Abstract
The World Health Organization suggests visual examination of stained sputum smear samples as a preliminary and basic diagnostic technique for diagnosing tuberculosis. The visual examination process requires much time of laboratorian, and also, it is prone to mistakes. For this purpose, this paper proposes a novel random forest (RF)-based segmentation and classification approaches for the automated classification of Mycobacterium tuberculosis in microscopic images of Ziehl–Neelsen-stained sputum smears obtained using a light-field microscope. The RF supervised learning method is improved to classify each pixel depending on local color distributions as a part of candidate bacilli regions. Therefore, each pixel is labeled as either a candidate tuberculosis (TB) bacilli pixel or not. The candidate pixels are grouped together using connected component analysis. Each pixel group is then rotated, resized and centrally positioned within a bounding box, respectively, in order to utilize appearance-based tuberculosis bacteria identification algorithms. Finally, each region is classified by using the proposed RF learning algorithm trained on manually marked TB bacteria regions in the training images. The algorithm produces results that agree well with manual segmentation and identification. Different two-class pixel and object classifiers are also compared to show the performance of the proposed RF-based pixel segmentation and bacilli objects identification algorithm. The sensitivity and specificity of the proposed classifier are above 75.77 and 96.97 % for the segmentation of the pixels, respectively. It is also revealed that the sensitivity increases over 93 % when the staining is performed in accordance with the procedure. Moreover, these measures are above 89.34 and 62.89 % for the identification of bacilli objects. The results show that the proposed novel method is quite successful when compared to the other applied methods.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Tuberculosis (TB)—one of the major health problems in the world—is an infectious disease caused by the bacillus Mycobacterium tuberculosis. The bacilli typically appear slightly curved or straight rods in microscopy. It has beaded and occasional branching form and also occurs singly, pairs or in small clumps. The dimensions of the bacilli are 1–10 \(\upmu \)m in length and 0.2–0.6 \(\upmu \)m in width [1]. Mycobacterium tuberculosis and similar microorganisms have acid-fast cell wall, which makes the cells impervious to acid–alcohol mixture. Therefore, acid-fast staining technique is used for detection of acid-fast bacilli (AFB). Ziehl–Neelsen (ZN) staining procedure is the most common method in acid-fast staining. AFB appears red–pink, while non-acid-fast region is stained blue after staining with ZN procedure, which is used by conventional microscopy [2]. Figure 1 shows an example of ZN-stained sputum smear image. Another staining procedure is fluorochrome staining in which bacilli are stained yellow fluorescence with dark background when observed with a fluorescence microscope [3]. Fluorochrome staining is more sensitive and requires lower work effort than ZN staining. However, the fluorescence microscopes are used in high-income countries because of greater cost of the equipment [4].
Patients complaints, physical examinations, chest radiographs and tuberculin tests are not sufficient for a definitive diagnosis in TB suspected cases. Microbiology diagnostic is required for a definitive diagnosis in such a case. In microbiology diagnostic, the tuberculosis is diagnosed by examining the stained sputum smear. The laboratory clinicians normally look for the presence of AFB in magnified microscopic images. Three specimens of sputum are drawn from the patient on two consecutive days and stained with ZN staining procedure. Experienced laboratory clinician needs to examine at least 100 field and spends at least five full minutes for each field [5]. If each slide is not examined carefully or is examined too short, AFB will be missed and the specimens result will be negative when it is actually positive. Therefore, manual screening is error-prone. Additionally, it is a labor-intensive task because the examination of each specimen requires visual inspection examination, which takes a long time [6]. In other words, since the visual examination with mental concentration is required, the number of specimens to be inspected is limited for reliable manual visualization. Consequently, automatic screening speeds up diagnosis, reduces the workload of laboratory technicians and decreases error by improving accuracy and sensitivity of the diagnosis [7].
