Abstract
Introduction
Lesion segmentation plays an important role in the diagnosis and follow-up of multiple sclerosis (MS). This task is very time-consuming and subject to intra- and inter-rater variability. In this paper, we present a new tool for automated MS lesion segmentation using T1w and fluid-attenuated inversion recovery (FLAIR) images.
Methods
Our approach is based on two main steps, initial brain tissue segmentation according to the gray matter (GM), white matter (WM), and cerebrospinal fluid (CSF) performed in T1w images, followed by a second step where the lesions are segmented as outliers to the normal apparent GM brain tissue on the FLAIR image.
Results
The tool has been validated using data from more than 100 MS patients acquired with different scanners and at different magnetic field strengths. Quantitative evaluation provided a better performance in terms of precision while maintaining similar results on sensitivity and Dice similarity measures compared with those of other approaches.
Conclusion
Our tool is implemented as a publicly available SPM8/12 extension that can be used by both the medical and research communities.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Magnetic resonance imaging (MRI) plays an important role in medical image analysis for both clinical and research studies. Inflammatory demyelinating diseases such as multiple sclerosis (MS) [15], which affects more than 2.5 million people worldwide, particularly women, presents plaques (lesions) of demyelination typically observed in conventional MRI. Detecting those lesions is a crucial task for MS diagnosis as stated in the 2010 revision of the McDonald criteria [18]. Thus, a fully automatic tool that can segment the lesions would prevent user variability and reduce the time consumption considerably. In the literature, there is not yet a standard tool feasible for daily clinical practice [14], although many attempts have been proposed thus far [6, 11, 13, 22, 27, 29, 31]. Automatic detection of MS lesions is a challenging problem [12, 14] that is hampered by factors such as diversity among devices, MRI acquisition protocols, and case of studies.
In this study, we present a new tool that follows the principles of a recently presented algorithm for MS lesion detection [5] that was configured and tested only for 1.5-T images. This algorithm is based on two main steps, initial brain tissue segmentation according to the gray matter (GM), white matter (WM), and cerebrospinal fluid (CSF), followed by a second step where lesions are segmented as outliers to the normal apparent GM brain tissue on the fluid-attenuated inversion recovery (FLAIR) image. To extend this tool for processing 3 T brain volumes, we have changed the bias normalization and tissue segmentation steps. Moreover, we have modified the lesion segmentation process to include two iterations. These changes allow the reduction of false-positive (FP) detection, maintaining a good true-positive (TP) rate. Compared with the previous approach, this new strategy only needs T1w and FLAIR while T1w, T2w, proton density-weighted (PDw), and FLAIR images were previously needed for the tissue segmentation process.
Evaluation of the tool is performed using three distinct databases and comparing the results with the annotations manually performed by expert radiologists, which are considered as a gold standard. Two different datasets were acquired using 3-T MRI scanners, while one was acquired with a 1.5-T scanner. To quantitatively evaluate our tool with respect to the state-of-the-art, we have compared our results on the Medical Image Computing And Computer-Assisted Intervention (MICCAI) MS Challenge 2008 training dataset with three recent works [13, 27, 31]. Moreover, we have also submitted onlineFootnote 1 the segmentation results for the testing dataset of the MICCAI Challenge 2008, obtaining at the time of submission the best results in the ranking using an unsupervised strategy.
Additionally, we integrate this novel tool as a SPM8/12Footnote 2 extension toolbox that is publicly available to the communityFootnote 3 and may be used in both 1.5-T and 3-T MRI.
Materials and methods
Data
We use data from the following sources.
-
1.
3-T dataset: 3-T Siemens (data from Hospital Vall d’Hebron, Spain). This (non-public) database comprised data from 70 patients with clinically isolated syndrome (CIS) and was a very challenging dataset where the lesion volume per patient was very small. The scanner used was the 3-T magnet with a 12-channel phased-array head coil (Trio Tim; Siemens, Germany). The following pulse sequences were obtained: (1) transverse proton density and T2-weighted fast spin-echo (TR = 2500 ms, TE = 16–91 ms, voxel size = 0.78 × 0.78 × 3 mm3); (2) transverse fast T2-FLAIR (TR = 9000 ms, TE = 93 ms, TI = 2500 ms, flip angle = 120, voxel size = 0.49 × 0.49 × 3 mm3); and (3) sagittal 3D T1 magnetization-prepared rapid gradient-echo (MPRAGE) (TR = 2300 ms, TE = 2 ms; flip angle = 9; voxel size = 1 × 1 × 1.2 mm). For 24 patients, the lesions were annotated by experts on FLAIR images with a lesion volume variation (mean ± standard deviation) and range (min–max) of 4.1 ± 4.7 [0.18–18] ml; however, for the rest of the cases, annotations were performed in PDw images with a lesion volume variation and range of 2.8 ± 2.5 [0.25–9.5] ml.
