(x) \) is the additive noise. Inhomogeneity correction methods are categorized into two groups: prospective methods and retrospective methods [6, 7]. The prospective methods group includes special sequences, multi-coil, and phantom-based methods. These methods are able to correct some intensity inhomogeneity caused by machine error, but not able to solve the inhomogeneity caused by patients. In contrast, the retrospective methods are able to correct both MR scanner-induced and patient-induced inhomogeneity.
The MICO algorithm has the capacity for doing bias field correction and tissue segmentation simultaneously based on an energy minimization mechanism. The energy function is expressed as follows:
where \( u_{i} (x) \) is the probability that voxel x is allocated in the ith tissue, N is the total number of tissues involved in the process, \( c_{i} \) is the weight of \( u_{i} (x) \), \( w^{\text{T}} \) is the linear coefficient vector, \( G(x) \) is the basis vector of bias and \( b\left( x \right) = W^{\text{T}} G(x) \).
By minimizing the energy function \( F(u,c,w) \), it is able to obtain the optimal tissue segmentation, denoted by vector \( \hat{u} = \left( {\hat{u}_{1} , \ldots , \hat{u}_{N} } \right)^{\text{T}} \) and \( \hat{w} = \left( {\hat{w}_{1} , \ldots ,\hat{w}_{M} } \right)^{\text{T}} \), respectively. Then, the bias field estimation can be computed by \( \hat{b}(x) = \hat{w}^{\text{T}} G(x) \) using the obtained vector \( \hat{w} \).
2.3 Segmentation of Multi-channel WMLs
The T1-w image is classified into CSF, GM and WM by the MICO algorithm, in which intensity value of 1, 2 and 3 represents CSF, GM and WM, respectively. Note that WMLs are generally classified as GM since their intensity value is similar to GM [25]. Figure 3 illustrates an example that the WMLs are classified into GM in T1-w image. In order to separate WMLs from GM, a prior probability-based atlas is used. Let \( {\text{I}}_{{{\text{T}}1\_{\text{PVEi}}}} = \left\{ {1, 2, 3} \right\} \) be the intensity for the ith voxel in the T1 PVE image, where i = 1, 2,…, 256 × 256 × 170. Hence, a new intensity distribution over the T1 PVE image can be presented as follows:
where Pri is the probability of the ith voxel belonging to WM in the atlas. Note that when the ith voxel belongs to a WML, its probability Pri is larger than that of GM voxel. Since the intensity value \( I_{{{\text{T1\_PVE}}i}} \) of the ith voxel in the T1 PVE image is equal to 2, which is the same as the intensity of GM voxel. Hence, the voxels belonging to WMLs can be identified using \( I_{{i{\text{new}}}} \). Likewise, the WMLs can be segmented using the threshold method in (3). Regarding the segmentation of WMLs in FLAIR image, there are two key tasks.
Task 1. The brightest tissue in the FLAIR image is classified into WMLs and artifacts two categories, where artifacts are usually caused by ventricle pulsation. As shown in Fig. 1, it is difficult indeed to make an accurate distinction between WMLs and artifacts in the FLAIR image. However, in the prior probability-based atlas the probability of artifact voxel is close to 0, while the probability of WML voxel is bigger than 2. From this point of view, the WML voxel can be identified using Eq. (3).
Task 2. When a FLAIR image, denoted as \( I_{\text{FL}} \), is segmented into CSF, WM and GM tissue using the MICO algorithm, a result of PVE image, denoted as \( L_{{{\text{FL}}\_{\text{PVE}}}} \) is acquired, in which intensity values of 1, 2, and 3 represent CSF, WM, and GM, respectively. It has been noticed that the borders of WM, GM and CSF are vague in FLAIR image [2]. In this case, the WM voxels, which are close to the border, may be incorrectly classified as GM, called “fake GM.” Likewise, the GM voxels, which are close to the border, may be incorrectly classified as WM, called “fake WM.” Since WML has a higher prior probability than WM has, which is allocated to GM, therefore, it is difficult indeed to make an accurate distinction between WMLs and “fake GM.” To resolve this problem, a two-step approach is proposed.
Step 1. One PVE image segmented from the FLAIR image and one accurate WM label image from the T1-w image are acquired, denoted as \( L_{{{\text{FL\_PVE}}}} \) and \( L_{{{\text{T1\_WM}}}} \), respectively. Then, a label image \( L_{{{\text{WML\_fake}}}} \), which contains both of WMLs and “fake GM” can be obtained by
where XOR is an exclusive OR operator.