1.1 Related work
The topic of analyzing microscopic images has become even more important in recent years. However, most of the previous approaches focused on microscopic images of fluorochrome-stained slide samples. Forero et al. [7–9] and Veropoulos et al. [10] proposed an approach of identification of TB in fluorochrome-stained sputum smear slide images. In [7–9], canny edge detection has been applied to microscopic images to segment TB bacilli. Then, closing and opening from mathematical morphology are used to complete broken edge contours in segmented objects. Several feature descriptors are obtained from the most frequent bacilli shapes, and decision based on classification tree, classification tree with feature selection and Gaussian mixture model are used for the identification stage, respectively. Veropoulos et al. [10] demonstrated edge pixel linkage to segment bacilli and used feed-forward neural network for classification. Besides these studies, a trend in using novel methods in images of ZN-stained sputum smear slides is available in the literature. Sadaphal et al. [11] proposed color-based segmentation by using Bayesian segmentation. After that, shape–size analysis is applied to segmented images to detect bacilli. Siena et al. [12] applied decorrelation stretching to microscopic images for segmentation and used back-propagation neural network for detection. Khutlang et al. [13] used two-class pixel classifiers such as Bayes, Euclidean distance linear, logistic linear and quadratic to segment candidate bacilli objects. Geometric transformation invariant features were extracted, and feature subset selection and Fisher transformation were used for optimization of the feature set. Two-class object classifiers such as kNN, Bayes, linear, quadratic, PNN and SVM were also used to show the performance of classifiers.
Among these related works, the segmentation process performs well. However, most of them are related to basis clustering and thresholding algorithms which use color differences in an image. Moreover, instead of using bacilli appearance, shape–size analysis is utilized in identification process, and some well-known and frequently used methods are applied to these extracted features. Therefore, this article will discuss how novel learning algorithms can be applied to microscopic images.
In addition, several very known appearance-based learning methods are implemented to compare the proposed approach. Gaussian probability density function (GPDF)- and support vector machine (SVM)-based pixel segmentation algorithms are separately performed onto same data set to compare the performance of the proposed RF-based tuberculosis bacilli pixels segmentation. For the comparison of the tuberculosis bacilli classification performance of the proposed RF-based learning algorithm, SVM and artificial neural network (ANN)-based pattern identification methods are also accomplished onto the tuberculosis bacilli patterns data. The comparative results of the segmentation and classification both obtained with the proposed algorithm and other methods are quantitatively presented using some quantitative measurements such as sensitivity, specificity and accuracy measures.
1.2 Proposed method
This paper presents novel RF-based method for the automated pixel segmentation and identification of tuberculosis bacilli in microscopic images of ZN-stained sputum smears obtained by using a light-field microscope. A data set including 116 images collected from five different slides taken from various patients was obtained to achieve the experimental results.
In each training image, the pixels belonging to regions of tuberculosis bacilli were manually labeled by medical technician. To minimize the number of pixels manually marked incorrectly in each image, noisy data elimination using Mahalanobis distance is also performed by comparing the RGB color components in the color space of each pixel with the color distributions. This data set was then divided as training and test sets for experimental studies. To achieve RF-based supervised learning algorithm for pixel segmentation, a training procedure is firstly employed on different two-class pixels. The first class pixels are constituted with \(3\times 3\) pixel windows centered on each pixel manually marked as the part of bacilli region. The other class pixels represented as non-bacilli pixels are extracted by randomly selecting \(3\times 3\) windows outside of the bacilli class pixels. Therefore, each pixel in the ZN-stained images in the test set is automatically labeled by using RF-based supervised learning algorithm either bacilli pixel or non-bacilli pixel. The tuberculosis bacilli pixels are then grouped into the regions by using connected component analysis. Each region is then rotated, resized and centrally positioned within \(30\times 30\) bounding box, respectively, in order to utilize appearance-based tuberculosis bacilli identification algorithms. As a result of the pixel segmentation, the bounding box can include background (white color pixels for non-bacilli) and foreground pixels (RGB color pixels for candidate bacilli region).