-
2.
3-T MS Challenge 2008 dataset: 3-T Siemens high-resolution images (data from the MICCAI Challenge 2008 dataset) [28].
-
(a)
Training dataset: This dataset was composed of 20 images from two different hospitals with 3-T scanners, 10 images from the Childrens Hospital Boston (CHB; 3-T Siemens), and 10 images from the University of North Carolina (UNC; 3-T Siemens Allegra). Each case was labeled using FLAIR images by an expert from the respective hospital. The protocol consisted of T1w, T2w, and FLAIR images. The T1w image was then rigidly co-registered to the standard Montreal Neurological Institute (MNI) atlas. The T2w and FLAIR images were rigidly registered to its corresponding T1w images. All images were re-sliced at an isotropic 0.5 × 0.5 × 0.5 mm resolution with cubic spline interpolation. This resolution was chosen, as most of the structural T1w, T2w, and FLAIR datasets had originally an in-plane resolution of 0.5 × 0.5 mm, and several images had an original slice thickness of 0.5 mm. The mean lesion volume and range for the CHB and UNC datasets were 9.85 ± 5.75 [4.4–19.6] ml and 1.60 ± 1.64 [0.1–4.5] ml, respectively.
-
(b)
Testing dataset: This dataset comprised 23 images from the same two hospitals, 14 cases from the CHB and 9 cases from the UNC. All of them followed the same scheme explained above. Manual annotations were not available for this dataset. Participants had to send the results to the MICCAI Challenge platform to obtain an evaluation and an overall score.
-
(a)
-
3.
1.5-T dataset: 1.5-T General Electric scanner (data from the Clínica Girona, Spain). This (non-public) database comprised data from 14 patients with clinically confirmed multiple sclerosis. The scanner used was a 1.5-T GE Signa HDxt with 3D fast spoiled gradient T1w (TR 30 ms, TE 9 ms), fast spin echo T2w (TR 5000–5600 ms, TE 74–77 ms), PDw (TR 2700 ms, TE 11.9 ms), and FLAIR (TR 9002 ms, TE 80 ms, and TI 2250 ms). All images were acquired in the axial view with a slice thickness of 3 mm and with a resolution of 1 × 1 × 3 mm3. MS lesions were annotated on PDw images with a lesion volume average and range of 10.72 ± 17.45 [0.43–67.8] ml.
Due to the variability of acquisition conditions and protocols for MS datasets and in order to simplify our pipeline, we used only T1w and FLAIR images.
Pre-processing
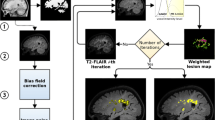
To address the MS lesion segmentation, a set of pre-processing steps are necessary in advance (left column of Fig. 1). First, the brain region is extracted from all of the images involved. There is a large literature covering the skull-stripping problem [1, 19, 21, 24]. Among the available approaches, the Brain Extraction Tool (BET) [26] from the FMRIB Software LibraryFootnote 4 is the most commonly used mainly for the satisfactory results provided [3]. Thus, we decided to use BET here.
Intensity inhomogeneities are inherent to MRI for various reasons: variable imaging parameters, overlapping intensities, noise, motion, echoes, blurred edges, normal anatomical variations, and susceptibility artifacts [23]. The effect of these inhomogeneities can be attenuated by first reducing the image noise and then correcting the intensities with the estimation of the multiplicative bias field. Here, we have first used the anisotropic diffusion filter provided by the Insight Toolkit (ITK) software library,Footnote 5 which implements an N-dimensional version of the classic Perona and Malik filter [17]. Thereafter, the inhomogeneities were corrected using the well-known, non-parametric, non-uniform intensity normalization (N3) methodFootnote 6 [25], which is becoming a standard in brain MRI pipelines. Although some works in the literature perform BET after N3 intensity correction [19], when optimizing non-uniformity correction using N3, the brain mask also helps with its improvement [4, 32]. The main goal of our work is the segmentation of MS lesions by using intensity features; therefore, the intensity normalization is a primary requirement. Other works with similar necessities also perform as we do here BET before intensity correction [24, 30]. The use of default parameters for this N3 method has been revised in recent works to adapt them to newer MRI scanners with higher field strength and multichannel receiver coils, reducing the smoothing distance parameter to 30–50 mm [4, 32].
