Abstract
Acca sellowiana (Myrtaceae) is a multipurpose species with edible fruits and ornamental value, native to Uruguay and southern Brazil. Domestication of the species in Uruguay is incipient although in other countries, it is widely cultivated. It is an evergreen out-crossing shrub, pollinated by birds and bees. For this reason, it is necessary to develop vegetative propagation strategies such as stem cuttings to reproduce outstanding genotypes for conservation or breeding programs. Adventitious root (AR) formation in cuttings is regulated by environmental and endogenous factors. Among phytohormones, indole-butyric acid (IBA) is the most widely exogenous auxin used to improve rooting of cuttings. Most studies on AR formation at the molecular level use model species; however, the conservation of these mechanisms in non-model plants has been little studied, consequently the effects of different factors and their interactions in A. sellowiana are not well understood. The identification and expression analysis of genes known to be involved in the regulation of the process is an important step to elucidate the molecular mechanisms that regulate AR differentiation in A. sellowiana cuttings. In this study, we compared two genotypes with contrasting rooting ability, and we identified and characterized three genes that might regulate the onset of AR development in A. sellowiana: AsPIN1, AsTIR1 and AsSHR. Their expression analysis showed that in the difficult-to-root genotype, AsTIR1 increases strongly in response to exogenous IBA, shortly after induction treatment. Relative expression of AsPIN1 and AsSHR also increases 24 h later. The biological significance of this gene expression pattern is discussed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Acca sellowiana (Berg.) Burret is an evergreen shrub of the Myrtaceae family which is cultivated for its fruits and is also valued as an ornamental plant for its flowers and foliage. Extracts taken from fruits and leaves contain antioxidant, antimicrobial, and pharmacological activities (Vuotto et al. 2000; Bontempo et al. 2007; Mosbah et al. 2018; Tortora et al. 2019). Domestication and breeding of this species depend on the ability of elite plant materials to be propagated. However, adventitious root differentiation of cuttings varies between 0 and 80% depending on the genotype set evaluated (Franzon et al. 2004; Guerra et al. 2012; Ross et al. 2017; Niella et al. 2018), and it is difficult to provide nurseries with mother plants of selected materials and particularly difficult-to-root genotypes.
It was hypothesized that these differences in AR formation among A. sellowiana genotypes are due to an earlier phase change in the difficult-to-root genotypes which can explain the loss of competence to form adventitious roots (Ross et al. 2021). In woody plants, the ability of cuttings to form adventitious roots declines with the age of donor plants (Wendling et al. 2014a; Aumond et al. 2017) and has been inversely linked with the xylogenesis program (de Almeida et al. 2012; Abarca et al. 2014; Vielba et al. 2016). Some genes of the GRAS family, such as SCR and SHR, are involved in the maturation-related decline of adventitious rooting (Pizarro and Díaz-Sala 2019). This might be explained by the different functions that the same gene regulatory network may play at different developmental stages of a cell type (De Lucas and Brady 2013). The lower rooting capacity associated with ageing of donor plants is little understood in woody plants (Aumond et al. 2017). Anatomy of stem cuttings in difficult-to-root genotypes of A. sellowiana shows an earlier development of the periderm associated with loss of competence to form adventitious roots (Ross et al. 2021). It is known that competence to form AR declines with phase change as a result of changes in auxin homeostasis (Rasmussen et al. 2014). The different rooting performance observed in genotypes with different rooting ability is mainly due to the regulation of endogenous active auxin accumulation in the AR source tissues after cutting excision, as well as differences in auxin sensitivity (Guan et al. 2015; Druege et al. 2016, 2019). This differential distribution of auxin seems to be a sufficient signal to trigger or modify the developmental program of a cell in model plants (Vanneste and Friml 2009; Negishi et al. 2011; Ruedell et al. 2015) although the mechanism has not been characterized in A. sellowiana specifically.