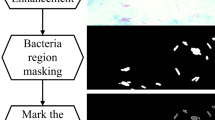
Once the image is segmented, only the region of pixels given same bacilli colors is retained. Figure 2 shows sample images manually segmented and classified by an expert. Subsequently, appearance-based tuberculosis bacilli identification process is then performed for determining which of them are true bacilli. To achieve the proposed appearance-based identification algorithm, the regions of the bacilli and non-bacilli given very similar colors and structures were also manually marked by technician as shown in Fig. 2b. For instance, the bacilli in the regions marked with black circles are not tuberculosis bacilli. The bacilli marked with red circles are also tuberculosis bacilli. Therefore, we are facing a two-class classification scheme: a single class of bacilli and a rejection class for all the rest of the pixel regions. Finally, the segmented and positioned region (pixels) into the bounding box is classified as either tuberculosis bacilli or not by using the proposed RF-based learning algorithm. The overall flowchart of the proposed algorithm is presented in Fig. 3.
Expert guided segmented and classified images of Fig. 1. a Manually segmented images; the red color pixels represent the candidate tuberculosis bacilli pixels. b The red circled objects are tuberculosis bacilli, and the black circled objects are non-bacilli regions (color figure online)
2 Methodology
2.1 Feature extraction for pixel segmentation
Training-based pixel segmentation algorithm is proposed for each pixel classification as either candidate tuberculosis bacilli or background pixels in the images. For that aim, the proposed RF-based classifier algorithm is trained on color pixels collected from the bacilli and non-bacilli regions. Each region consists of nine neighborhood pixels in a \(3\times 3\) window. For bacilli region, each \(3\times 3\) window is centered around a pixel, which is manually marked as a bacilli pixel. Non-bacilli regions are also randomly selected from the outside of the bacilli class pixels. Therefore, red, green and blue components of each \(3\times 3\) window region are used to produce color distributions for the bacilli and non-bacilli pixels.
In addition, a noisy pixel elimination is required on the pixels manually marked. In microscopic images, bacilli seem like tiny objects when they are compared with original image size. Although the position of the cursor in the image was magnified up to a specified ratio, laboratory technician might click on non-bacilli objects as bacilli objects by mistake. Therefore, it is required an automated data variation analysis to eliminate the pixels given more unfamiliar data than averaged color distribution of the selected pixels. For this reason, Mahalanobis distances between samples to be used for training are calculated, and then, noisy data are eliminated. The noisy data are identified by calculating the Mahalanobis distance of overall data and then determining a threshold value which is close the maximum distance.
Mahalanobis distance is a measure of distance between two n-dimensional random vectors, X and Y. This metric is defined as follows:
where \(T\) denotes matrix transpose, and \(\epsilon \) denotes the common covariance matrix. Unlike other distance metrics, it takes into account the data distribution, in other words covariance between variables. Also, it maximizes the distances between variables with different labels, while minimizing the distance between variables labeled similarly. Therefore, it is chosen as an appropriate distance metric [14].
In order to make the coefficient of each element in the mask different, the bivariate GPDF is fitted to \(3\times 3\) mask, and so, the numerical coefficient of each element begins to decrease with distance from the center. Finally, twenty-seven dimensional feature vector is obtained for each pixel manually marked and randomly selected because of using RGB color model.
2.2 Feature extraction for bacteria classification
The main idea of the appearance-based approach is to learn template characteristics. Therefore, each pixel of the objects in the segmented image is quite significant for this approach. For this reason, the laboratory technician manually enclosed the bacilli and non-bacilli objects with a rectangle box to produce a training set for tuberculosis bacilli regions. The proposed RF learning algorithm is then applied on this set to achieve an appearance-based training stage.
After each pixel is assigned as either bacilli or background pixels by using the proposed RF-based pixel segmentation algorithm, the RF-based bacteria identification is then performed for learning the appearance of the bacilli and non-bacilli objects. The segmented tuberculosis bacilli pixels are firstly grouped into the regions by using connected component labeling method [15]. Each region is then rotated, resized and centrally located within \(30\times 30\) sized image, respectively, in order to utilize appearance-based tuberculosis bacilli identification algorithms. As the results of the pixel segmentation process, the \(30\times 30\) sized image contains RGB color pixels belong to candidate bacilli region and white color pixels for background (non-bacilli pixels). This is repeated for each pixel region labeled as tuberculosis bacilli pixel. The direction of each pixel region is determined by using moment invariant method [16]. The angle of direction formula is given as follows;
where \(\mu \) is second-order moments. These central moments are defined for a raw image as follows:
where \(\bar{x}\) and \(\bar{y}\) are centroid coordinates and calculated using following equations.