Finally, we performed intra-subject registration, which consists of the co-registration of T1w and FLAIR images of each patient to the MNI space [16], which is the reference standard space in SPM8/12. The target of the registration process is the T1w image due to the higher resolution, thus the voxel size used is the one from T1w, which depends on each database. Different registration software programs can be found in the literature [8]; however, because we aimed to develop a toolbox integrated on SPM8/12, we used the spatial co-registration, estimate, and re-slice method from SPM8/12, based on the work described in [7]. This is an affine registration process using the normalized mutual information as an objective function and trilinear interpolation with no wrapping. This step was unnecessary for the 3-T MS Challenge 2008 dataset because images were already registered to this space.
Lesion segmentation tool
Once the images were pre-processed, they could be analyzed accurately. The lesion detection tool was based on identifying the hyperintense regions in the FLAIR image, which we considered to be intensity outliers. Because the GM was the brighter healthy tissue in FLAIR images, we used its distribution to define the hyperintense outliers. Hence, before performing the lesion detection, a previous step consisting of tissue segmentation was necessary.
There are different approaches for brain tissue segmentation [30]. In the approach of Cabezas et al. [5], this step was performed by an expectation maximization segmentation method combining T1w, T2w, and PDw images to address partial volume effects. However, because our aim was to integrate our tool into the Statistical Parametric Mapping (SPM) framework, we simplified this step using the well-known SPM8/12 segment algorithm [1] that relies only on T1w images. Because the result of this segmentation was a probability map per tissue type, we assigned GM, WM, or CSF class according to the maximum value.
Subsequently, this GM mask was used to compute the intensity distribution of the GM in the FLAIR image. This distribution should represent the highest intensities of the image, but the lesions were still brighter; hence, their intensities were considered to be outliers of this distribution. To detect the outliers, we used the full width at half maximum (FWHM) of the main peak to determine the standard deviation. The threshold Thr is computed as follows:
where μ is the mean intensity of the GM distribution and σ is the standard deviation determined using the FWHM. This method provides a high robustness for determining the variability of a distribution, minimizing the effect of outliers (in our case, lesions misclassified as GM). The parameter α is used to adjust the detected candidate lesions.
Thereafter, we applied the following method to remove FP lesions that remained after thresholding the FLAIR volume:
-
1.
Tissue of the lesions (λ ts). The lesions should be classified as WM, but registration limitations could lead to some misalignments, and the tissue segmentation process could classify them as GM. The percentage of voxels belonging to the WM and GM over CSF was computed for all detected lesions. When the threshold λ ts was higher than this percentage, the lesion was then refused.
-
2.
Neighborhood of the lesions (λ nb). Because the lesions should appear in the WM (although the lesions could be classified as the GM), the surrounding voxels must strictly belong to the WM. Therefore, another threshold was defined to limit the proportion of the WM over GM and CSF in the lesion neighborhood.
-
3.
Lesion size. As stated in different works [2, 9], a brain region could be considered as an MS lesion if its volume is larger than 3 mm3. This rule allows the elimination of hyperintense voxels or group of voxels smaller than 3 mm3.
In summary, the two first post-processing steps are as follows:
where λ ts represents the ratio of lesion voxels belonging to the WM (Les iWM ) or the GM (Les iGM ) and λ nb represents the ratio of lesion neighbor voxels belonging to the WM (Les iWM ), both over all lesion voxels (Lesi). Superindex i indicates the actual lesion candidate.
Depending on the image acquisition machine and due to the high heterogeneity in the lesion intensities, we noticed that some of the lesions may fall under the automated threshold determined in Eq. 1. However, setting this threshold at a lower value enlarges real lesions at the expense of also introducing much more FP, even after the refinement. To address this issue, our segmentation tool allows the possibility to perform the thresholding and refinement step twice, the second iteration after discarding the voxels segmented in the previous step that is performed at a higher threshold being more restrictive. The advantage of this iteration instead of directly using a less restrictive threshold is to avoid the oversegmentation of lesions and ensure a more accurate application of the post-processing steps. This simple iterative strategy increases the performance, particularly for 3-T images and challenging images that present more artifacts and abnormal intensities [10].