Step 2. Let \( I_{\text{newFL}} \) represent the region of interest (ROI) in the FLAIR image. Then \( I_{\text{newFL}} \) can be presented as
where i = 1, 2,…, 256 × 256 × 170 is the index of voxel in the FLAIR image. Substituting (4) to (5), it is clear \( I_{\text{newFL}} \) only consists of the WMLs and “fake GM” voxels. Applying the MICO algorithm to \( I_{\text{newFL}} \), WMLs voxels can be acquired because of the intensity of WMLs has the significant difference from the intensity of “fake GM” in \( I_{\text{newFL}} \). It can be seen that this proposed 2-Step approach based on the MICO algorithm associated by Eqs. (4) and (5) is able to effectively reduce segmentation errors and acquire high-quality WMLs segmentation in FLAIR image.
2.4 Label fusion
In the multi-atlas-based segmentation, each atlas involved in the process is able to create a label image. In this case, a novel approach is proposed focusing on how to optimally integrate the label images obtained from different atlases into a final label image. This novel approach computes the normalized mutual information (NMI) between each individual atlas and the target image. Then a unique weight coefficient based on the calculated NMI value is assigned to the label image, which is obtained from each individual atlas.
Define that
where x is a voxel in the target image, in which has n segmentation labels corresponding n atlases. The probability of voxel x to be a WMLi in the final label image is given by
where \( L_{\text{final}} (x) = 1 \) represents the xth voxel to be a WML, \( {\text{MI}}_{i} \) is the NMI value between the ith atlas and target image, and n is the number of atlases involved in the multi-atlas segmentation process.
As shown in Fig. 1a, it can be observed that the lesion is more distinguishable in FLAIR image than that in T1-w image because of the lesion volume in T1-w image is smaller than that in FLAIR image. For example, as shown in Fig. 6b, considering the UNC case 07, the total lesion volume in FLAIR image is 0.341 ml. In contrast, under the same condition, the total lesion volume in T1-w image is 0.209 ml. In this case, a weight coefficient, denoted as τi, is assigned to the corresponding atlas in FLAIR image. The induced final segmentation result for FLAIR image is given by
Note that the weight coefficient for each atlas in Eq. (8) only takes into account NMI and the effects of multi-channel images, but it ignores the across-correlation between the atlases. However, when atlas is duplicated or some atlases are highly similar to each other, then the quality of segmentation can be affected. In this case, the duplicated atlases are aligned into one atlas. Likewise, the atlases similar to each other are given small weight coefficient \( \sigma_{i} \) in order to reduce their effect. The weight of coefficient for atlas can be expressed as follows:
where \( {\text{NMI}}_{ij} \) represents the NMI between the ith and jth atlases and n is the number of atlases in the FLAIR image. Substituting (9) to (8), the modified final segmentation result for FLAIR image is presented as follows:
Note that Eq. (10) takes into account not only the NMI between the atlas and target image, but also the effect of similarities among atlases in the final label image.
3 Numerical results and discussion
Since there is no evaluation standard available for validating the segmentation of WMLs, the performance evaluation for the proposed MAMC approach is based on the MR image datasets provided by the MICCAI 2008 MS Lesion Segmentation Challenge. The MR image dataset being used in the evaluation includes 20 training cases, in which 10 cases are from the Children’s Hospital of Boston (CHB) and the other 10 cases are from the University of North Carolina (UNC) [28]. All 20 training cases have been manually segmented by expert raters, so that they can be used as the ground truth for the evaluation. The output data collected from the experiment include true positives (TP), true negatives (TN), false positives (FP) and false negatives (FN). TP and TN measure the number of voxels being segmented as WMLs and non-WMLs by the MAMC approach, respectively. Likewise, FP and FN measure the number of voxels being segmented as WMLs and non-WMLs, respectively, by the MAMC approach only. The performance of the proposed MAMC approach is evaluated in terms of Dice similarity coefficient (DSC), specificity, true positive rate (TPR), accuracy, absolute volume difference (VD) and positive predictive value (PPV). DSC measures the degree of coincidence between the MAMC approach and the ground truth data provided by MICCAI 2008 dataset. Specificity measures the proportion of negatives, which are correctly identified as non-WMLs. TPR is given by TP/(TP+ FN). This value represents a good compromise combining of sensitivity, specificity and efficiency. The evaluation metrics are defined in Table 1.