2.3 Gaussian probability density function
A random vector \(X=[X_{1},X_{2},\ldots ,X_{n}]^{T}\) is said to multivariate normally distributed if its probability density function is defined as follows:
where \(\mu \) is mean vector, \(\epsilon \) is covariance matrix and n is the dimension of random vector [17]. The mean vector is calculated by averaging each random variable \(X_{i}\). It is the centroid of the probability density function, or it is known as the point at which the probability density function is maximum.
2.4 Support vector machines
Support vector machine (SVM) is very popular learning method for classification and regression analysis. The basic idea behind it is to construct a maximum-margin hyperplane. So it means that SVM calculates the best hyperplane which separate the classes from each other. By using kernel functions, it maps pattern vectors to high dimensional feature space and separates data linearly in this space [18].
Decision function that uses the kernel function is defined as follows:
where \(x\) is input vector, \(y\) is target value and \(K(x,x_{i})\) is the kernel function. The coefficients \(a_{i}\) and \(b\) are obtained from the following formula (9) which is required to maximize with respect to the \(a_{i}\) subject to (10).
where \(C>0\) expresses the strength of penalty errors.
This decision machine method was applied to the training data acquired from microscopic images as follows:
-
1.
A simple scaling was performed on the training data because of eliminating the computational complexity and transforming large numerical data into small numerical data.
-
2.
Radial basis function was chosen as the kernel function. This function handles the situation when the relation between the features and labels is nonlinear and nonlinearly maps the data into higher dimensional space. The other reason why this function was chosen is that the number of hyper parameters which affects the complexity of model is less than other kernel functions.
-
3.
In order to determine optimum \(C\) and \(\gamma \) hyper parameter, k-fold cross-validation technique was used. Cross-validation accuracy is calculated as the percentage of correctly classified samples. The grid search approach was used to determine optimum \(C\) and \(\gamma \) parameters using cross-validation. In this approach, various pairs of these parameters were tried and the pair which gives the best cross-validation accuracy was chosen as optimum parameter.
-
4.
The training data were trained by using parameters determined in step (3).
2.5 Random forest
Ensemble learning is a machine learning model where multiple classifiers are trained to solve a problem instead of a single classifier. It means that this model generates a set of assumptions and aggregates their results. Bagging [19] is the first simple and efficient method of ensemble learning models. This method uses the combination of multiple bootstrap samples of a training data set. Each of sample sets constructs a tree, and a majority vote is taken for class prediction. Boosting [20] is the other well-known ensemble learning method. In boosting, a set of weights which are initially equal is assigned to training set. The weights are updated for misclassified samples. The final classifier is constructed from weighted majority voting of each classifier. Random forest is obtained by adding randomness to bagging method and [21] have an impact on proposal of this method by Breiman.
Random forest (RF) [22] is an ensemble learning method which consists of a collection of tree classifiers \(h(x,\varphi _{k}),k=1,\ldots \). Each tree is built by a random vector \(\varphi _{k}\) where \(\varphi _{k}\) is sampled independently but with same distribution for all random vector \(\varphi _{1},\ldots , \varphi _{k-1}\) and casts a vote for the most popular class at input x.