Figure 2 shows the results of the strategy when using only one iteration and when using two iterations. We also show the differences (g, h) with the ground truth and (i) between strategies. One can see how (c) the strategy with a single iteration leads to oversegmentation due to the necessity of fixing a smaller global threshold compared to the strategy with two iterations. The intensity of the dirty white matter (inflammation) of higher hyperintense lesions sometimes may be higher than small focal lesions; therefore, a single threshold may lead to misclassification. Instead, the use of two thresholds combined with the post-processing rules (λ ts and λ nb) allows the definition of an initially higher threshold ((d) first iteration) in order to avoid surrounding voxels belonging to the lesion inflammation; however, in e, the second iteration threshold can be smaller to detect lower intensity lesions (in the example, two new small lesions are added in this step). Note that the adjacent voxels of the initial segmented lesions will be discarded through the post-processing rules (f).
Images representing a the original FLAIR image, b manual annotation, c single-iteration strategy with only one threshold applied, d–f the first threshold, second threshold, and final result, respectively, of the two iteration strategies, g differences between the ground truth and first strategy, h differences between the ground truth and second strategy, and i differences between the first and second strategy
Evaluation
Quantitative evaluation has been performed using three different well-known measures, the Dice similarity coefficient (DSC), true-positive rate (TPR), and positive predictive value (PPV). All of those measures have been computed between the ground truth (lesions manually segmented by the experts) and automatic segmentation. The DSC is the most common measure used to validate segmentation methods and compute segmentation accuracy. This measure considers the TP, FP, and false-negative (FN) voxels. It ranges between 0 and 1 and accounts for the presence or absence of voxels in both annotations:
The TPR measures the sensitivity of the method and ranges between 0 and 1. The lower the false negatives (lesions missed) were, the better the measure becomes. We computed TPR region-wise, therefore rating the detection accuracy:
Finally, PPV computes the precision of the method. The lower the false positives (healthy tissue classified as lesion), the bigger the PPV. PPV ranges from 0 to 1 and is also computed region-wise:
Experimental results
3-T dataset
We have divided this dataset of 70 patients into two different groups depending on the image modality used to annotate the MS lesions (FLAIR or PDw). Notice that most patients in this dataset have a very low lesion load.
We set up the parameters of the algorithm using only the first group of patients. An exhaustive analysis was performed to find both λ ts and λ nb from 0 to 1 each 0.05 in both the first and second iterations. This optimization is highly dependent on the parameter α which was also tested in both iterations from 1 to 3 each 0.1. In this dataset, the tissue classification classifies almost all detected candidate lesions—i.e., the hyperintensities in FLAIR images, either as WM or GM. Therefore, λ ts does not have a particular influence in the results.
In this test, the parameters of our proposal for the first iteration were set conservatively, allowing only voxels over 97 % of the GM distribution to obtain a low number of FP: α = 3, λ ts = 0.7, and λ nb = 0.6, although λ ts was stable from [0–0.60], there was a weak improvement when 70 % of the voxels belongs to the WM. In the second iteration, those parameters were modified to allow more regions to be considered (α = 1.5, λ ts = 0.7 and λ nb = 0.65, although similar results were found for ranges of λ ts = [0.40–0.80] and λ nb = [0.60–0.70]). Because the second dataset was acquired using the same scanner and using the same protocol, we maintained the same parameter configurations for the testing. Table 1 shows how, using the two iterations strategy, we could improve the averages of DSC Δ0.03 and TPR Δ0.12 maintaining the same PPV.
When analyzing the first group of patients (see Fig. 3), we obtained a mean Dice of 0.30 ± 0.19, a mean TPR of 0.36 ± 0.21, and a mean PPV of 0.53 ± 0.29. While evaluating the second group of patients (annotations in PDw), the measures were a mean DSC of 0.33 ± 0.18, a mean TPR of 0.50 ± 0.20, and a mean PPV of 0.62 ± 0.23. Thus, the focus was to reduce the FP detection trying to detect as much TP as possible.
A qualitative analysis of the segmentation results is illustrated in Fig. 4, where we show 2D and 3D views of four different cases with different lesion loads. The trend observed in the results showed that the higher the lesion volume was, the higher the accuracy obtained, although 12 ml is still a low volume. The slices shown in Fig. 4 illustrate clear examples of MS lesions that our tool could easily detect. However, the segmentations were usually slightly smaller than the ground truth (GT), and some FPs were found, particularly in confusing areas where the intensities were similar to MS lesions, and the tool classified them as GM outliers (for example, the first slice of case 0.5 ml or third slice of case 1.2 ml).