Figure 4 illustrates a group of bias-corrected FLAIR image, in which the first column shows the original image without segmentation, the second column shows the lesion segmentation in green color obtained by the proposed MAMC approach, and third column shows the lesion segmentation in red color obtained by the CHB expert manual approach. From the second column in Fig. 4, it can be seen that the proposed MAMC approach is able to accurately discriminate artifacts from WMLs as indicated using green circle. However, the MAMC approach has overestimated lesions indicated using yellow circle. In contrast, the expert manual approach has a number of underestimated and missed subtle lesions, but these subtle lesions can be identified using MAMC approach as indicated using red circle. Furthermore, it can be seen that the WMLs segmented using the MAMC approach is much more accurate than that of expert manual approach. Therefore, the MAMC approach has a higher segmentation precision than manual expert approach has in the subtle lesion detection.
The experimental results from MAMC approach and CHB expert manual approach. The bias-corrected FLAIR images, the segmentation in green is using MAMC approach, the CHB expert manual approach in red are shown from column 1, 2 and 3, respectively. The datasets come from CHB 10 training cases (color figure online)
Figure 5 illustrates a lesion segmentation comparison of using CHB manual approach, UNC manual approach and proposed MAMC approach, respectively.
The experimental results using MAMC approach, CHB expert manual and UNC expert manual approach. The bias-corrected FLAIR images, the segmentation in green is using MAMC approach, the CHB expert manual approach in red, the UNC expert manual approach in blue are shown from column 1, 2, 3 and 4, respectively. The datasets come from UNC 10 training cases (color figure online)
In Fig. 5, the first column is the original MR images without segmentation, the lesion segmentation images in second column are using the proposed MAMC approach, the third column by the CHB expert manual approach and the last column by the UNC expert manual approach. In Fig. 5, the yellow circle shows the overestimated lesion by the MAMC approach, the blue circle shows the WMLs that are only partially delineated by the CHB expert manual approach. Furthermore, the CHB expert manual approach has a number of lesions either underestimated or missed, but these lesions can be identified using the MAMC approach in red circle and the UNC expert manual approach in green circle, respectively. From Fig. 5, it can be seen that the both of UNC expert manual approach and CHB expert manual approach have minor errors in lesion segmentation. Likewise, the MAMC approach is able to achieve reasonable good output of lesion segmentation comparing to that of using the expert manual approach of UNC and CHB, respectively.
Table 2 presents the numerical results in terms of accuracy, specificity, and absolute VD using the MAMC approach based on the training cases of CHB and UNC, respectively. It can be seen that excellent accuracy and specificity ranging from 0.9955 to 0.9997 and from 0.9976 to 0.9999, respectively, can be achieved by the proposed MAMC approach. This is a strong evidence to demonstrate that the proposed MAMC approach is able to identify the lesion using MR image for patients who have been affected by MS lesion disease.
Figure 6a illustrates the total lesion volume (ml) estimated using the CHB expert manual approach, the UNC expert manual approach and the MAMC approach in FLAIR and T1-w image, respectively. Comparing the total lesion volumes obtained by the CHB expert manual approach and the UNC manual approach, as shown in Fig. 6a, b, it can be seen that the total lesion volume of using the CHB expert manual approach is 0.605 ml, which is 3.8% larger than that of using the UNC expert manual approach. Furthermore, it also can be found the total lesion volume obtained by the MAMC approach is more close to that of the CHB expert manual approach than that of using the UNC expert manual approach. This indicates that the MAMC approach is comparable to manual delineation.
Table 3 shows numerical results in terms of TPR, PPV and DSC for a comparison of the proposed MAMC approach and the other two of conventional approaches, including “Automatic Segmentation of T2 FLAIR Multiple Sclerosis Lesions” by Souplet et al. [23] and “Multiple Sclerosis Lesion Segmentation Using Dictionary Learning and Sparse Coding” proposed by Weiss et al. [24]. TPR is defined as \( \frac{\text{TP}}{{{\text{TP}} + {\text{FN}}}} \), PPV is defined as \( \frac{\text{TP}}{{{\text{TP}} + {\text{FP}}}} \) and DSC is defined as \( \frac{{2 \times {\text{TP}}}}{{{\text{FP}} + {\text{FN}} + 2 \times {\text{TP}}}} \). Therefore, TPR, PPV and DSC are the parameters that compromise the combination of TP, TN and FP to measure the efficiency of the approach. From Table 3, it can be seen that mean values of TPR, PPV and DSC for the proposed MAMC approach are 0.34, 0.45 and 0.38, respectively. The gain of the MAMC approach comparing to that in [24, 25] is 14%, 4% and 13%, and 0%, 10% and 13%, respectively. This indicates that the proposed MAMC approach certainly has the advantages comparing with these two conventional approaches.