Number of trees, \(N\), and number of variables used to split each node, \(m\), are defined by the user in this technique. \(N\) bootstrap samples are randomly chosen from the training data set. In bootstrap sampling, a new data set is formed by random sampling with replacement from the existing data set. The majority of the bootstrap samples are used to build the tree, in-bag data, and the rest of the samples which are called out-of-bag (OOB) data are used to estimate the error of the tree. They correspond to two-third and one-third of the training data set, respectively. After choosing the in-bag data, the tree is constructed according to the CART algorithm [23], which consists of followings. For each node of the tree, the best split among \(m\) attributes is chosen by using information gain. After decided at which variable that is split, the value of the mentioned variable that is branched is determined by using Gini index. The recommended value of \(m\) is equal to \([\sqrt{k}]\) where \(k\) is the total number of features. A weight is assigned to the constructed tree according to the OOB error; the most the OOB error, the least the weight. While classifying the test data, each tree casts a vote at its terminal nodes. The votes are counted up separately; a class of which the sum of the votes is higher is determined, and test data are assigned to this class. A diagram describing the process of random forest is presented in Fig. 4.
2.6 Artificial neural network
Artificial neural network [24] models the way biological brains work. In other words, it allows the machine to learn in the same ways that humans do. In this work, a three-layer feed-forward neural network was implemented with \(n\) input, \(m\) hidden layer and \(1\) output. This output classifies the segmented objects as the bacilli or non-bacilli. The learning rule was determined as the generalized delta learning rule, also known as the error back-propagation algorithm, which belongs to supervised learning. The log-sigmoid activation function was used for hidden and output layers.
3 Experimental results
3.1 Dataset
The performance of the proposed approaches was evaluated using database consisting of microscopic images. ZN-stained sputum smear slides were prepared by Mycobacteriology Laboratory at Faculty of Medicine in Karadeniz Technical University. Five smear-positive slides from five subjects were used. Different number of color images were acquired from them. Image acquisition system was set up in our computer vision and pattern recognition laboratory [25]. The system consists of a standard personal computer, a conventional light microscopy and a digital camera. Sample slides were scanned by using Nikon Eclipse 80i microscopy at 100\(\times \) magnification. A Premiere Digital Microscope Eyepiece MA88-300 digital camera was attached to the ocular on a microscope for image acquisition. The taken images were stored in bitmap file format with 24 bit depth in color, and the pixel resolution of an image was \(640\times 480\).
The whole data set consists of 116 positive images. The numerical data about the data set are given in Table 1. To develop segmentation and classification process, about one-third of these images were used for training and the rest of the images were employed to test the proposed approaches.
A few images acquired from each subject are shown in Fig. 5. The images of the first subject are divided into four class because of the complicated background (i.e., unexpected changes in intensity). Also, the images acquired from the third and the fourth subjects are blurred images. The reason is that the staining procedure was not performed correctly. The contrast between background and foreground colors is clearly seen in the second and the fifth sample slide images.
All images were analyzed by an expert laboratory technician to decide which objects are bacilli. Also, it was decided whether each pixel of the objects looks like bacilli in color. One of these expert guided segmented and classified images is shown in Fig. 2. In Fig. 2a, red-painted pixels belong to candidate bacilli regions, and also in Fig. 2b, the red circled objects are tuberculosis bacilli, and the black circled objects are non-bacilli regions but have similar color distributions.
3.2 The quantitative measurements
The performance of the proposed algorithm is estimated by using some criteria such as sensitivity, specificity and accuracy. For this reason, the number of true positives (TP), false positives (FP), true negatives (TN) and false negatives (FN) was obtained for each classifier. TP is the number of positive cases correctly identified, FP is the number of negative cases incorrectly identified, TN is the number of negative cases correctly identified, and finally, FN is the number of positive cases incorrectly identified. Sensitivity measures the proportion of actual positive cases which are correctly identified, specificity measures the proportion of actual negative cases, which are correctly identified, and accuracy is the proportion of the number of correctly classified cases to the number of cases. These measures are given as follows:
3.3 Parameter selection
Most of the parameters used in the proposed method were estimated automatically except a few of them which were selected empirically.
The first step in this work is to classify (segment) each pixel in the image as a foreground (candidate bacilli) or a background pixel. For that purpose, GPDF and SVM methods were also performed for the pixel segmentation experiments on the same images to compare the performance of the proposed RF-based tuberculosis bacilli segmentation algorithm. The parameters of the methods were adjusted as follows:
-
A Gaussian curve was fitted to training data. The range of Gaussian curve which indicates the distance from the mean value was selected empirically.