The fact that IAA is involved in the early events of AR formation has been well established (Blakesley 1994; Della Rovere et al. 2013) and the concentration of endogenous IAA is considered to play a central role in the control of AR initiation and development in various plant species (Ford et al. 2001; Kelen and Ozkan 2003; Davies 2010; Bellini et al. 2014; Pacurar et al. 2014; Wendling et al. 2015; Vilasboa et al. 2018; Gonin et al. 2019; Druege et al. 2019; de Almeida et al. 2020). The concentration of auxin in specific tissues is controlled by its biosynthesis, metabolism, and transport, and these processes are regulated by multiple mechanisms (Han et al. 2009). The differential accumulation of endogenous IAA in easy and difficult-to-root genotypes might be explained by the different expression of IAA biosynthesis and transport genes (de Almeida et al. 2015; Druege et al. 2019) as well as higher expression of repressors of auxin-responsive genes in hard to root species (Ruedell et al. 2015). AR formation depends on the early accumulation of IAA at the base of the cutting, via polar transport (Negishi et al. 2014; Pacurar et al. 2014; Druege et al. 2016). Quantification of indole 3 acetic acid (IAA) is not extensively used in AR studies because of its low concentration in tissues and the interference of other compounds in the analysis (Stuepp et al. 2016).
An alternative strategy to understand AR formation would be to identify and study the expression of genes involved in auxin homeostasis and root meristem patterning in the excised tissues of cuttings. Sensitivity to auxin is determined by the presence and affinity of auxin receptors. The auxin receptor TRANSPORT INHIBITOR RESPONSE 1 (TIR1) and its homologous gene PagFBL1 increased their expression a few hours after auxin treatment (de Almeida et al. 2015; Shu et al. 2019). Once auxin sensitivity is established, other genes in the auxin response network which act downstream in the AR signaling pathway are bound to activate (Teale et al. 2006; Pierre-Jerome et al. 2013; Wachsman et al. 2015). Changes in gene expression are detected during the first hours after cutting excision in response to the stress caused by wounding mainly during the first 24 h (Druege et al. 2016).
Several genes are induced by exogenous auxin during the process of AR formation (Ludwig-Müller 2000; Guan et al. 2015; Druege et al. 2019). Auxin-induced AR formation in cuttings involves transcription factors of the GRAS family, particularly in the context of maturation of woody plants (Druege et al. 2016). Root meristem patterning and stem cell specification in response to auxin require of the GRAS family transcription factors such as SHORTROOT (SHR) (Blilou et al. 2005). These are also involved in the maturation-related decline of adventitious root formation in distantly related forest species, and the switch between the developmental programs of xylogenesis and AR formation in A. thaliana (Xuan et al. 2014; Abarca et al. 2014; Stevens et al. 2018; Pizarro and Díaz-Sala 2019). In other species, these genes are upregulated during AR formation and null mutants exhibit reduced AR formation (Druege et al. 2019). The expression of the auxin efflux carrier gene PINFORMED 1 (PIN1) increases in response to exogenous auxin; short auxin treatments activate the transcription of PIN genes in different tissues regulating its own distribution (Vanneste and Friml 2009; Fett-Neto et al. 2011).
AR formation in Acca sellowiana cuttings is strongly affected by genotype. Among several treatments used to induce rooting, difficult-to-root genotypes (NR) respond only to exogenous IBA and reach rooting levels similar to the easy-to-root genotypes (R) without exogenous hormone (Ross et al. 2017). Our hypothesis is that exogenous IBA improves AR formation in NR cuttings of A. sellowiana by modifying the expression of genes involved in auxin perception and homeostasis during the first stages of the process, leading to the acquisition of competence of some cells to form AR. In order to contribute to the understanding of AR formation and the causes of intraspecific variability specifically in A. sellowiana, the purpose of our study was to identify some of those genes in model species, validate them in A. sellowiana and study their expression pattern in response to exogenous IBA. In order to provide evidence that supports this interpretation of the differences among genotype, we selected a few but critical genes that have been shown to be involved in auxin homeostasis (PIN1; TIR1) and root meristem patterning (SHR) in E. grandis and A. thaliana, to study their expression in micro-cuttings of two A. sellowiana genotypes with contrasting rooting ability.