Qualitative example of the obtained results using our automatic tool. The GT is annotated on PDw images for cases 7.8 and 1.2 ml, and FLAIR images for cases 12 and 0.5 ml). The first row of each case shows the original FLAIR image, while the second rows show the automatic segmentation results (green = TP, red = FP, and yellow = FN)
It is important to highlight that the lesion volume of the segmentation correlates with the GT. In the 2D views, when there are no lesions, the tool does not find anything; however, when a significant lesion volume is present, the method can detect it. The 3D view also helps to summarize the overall performance, location, volume, and accuracy. Figure 5 plots the linear correlation for the different datasets. The upper left plot shows the result obtained for the FLAIR dataset, where a high Pearson’s coefficient was obtained (r = 0.95). By contrast, for the PDw dataset (Fig. 5b), the presence of a few outliers reduced the correlation coefficient to r = 0.80 (r = 0.91 will be obtained if the two most significant outliers are avoided). In both datasets, the p value <<0.01 confirms that the correlation is significant.
However, it is important to notice the crucial issue regarding the evaluation of the segmentation approaches, which is the actual dataset used. This dataset contains 70 cases, which comprise clinically isolated syndrome (CIS) patients presenting a small lesion load. This fact implies lower DSC values than previously published results in the state-of-the-art [14] due particularly to the severe punishment of small errors in the segmentation.
3-T MS challenge 2008 dataset
The second database used to test our tool was the MICCAI MS Challenge 2008 dataset, a high-resolution 3-T dataset. This database is becoming a benchmark in the field, allowing researchers to compare their results. Here, we present a comparison of our results on the training dataset with the ones obtained by three recent works of the state-of-the-art [13, 27, 31]. The training results of Souplet et al. [27] are the ones reported in Geremia et al. [13] and Weiss et al. [31]. Moreover, results obtained on the online testing dataset are also discussed below and compared to the results of Souplet et al. and Geremia et al. extracted from the MS Challenge webpage.
Training dataset
Table 2 shows the results obtained when testing the training data from the Children’s Hospital Boston. The parameters for the first iteration were set following the same optimization strategy explained above, allowing only voxels over 97 % of the GM distribution (α = 3, λ ts = 0.60, and λ nb = 0.60). In the second iteration, those parameters were modified to allow larger regions to be considered (α = 2.5, λ ts = 0.60, and λ nb = 0.55, obtaining similar results within ranges of λ ts = [0.40–0.80] and λ nb = [0.50–0.60]). Notice that while the first iteration uses almost the same configuration as the previous dataset (Section “3T dataset”), the second requires new values for a better adjustment to the high-resolution data.
Looking at the obtained results, our approach showed higher values in almost every case for all TPR and PPV. DSC values were only provided in the work of Weiss et al. [31]. A higher performance is obtained with our proposal than that of Weiss et al. with an average of 0.38 ± 0.15 and 0.29 ± 0.15, respectively. In Table 3, we show again the mean results of each measure for the three segmentations (first and second iteration and one iteration maintaining the PPV). The improvement of the second iteration for this dataset was 0.04 in DSC and 0.05 in TPR.
Table 4 shows the performance of our tool when using the training data from the University of North Carolina. In this test, we used the same configuration obtained when optimizing the previous dataset (CHB). The performance of the algorithms followed a similar trend (TPR increased and PPV decreased). We obtained a TPR average of 0.68 ± 0.19 and a PPV average of 0.38 ± 0.21. However, in this case, the DSC values were more similar, obtaining 0.30 ± 0.14 in our proposal and 0.29 ± 13 with the one by Weiss. The different performances of all the algorithms between both datasets may be due to the fact that lesion volume being different.
Testing dataset
Using the previous parameter configuration, we ran our tool over the 23 MS patients of the testing dataset. The results were submitted to the MICCAI MS Challenge 2008, which provides an automatic evaluation of the segmentation, allowing a comparison with other participants. In Table 5, we show the average TPR, FPR, and overall score of the approaches of [13, 27] and our tool. Our results are slightly lower in terms of TPR, while the improvement in terms of FPR with respect to the other approaches is clearly evident, with a better overall score (82.344). At the time of submission, our overall score was the first that used an unsupervised lesion detection approach.