The proposed MAMC approach has the great potential to be applied to clinical practice. For example, the segmentation of WMLs using the MAMC approach for a patient can be measured at different time periods. A comparison of these obtained WM lesion images is able to predict and model the development of MS disease, to provide an additional dataset in support of the medical diagnosis. A variety of algorithms have recently been applied to the automatic segmentation of MS lesions, such as surrogate training metrics [24] and the constrained Gaussian Mixture Model (GMM) [25]. The comparison shown in Table 3 has demonstrated that the proposed MAMC approach is better than the approaches presented in [24, 25].
The proposed algorithm represents a novel approach to MS lesion segmentation and takes advantage of the benefits provided by the MICO algorithm. It is even more important that the MR image is decomposed into two multiplicative components for the characterization of physical tissue and the bias field, which take into account intensity inhomogeneity. This technique could be a valuable new methodology to be used in the diagnosis for monitoring of MS lesion development.
Future work will focus on the application of the MAMC approach to other non-MS lesion data since the MAMC approach focuses on MR image processing, which can be typically used for diagnosis of soft tissue contrast such as the segmentation of other anatomical areas in MR image.
4 Conclusion
This paper focuses on an automatic approach for WM lesion segmentation using multi-channel and multi-atlas brain MR images, called MAMC approach. It has two novel elements. The first one is the segmentation of MR image into GM, WM and CSF using the proposed MAMC approach, which can be effectively applied to lesion segmentation. The second one is the identification of artifacts from WMLs. The MAMC approach is evaluated in terms of accuracy, specificity and absolute VD using MS lesion dataset obtained from the MICCAI 2008. The numerical results have demonstrated the advantages in both accuracy and specificity of ranging from 0.9955 to 0.9997 and 0.9976 to 0.999, respectively. The absolute VDs are small enough except in Case 05 and Case 09 due to the missing of some very tiny lesions in these two cases.
Furthermore, the efficiency in terms of TPR, PPV and DSC is evaluated through a performance comparison with conventional approach presented in [24] [25], respectively. The numerical results show that the reasonable gains in efficiency for the MAMC approach comparing with the approach in [24, 25] are achieved. Therefore, the proposed MAMC approach is a novel approach for WM lesion segmentation using MR images.
References
Li, C., Gore, J.C., Davatzikos, C.: Multiplicative intrinsic component optimization (MICO) for MRI bias field estimation and tissue segmentation. Magn. Reson. Imaging 32(7), 913–923 (2014)
Lladó, X., Oliver, A., Cabezas, M., Freixenet, J., et al.: Segmentation of multiple sclerosis lesions in brain MRI: a review of automated approaches. Inf. Sci. 186(1), 164–185 (2012)
Gaser, C., Schmidt, P., Arsic, M., et al.: An automated tool for detection of FLAIR-hyperintense white-matter lesions in multiple sclerosis. NeuroImage 59(4), 3774–3783 (2012)
Tohka, J.: Partial volume effect modeling for segmentation and tissue classification of brain magnetic resonance images: a review. World J. Radiol. 6(11), 855–864 (2014)
Cabezas, M., Oliver, A., Valverde, S., Beltran, B., et al.: BOOST: a supervised approach for multiple sclerosis lesion segmentation. J. Neurosci. Methods 237, 108–117 (2014)
Vovk, U., Pernus, F., Likar, B.: A review of methods for correction of intensity inhomogeneity in MRI. IEEE Trans. Med. Imaging 26, 405–421 (2007)
Balafar, M.A., Ramli, A.R., Saripan, M.I., Mashohor, S.: Review of brain MRI image segmentation methods. Artif. Intell. Rev. 33(3), 261–274 (2010)
Pouwels, P.J.W., Steenwijk, M.D., Daams, M., et al.: Accurate white matter lesion segmentation by k nearest neighbor classification with tissue type priors (kNN-TTPs). NeuroImage Clin. 3, 462–469 (2013)
Cerasa, A., Bilotta, E., Augimeri, A., Cherubini, A., et al.: A cellular neural network methodology for the automated segmentation of multiple sclerosis lesions. J. Neurosci. Methods 203(1), 193–199 (2012)