-
Scaling values of SVM were chosen \(-\)1 as minimum and \(+\)1 as maximum. The training data set was divided into five subset which shows the \(k\) parameter. In the grid search approximation, \(C\) and \(\gamma \) parameters were tried in exponentially growing sequences. (e.g., \(C=2^{15}, 2^{13},\ldots , 2^{-5},\,\gamma =2^{-6}, 2^{-5.5},\ldots , 2^{-1}\)).
-
In the process of RF, the number of trees \((N)\) and the number of variables \((m)\) affect the accuracy. Therefore, the experiments were carried out through setting the \(N\) to 100, 150, 200, 300, 400, 500 and \(m\) to 4, 5, 6, 7, 8, 9, 10.
Once segmentation process completed, the candidate pixel regions in the segmented images were classified as bacilli or non-bacilli objects. In this step, to analyze the results of the proposed RF-based tuberculosis bacilli classification method, the ANN- and SVM-based object identification algorithms were also applied to classify each candidate pixel region, as follows:
-
Error back-propagation training algorithm was performed for the three-layer neural network of which the hidden layer neurons were set to 100, 200, 300, 400, 500.
-
The parameters of SVM were selected as mentioned in the segmentation process.
-
\((m)\) and \((N)\) parameters were chosen as 2,000, 2,250, 2,500 and 250, 500, 750, 1,000, respectively, due to the large feature vector.
3.4 Segmentation experiments
Three different segmentation methods were applied on microscopic image database to evaluate the success ratio of the segmentation methods explained in Sects. 2.3, 2.4 and 2.5. Figure 6 depicts tuberculosis bacilli pixel segmentation results achieved on the image shown in Fig. 1 by using the mentioned methods. A visual comparison on the pixel segmentation performance of the algorithms can easily be made by considering the results shown in Fig. 6. The pixels manually segmented for the same image are also shown in Fig. 2a.
Segmentation results obtained on the image shown in Fig. 1 by independently using GPDF, SVM and the proposed RF-based tuberculosis bacilli pixel segmentation algorithms. a GPDF, b SVM, c RF
Based on these schemes, the segmentation results obtained by GPDF are listed in Tables 2, 3, 4, 5 and 6, respectively. During the experiments, different threshold values empirically selected were used to segment each pixel. As shown in the tables, the sensitivity performance of the GPDF shows an increase with a smaller threshold value. Moreover, when the sensitivity rate increases, specificity rate decreases. This trade-off causes the need of optimum threshold values decision. Threshold values were selected as 98.00, 98.90, 99.70, 99.50 and 99.80 % due to the sharp drop in the specificity rate. Therefore, the best sensitivity rates for these database were achieved with 60.12, 75.05, 34.50, 40.72 and 55.94 %, respectively. The calculated sensitivity, specificity and accuracy rates based on the selected threshold values are italicized in Tables 2, 3, 4, 5 and 6.
The parameters of SVM were estimated automatically using cross-validation technique. The sensitivity rates were calculated as in Tables 7, 8, 9, 10 and 11, respectively, when the estimated parameters were used.
Segmentation performance of the proposed RF method depends on user-defined parameters. OOB error estimation graph is used to evaluate the effects of different settings of these parameters, \(m\) and \(N\). These graphs show the error rates and stabilities of the constructed models. The correctness of the constructed model is estimated using them. Also, using the OOB error estimate removes the need for a set aside test set. Figure 7 shows that the OOB estimates are remarkably accurate. On the whole, the average OOB error values are about in the range of 1 and 2 % which reflects the correctness of the model. Tables 12, 13, 14, 15 and 16 present the segmentation performance of proposed RF method utilized the two parameters (\(N\) and \(m\)). The each cell in the tables which corresponds the various pairs of these parameters provides the calculated sensitivity, specificity and accuracy rates, respectively. Higher sensitivity rates, i.e., 82.31, 94.41, 90.63, 75.77 and 93.05 % for each subject respectively, are italicized.