1.5-T dataset
Images (1.5 T) are the most widely used clinically [20]. Therefore, we included a dataset of 14 MS patients who were acquired at 1.5 T in the evaluation of our segmentation tool. Notice, that in this dataset, we have three cases with a lesion load of 16, 21, and 68 ml.
Due to the low number of patients, we ran a non-exhaustive cross-validation scheme. We divided the database into four random groups (two of four patients and two of three patients) using only one set at each time for training, and the remaining ones for testing. Following the same strategy used in the previous datasets, we observed that the best configuration was obtained when setting a conservative first iteration (α = 3, λ ts = 0.6, λ nb = 0.7) and a more permissive configuration for the second iteration (α = 2, λ ts = 0.6, λ nb = 0.5, providing a similar performance within ranges of λ ts = [0.40–0.80] and λ nb = [0.55–0.65]). The comparison of different iterations is illustrated in Table 6, where no improvement is appreciable with the second iteration when maintaining the same PPV.
The quantitative evaluation in terms of DSC, TPR, and PPV is illustrated in the last three boxplots of Fig. 3. Within this dataset, all measures increased compared with the 3-T measures (mean DSC of 0.43 ± 0.14, mean TPR of 0.51 ± 0.17, and mean PPV of 0.79 ± 0.19). Furthermore, the boxplots were more compressed, indicating that the obtained results for those 14 patients were consistent independently of its lesion load.
A qualitative evaluation is also illustrated in Fig. 6, where we selected two extreme cases. Although the FP detections have increased, quantitative results remain high due to the high number of TPs detected. Regarding the volume correlation, Pearson’s coefficient for this dataset was 0.99 with a significant p value <<0.01 (see Fig. 5c), obtaining a very good linear correlation in terms of the lesion volume.
Discussion
We have developed an automatic tool for segmenting MS lesions using only T1w and FLAIR images. We used T1w images to obtain brain tissue segmentation that is subsequently used to find the GM distribution in FLAIR images and obtain the lesion mask as hyperintense outlier voxels of this distribution. To improve the performance, the tool allows the application of this threshold in two steps: first, in a conservative way to avoid a large number of FPs; and second, in a less restrictive way, with the aim to increase the TP detections. This simple iterative strategy increases the performance of the tool specifically for 3-T images and for challenging images with artifacts and abnormal intensities. By contrast, we cannot prove that same behavior in the 1.5-T dataset. This cause may be due to the small number of patients in this dataset.
In this work, we have presented results evaluated in three different datasets, analyzing more than 100 patients in total. They were acquired using different scanners; thus, different protocols and different resolutions were tested. The variability of the data and the obtained results in all of the datasets confirmed the robustness of our proposal. The general trend of the study shows an improvement in the FP reduction over the current state-of-the-art works, maintaining similar TPR in both the detection and segmentation accuracy. Furthermore, the rates in terms of FP detection present a regular performance in all cases. In terms of DSC, we believe that the lower values observed in the 3-T dataset may be a bias of the DSC measure because in those cases with an almost imperceptible lesion load, small voxel-wise errors have more influence into the DSC computation than in cases with a higher lesion load. Because the patients of this database have CIS, which may be early-MS but they have not been yet diagnosed, they present with a low lesion volume. Notably, some patients were annotated in FLAIR and others in PDw images. In the 3-T dataset, the tool was optimized with FLAIR annotations because the tool uses FLAIR images for the lesion segmentation, resulting in better lesion volume correlation with FLAIR annotation masks and oversegmentation when evaluated with PDw masks (see Fig. 5).
We want to remark that maintaining the two λ parameters with default values at λ ts = 0.6 and λ nb = 0.6 for all of the analyzed datasets and two iterations (therefore without a specific optimization per dataset) also provides very similar results. Instead, the α parameter has a strong influence on the performance of our tool, i.e., the more permissive it is, the more restrictive the two λ parameters must be. Therefore, the α parameter configuration plays a very important role and must be set for each scanner. Indeed, in an ideal configuration, λ ts could be avoided because all of the hyperintensities will be labeled as WM tissue by the SPM8/12 tissue segmentation. The MICCAI MS Challenge 2008 dataset presented images with a challenging bias and poor quality in some areas; therefore, λ ts had to be fixed accurately enough to reduce the number of artifacts misclassified as lesions.