Hadrich, A., Zribi, M., Masmoudi, A.: Bayesian expectation maximization algorithm by using B-splines functions: Application in image segmentation. Math. Comput. Simul. 120(Supplement C), 50–63 (2016)
Svensson, C.M., Bondoc, K.G., Pohnert, G., Figge, M.T.: Segmentation of clusters by template rotation expectation maximization. Comput. Vis. Image Underst. 154(Supplement C), 64–72 (2017)
Mitra, J., Bourgeat, P., Fripp, J., Ghose, S., et al.: Lesion segmentation from multimodal MRI using random forest following ischemic stroke. NeuroImage 98, 324–335 (2014)
Clatz, O., Geremia, E., Menze, B.H., et al.: Spatial decision forests for MS lesion segmentation in multi-channel magnetic resonance images. NeuroImage 57(2), 378–390 (2011)
Roy, P.K., Bhuiyan, A., Janke, A., Desmond, P.M., et al.: Automatic white matter lesion segmentation using contrast enhanced FLAIR intensity and Markov random field. Comput. Med. Imaging Graph. 45, 102–111 (2015)
Bazin, P., Shiee, N., Pham, D.L.: Multiple sclerosis lesion segmentation using statistical and topological atlases. In: Grand Challenge Work: Mult. Scler. Lesion Segm. Challenge, pp. 1–10 (2008)
Freire, P.G.L., Ferrari, R.J.: Automatic iterative segmentation of multiple sclerosis lesions using Student’s t mixture models and probabilistic anatomical atlases in FLAIR images. Comput. Biol. Med. 73(C), 10–23 (2016)
Zhao, Y., Guo, S., Luo, M., Liu, Y., et al.: An energy minimization method for MS lesion segmentation from T1-w and FLAIR images. Magn. Reson. Imaging 39, 1–16 (2017)
García-Lorenzo, D., Francis, S., Narayanan, S., Arnold, D.L., et al.: Review of automatic segmentation methods of multiple sclerosis white matter lesions on conventional magnetic resonance imaging. Med. Image Anal. 17(1), 1–18 (2016)
Artaechevarria, X., Munoz-Barrutia, A., Ortiz-de-Solorzano, C.: Combination strategies in multi-atlas image segmentation: application to brain MR data. IEEE Trans. Med. Imaging 28(8), 1266–1277 (2009)
Sabuncu, M.R., Yeo, B.T.T., Van Leemput, K., Fischl, B., et al.: A generative model for image segmentation based on label fusion. IEEE Trans. Med. Imaging 29(10), 1714–1729 (2010)
Isgum, I., Staring, M., Rutten, A., Prokop, M., et al.: Multi-atlas-based segmentation with local decision fusion—application to cardiac and aortic segmentation in CT scans. IEEE Trans. Med. Imaging 28(7), 1000–1010 (2009)
Wang, H., Suh, J.W., Das, S.R., Pluta, J.B., et al.: Multi-atlas segmentation with joint label fusion. IEEE Trans. Pattern Anal. Mach. Intell. 35(3), 611–623 (2013)
Souplet, J.C., Lebrun, C., Ayache, N., Malandain, G.: An automatic segmentation of T2FLAIR multiple sclerosis lesions. In: MICCAI-Multiple Sclerosis Lesion Segmentation Challenge Workshop (2008)
Weiss, N., Rueckert, D., Rao, A.: Multiple sclerosis lesion segmentation using dictionary learning and sparse coding. In: Mori K., et al. (eds.) Medical Image Computing and Computer-Assisted Intervention—MICCAI 2013: 16th International Conference, Nagoya, 22–26 Sept 2013, Proceedings, Part I, pp. 735–742. Springer, Berlin (2013)
Jain, S., Sima, D.M., Ribbens, A., Cambron, M., et al.: Automatic segmentation and volumetry of multiple sclerosis brain lesions from MR images. NeuroImage Clin. 8, 367–375 (2015)
Doshi, J., Erus, G., Yangming, O., Gaonkar, B., et al.: Multi-atlas skull-stripping. Acad. Radiol. 20(12), 1566–1576 (2013)
Pechaud, M., Jenkinson, M., Smith, S.: BET2: MR-based estimation of brain, skull and scalp surfaces. In: Eleventh Annual Meeting of the Organization for Human Brain Mapping (2005)
Acknowledgements
This work was jointly supported by the Shandong University Science and technology project (No. J17KA082), the National Natural Science Foundation of China (No. 61401259), the China Postdoctoral Science Foundation (No. 2015M582128) and Shandong Province Key Laboratory of Medical Physics and Image Processing Technology, School of Physics and Electronics, Shandong Normal University.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Wang, J., Hu, C., Xu, H. et al. A novel multi-atlas and multi-channel (MAMC) approach for multiple sclerosis lesion segmentation in brain MRI. SIViP 13, 1019–1027 (2019). https://doi.org/10.1007/s11760-019-01440-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11760-019-01440-5