We have first studied the influence of the parameter \(N\), i.e., the number of trees. The sensitivity rates with respect to the number of trees for fixed values of the number of variables show that these values remain constant at about the same integer value. The reason is that the minimum number of trees is selected as 100, and so, the other selected values are close to this value. Then, we have focused on the \(m\) parameter, i.e., the number of variables. The sensitivity rates begin to raise for an increasing number of features, but then begin to decrease except for \(m=10\). According to the [22], too much portion of features causes this decrease, and the number of variables has to be \(>\)1 and does not have to increase so much.
In order to put the given results more explicitly, the following comments can be made clearly: RF has better performance than SVM and GPDF when the sensitivity rates are considered except for the second subject where a trade-off exists, i.e., the sensitivity rate of SVM is higher than RF, whereas the specificity rate is less than RF. When a comparison between accuracy rates for second subject is made, it is seen that RF has higher performance than SVM.
So far, all experiments carried out on microscopic images were subjected to each sputum slide sample, i.e., each image was examined by using only images obtained from same slide. Therefore, another experiment was also performed to understand the power and robustness of the proposed RF-based tuberculosis bacilli pixel segmentation method. The training set was constructed with using the images obtained from the second subject only. Then, the images collected from the fifth subject were also employed for test set.
Based on these schemes, the pixel segmentation results independently achieved by GPDF, SVM and the proposed RF-based learning methods are summarized in Tables 17, 18 and 19, respectively. The best sensitivity rate is italicized in Table 19. It is clearly seen that RF-based pixel segmentation algorithm has given better performance than SVM- and GPDF-based algorithms, as in other experiments.
3.5 Classification experiments
To compare the performance of the proposed RF-based bacilli identification, the classification of segmented pixel regions was independently performed using classification methods explained in Sects. 2.4, 2.5 and 2.6. The obtained results of the classifications are given in Tables 20, 21 and 22 for ANN, SVM and the proposed RF-based learning methods, respectively.
The optimum hidden layer neuron number was selected according to the specificity rates, which increase up to a level and then begin to decrease. Hence, the number of hidden layer neuron was decided as 300, and sensitivity rate was equal to 76.82 %. This rate increases to 86.71 % when SVM is used. The best sensitivity which is resulted in 89.34 % with \(m=2{,}250\) and \(N=250\) was obtained by the proposed RF-based tuberculosis bacilli identification and also this rate is italicized in Table 22.
4 Conclusion
This paper presented novel random forest (RF)-based tuberculosis bacilli pixel segmentation and appearance-based pixel region classification approaches for the automated identification of Mycobacterium tuberculosis bacilli in microscopic images of ZN-stained sputum smears obtained using a light-field microscope. The performance of the proposed RF-based learning method was analyzed on the novel database includes ZN-stained sputum smear slide images obtained using our microscopic image acquisition system. For the performance measurement, three known quantitative measurements, i.e., sensitivity, specificity and accuracy, are used. To compare the results of the proposed pixel segmentation and pixel region identification of tuberculosis bacilli, two other very popular learning-based segmentation and classification algorithms were also implemented on this data set. The experimental results indicate that the proposed RF-based learning algorithm for TB bacteria classification has achieved higher performance than other very known learning methods which are GPDF, SVM and ANN. The proposed RF-based learning method, as well as future studies, will be incorporated into an automated microscope for tuberculosis bacilli identification, which would also feature automatic focusing and microscope stage control. Therefore, the automation in the context of TB screening will be very useful task for tuberculosis bacilli diagnosis with light-field microscope in order to speed up diagnosis, improve the accuracy and reduce the workload of laboratory technician.