Our tool is publicly available as an SPM8/12 extension toolbox, being easily adaptable and with a default configuration to be used straightaway. However, limitations in the performance of the tool can be found if different tools than the proposed here are used in the pre-processing steps. We recommend strictly following the pipeline presented in this work to maximize the performance of the tool.
References
Ashburner J, Friston KJ (2005) Unified segmentation. NeuroImage 26(3):839–851
Battaglini M, Rossi F, Grove RA, Stromillo ML, Whitcher B, Matthews PM, De Stefano N (2013) Automated identification of brain new lesions in multiple sclerosis using subtraction images. J Magn Reson Imaging
Boesen K, Rehm K, Schaper K, Stoltzner S, Woods R, Lüders E, Rottenberg D (2004) Quantitative comparison of four brain extraction algorithms. NeuroImage 22(3):1255–1261
Boyes RG, Gunter JL, Frost C, Janke AL, Yeatman T, Hill DL, Bernstein MA, Thompson PM, Weiner MW, Schuff N, Alexander GE, Killiany RJ, DeCarli C, Jack CR, Fox NC (2008) Intensity non-uniformity correction using N3 on 3-T scanners with multichannel phased array coils. NeuroImage 39(4):1752–1762
Cabezas M, Oliver A, Roura E, Freixenet J, Vilanova JC, Ramió-Torrentà L, Rovira À, Lladó X (2014) Automatic multiple sclerosis lesion detection in brain MRI by FLAIR thresholding. Comput Methods Prog Biomed 115(3):147–161
Cabezas M, Oliver A, Valverde S, Freixenet J, Beltran B, Vilanova JC, Ramió-Torrentà L, Rovira À, Lladó X (2014) BOOST: a supervised approach for multiple sclerosis lesion segmentation. J Neurosci Methods 237:108–117
Collignon A, Maes F, Delaere D, Vandermeulen D, Suetens P, Marchal G (1995) Automated multi-modality image registration based on information theory. In: Bizais
Diez Y, Oliver A, Cabezas M, Valverde S, Martí R, Vilanova J, Ramió-Torrentà L, Rovira A, Lladó X (2013) Intensity based methods for brain MRI longitudinal registration. A study on multiple sclerosis patients. Neuroinformatics pp 1–15
Ganiler O, Oliver A, Diez Y, Freixenet J, Vilanova J, Beltran B, Ramió-Torrentà L, Rovira A, Lladó X (2014) A subtraction pipeline for automatic detection of new appearing multiple sclerosis lesions in longitudinal studies. Neuroradiology pp 1–12
García-Lorenzo D, Prima S, Collins DL, Arnold L Douglas, Morrissey SP, Barillot C (2008) Combining robust expectation maximization and mean shift algorithms for multiple sclerosis brain segmentation. In: MICCAI workshop on Medical Image Analysis on Multiple Sclerosis (validation and methodological issues) (MIAMS’2008), New York, United States, pp 82–91
García-Lorenzo D, Lecoeur J, Arnold D, Collins D, Barillot C (2009) Multiple sclerosis lesion segmentation using an automatic multimodal graph cuts. Lect Notes Comput Sci 5762(PART 2):584–591
García-Lorenzo D, Francis S, Narayanan S, Arnold DL, Collins DL (2013) Review of automatic segmentation methods of multiple sclerosis white matter lesions on conventional magnetic resonance imaging. Med Image Anal 17(1):1–18
Geremia E, Clatz O, Menze BH, Konukoglu E, Criminisi A, Ayache N (2011) Spatial decision forests for MS lesion segmentation in multi-channel magnetic resonance images. NeuroImage 57(2):378–390
Lladó X, Oliver A, Cabezas M, Freixenet J, Vilanova JC, Quiles A, Valls L, Ramió-Torrentà L, Rovira A (2012) Segmentation of multiple sclerosis lesions in brain MRI: a review of automated approaches. Inf Sci 186(1):164–185
Love J (2006) Demyelinating diseases. J Clin Pathol 59(11):1151–1159
Mazziotta JC, Toga AW, Evans A, Fox P, Lancaster J (1995) A probabilistic atlas of the human brain: theory and rationale for its development. NeuroImage 2(2A):89–101
Perona P, Malik J (1990) Scale-space and edge detection using anisotropic diffusion. Pattern Anal Mach Intell IEEE Trans 12(7):629–639