References
Palomino, J.C., Leao, S.C., Ritacco, V.: Tuberculosis 2007—From Basic Science to Patient Care. http://www.tuberculosistextbook.com/index.htm. Accessed June 2007
International Union Against Tuberculosis and Lung Disease: Sputum Examination for Tuberculosis by Direct Microscopy in Low Income Countries. France (2000)
Auramine-rhodamine Fluorescence-Acid Fast Bacteria. http://library.med.utah.edu/WebPath/webpath.html
Steingart, K., Hnery, M., Ng, V., Hopewell, P., Ramsay, A., Cunningham, J., Urbaczik, R., Perkins, M., Aziz, M., Pai, M.: Fluorescence versus conventional sputum smear microscopy for tuberculosis: a systematic review. Lancet Infect. Dis. 6(9), 570–581 (2006)
Revised National Tuberculosis Control Programme: Module for Laboratory Technicians. Central TB Division, New Delhi (2005)
Nguyen, T.N.L., Wells, C.D., Binkin, N.J., Pham, D.L., Nguyen, V.C.: The importance of quality control of sputum smear microscopy: the effect of reading errors on treatment decision and outcomes. Int. J. Tuberc. Lung Dis. 3(6), 483–487 (1999)
Forero, M.G., Sroubek, F., Cristobal, G.: Identification of tuberculosis bacteria based on shape and color. Real-Time Imaging 10, 251–262 (2004)
Forero, M.G., Cristobal, G.: Automatic identification techniques of tuberculosis bacteria. In: Proceedings of the SPIE, vol. 5203, pp. 71–81 (2003)
Forero, M.G., Cristobal, G., Desco, M.: Automatic identification of Mycobacterium tuberculosis by Gaussian mixture models. J. Microsc. 223, 120–132 (2006)
Veropoulos, K., Learmonth, G., Campbell, C., Knight, B., Simpson, J.: Automated identification of tubercle bacilli in sputum a preliminary investigation. Anal. Quant. Cytol. Histol. 21(4), 277–281 (1999)
Sadaphal, P., Rao, J., Comstock, G.W., Beg, M.F.: Image processing techniques for identifying Mycobacterium tuberculosis in Ziehl–Neelsen stains. Int. J. Tuberc. Lung Dis. 12(5), 579–582 (2008)
Siena, I., Adi, K., Gernowo, R., Mirnasari, N.: Development of algorithm tuberculosis bacteria identification using color segmentation and neural networks. Int. J. Video Image Process Netw. Secur. 12(4), 9–13 (2012)
Khutlang, R., Krishnan, S., Dendere, R., Whitelaw, A., Veropoulos, K., Learmonth, G., Douglas, T.S.: Classification of Mycobacterium tuberculosis in images of ZN-stained sputum smears. IEEE Trans. Inf. Technol. Biomed. 14(4), 949–957 (2010)
Alpaydn, E.: Introduction to Machine Learning. The MIT Press, London (2010)
Di Stefano, L., Bulgarelli, A.: A simple and efficient connected components labelling algorithm. In: International Conference on Image Analysis and Processing, pp. 322–327 (1999)
Hu, M.: Visual pattern recognition by moment invariants. IRE. Trans. Inf. Theor. 8, 179–187 (1962)
Do, C.B.: The multivariate Gaussian distribution. http://cs229.stanford.edu/section/gaussians (2008)
Cortes, C., Vapnik, V.: Support vector machines. Mach. Learn. 20, 273–297 (1995)
Breiman, L.: Bagging predictors. Mach. Learn. 24, 123–140 (1996)
Freund, Y., Schapire, R.E.: A short introduction to boosting. J. Jpn. Soc. Artif. Intell. 14(5), 771–780 (1999)
Ho, T.K.: The random subspace method for constructing decision forests. IEEE Trans. Pattern Anal. Mach. Intell. 20(8), 197–227 (1990)
Breiman, L.: Random forests. Mach. Learn. 45, 5–32 (2001)
Breiman, L., Friedman, J., Olshen, R., Stone, J.: Classification and Regression Trees. Chapman and Hall, Wadsworth, CA (1984)
Zurada, J.M.: Introduction to Artificial Neural Systems. West Publishing Co., St. Paul (1992)
KTU-CVPR Lab http://ceng2.ktu.edu.tr/cvpr/
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Ayas, S., Ekinci, M. Random forest-based tuberculosis bacteria classification in images of ZN-stained sputum smear samples. SIViP 8 (Suppl 1), 49–61 (2014). https://doi.org/10.1007/s11760-014-0708-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11760-014-0708-6