Polman CH, Reingold SC, Banwell B, Clanet M, Cohen JA, Filippi M, Fujihara K, Havrdova E, Hutchinson M, Kappos L, Lublin FD, Montalban X, O’Connor P, Sandberg-Wollheim M, Thompson AJ, Waubant E, Weinshenker B, Wolinsky JS (2011) Diagnostic criteria for multiple sclerosis: 2010 revisions to the McDonald criteria. Ann Neurol 69(2):292–302
Popescu V, Battaglini M, Hoogstrate W, Verfaillie S, Sluimer I, van Schijndel R, van Dijk B, Cover K, Knol D, Jenkinson M, Barkhof F, de Stefano N, Vrenken H (2012) Optimizing parameter choice for FSL-brain extraction tool (bet) on 3D T1 images in multiple sclerosis. NeuroImage 61(4):1484–1494
Pratt W (1 May 2014) Magnetic resonance in medicine. The basic textbook of the European magnetic resonance forum, 8th edn. Electronic version
Roura E, Oliver A, Cabezas M, Vilanova JC, Rovira A, Ramió-Torrentà L, Lladó X (2014) MARGA: multispectral adaptive region growing algorithm for brain extraction on axial MRI. Comput Methods Prog Biomed 113(2):655–673
Schmidt P, Gaser C, Arsic M, Buck D, Fẗorschler A, Berthele A, Hoshi M, Ilg R, Schmid VJ, Zimmer C, Hemmer B, Mühlau M (2012) An automated tool for detection of FLAIR-hyperintense white-matter lesions in multiple sclerosis. NeuroImage 59(4):3774–3783
Sha DD, Sutton JP (2001) Towards automated enhancement, segmentation and classification of digital brain images using networks of networks. Inf Sci 138(1–4):45–77
Shattuck DW, Sandor-Leahy SR, Schaper KA, Rottenberg DA, Leahy RM (2001) Magnetic resonance image tissue classification using a partial volume model. NeuroImage 13(5):856–876
Sled J, Zijdenbos A, Evans A (1998) A nonparametric method for automatic correction of intensity nonuniformity in MRI data. IEEE Trans Med Imaging 17(1):87–97
Smith SM (2002) Fast robust automated brain extraction. Hum Brain Mapp 17(3):143–155
Souplet JC, Lebrun C, Ayache N, Malandain G (2008) An automatic segmentation of T2-FLAIR multiple sclerosis lesions. In: Multiple Sclerosis Lesion Segmentation Challenge Workshop (MICCAI-2008), New York, NY, USA, United States, pp 1–8
Styner M, Lee J, Chin B, Chin M, Commowick O, Tran H, Jewells V, Warfield S (2008) Editorial: 3D segmentation in the clinic: a grand challenge II: MS lesion segmentation. In: Grand Challenge Workshop: Multiple Sclerosis Lesion Segmentation Challenge, pp 1–8
Sushmita D, Ponnada AN (2013) A comprehensive approach to the segmentation of multichannel three-dimensional MR brain images in multiple sclerosis. NeuroImage Clin 2:184–196
Valverde S, Oliver A, Cabezas M, Roura E, Lladó X (2014) Comparison of 10 brain tissue segmentation methods using revisited IBSR annotations. J Magn Reson Imaging 41(1):93–101
Weiss N, Rueckert D, Rao A (2013) Multiple sclerosis lesion segmentation using dictionary learning and sparse coding. Med Image Comput Comput Assist Interv MICCAI 8149:735–742
Zheng W, Chee MW, Zagorodnov V (2009) Improvement of brain segmentation accuracy by optimizing non-uniformity correction using N3. NeuroImage 48(1):73–83
Acknowledgments
E. Roura holds a BRUdG2013 grant. S. Valverde holds a FI-DGR2013 grant from the Generalitat de Catalunya. This work has been partially supported by “La Fundació la Marató de TV3” and by Retos de Investigación TIN2014-55710-R.
Ethical standards and patient consent
We declare that all human studies have been approved by the appropriate Ethics Committee and have therefore been performed in accordance with the ethical standards laid down in the 1964 Declaration of Helsinki and its later amendments. We declare that patient consent was waived due to the retrospective nature of this study.
Conflict of interest
We declare that we have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Roura, E., Oliver, A., Cabezas, M. et al. A toolbox for multiple sclerosis lesion segmentation. Neuroradiology 57, 1031–1043 (2015). https://doi.org/10.1007/s00234-015-1552-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00234-015-1552-2