Materials and methods
Plant material and culture conditions
Mother plants of A. sellowiana were provided by a local breeding program of the species (INIA-Facultad de Agronomía-MGAP; Uruguay). Two selected genotypes with contrasting rooting ability were grown in the greenhouse under controlled conditions, and treated periodically with fungicide (Benlate®, 0.2%) and Phostrogen® [NPK(MgO3-SO3): 14–10-27 (2.5–7.5)]. Genotypes were identified as R (easy-to-root) and NR (difficult-to-root), according to their rooting performance ex vitro (more than 60% and less than 20% rooting, respectively). To minimize the phenotypic differences in growth habit that might exist between genotypes, sprouts were collected from the basal branches of 4-year-old, vigorous healthy plants, in the same position in the branch. Apical segments (1.5–2.0 cm long) were surface-disinfected with 2% NaOCl for 15 min, washed three times with distilled water and introduced in vitro on WPM medium (Lloyd and McCown 1980) supplemented with MS vitamins (Murashige and Skoog 1962). Rooting of micro-cuttings was induced by adding IBA (9.8 µM) to the culture medium as previously described (Ross et al. 2017). Micro-shoots without IBA treatment were used as control. Cultures were incubated at 25 ± 2 °C, provided with a photon flux of 30 µmol m−2 s−1 and 16:8-h photoperiod.
Bioinformatics analysis and primer design of candidate genes and reference genes
Three candidate genes and three reference genes were chosen for further gene expression analysis. Candidate genes were selected based on literature for other species, in which they are known to be involved in auxin transport (PIN1), auxin perception (TIR1), and root patterning (SHR). Reference genes were chosen among genes previously validated for their use during adventitious rooting in Eucalyptus globulus (EF2, H2B, UBI) (de Almeida et al. 2010). The primers used for reference genes were those validated for E. globulus by the Almeida et al. (2010), shown in Table 1.
For primer design, sequences of the three candidate genes in A. thaliana were taken from the GenBank database of the National Center for Biotechnological Information (NCBI) (http://www.ncbi.nlm.nih.gov/genbank/) and Plant Transcription Factor Database (PlantTFDB) http://planttfdb.gao-lab.org/). A sequence similarity search within E. grandis genome was performed by BLAST analysis using the Phytozome12 platform (https://phytozome.jgi.doe.gov/pz/portal.html). The predicted amino acid sequences were compared and conserved amino acids are colored by the Jalview multiple alignment editor (Clamp et al. 2004).
Conserved regions were obtained by the alignment of the encoding sequences of both species (A. thaliana and E. grandis), using BioEdit© Sequence Alignment Editor software (Hall 1999). Primers were designed from these conserved regions using Primer3Plus (https://primer3plus.com/) and sequenced at Macrogen Inc. (Seoul, Korea). If differences in the sequences of both species were observed, the designed primers were biased towards the sequence corresponding to E. grandis. Two pairs of specific primers were designed for each candidate gene of interest (PIN1, TIR1, and SHR) based on the sequence alignment of conserved regions of the homologous genes in A. thaliana and E. grandis (Table 2).
Amplification and sequence analysis of candidate genes in A. sellowiana
To confirm whether the primers showed homology with genomic regions of A. sellowiana, genomic DNA was extracted from leaves of both genotypes of A. sellowiana, using the cetyl-tri-methyl-ammonium bromide (CTAB) method (Doyle and Doyle 1987). DNA samples of E. grandis and A. thaliana were included as positive controls, using the same method. DNA quantity and quality were assessed by electrophoresis in agarose gel (0.8%) and spectrophotometry with a NanoDrop ND-1000 (Thermo Scientific©).
PCR amplification was performed in a 20-µl reaction containing: 1 × reaction buffer with 2-mM MgCl2, 1-mM dNTPs, 0.5 µM of each primer, 100-ng template DNA, and 0.5 U Taq DNA polymerase (Thermo Scientific©). The PCR program was as follows: 94 ºC for 5 min, followed by 35 cycles of 94ºC for 30 s, 52–58 ºC for 30 s, and 72 ºC for 40 s, using a Gene Touch Thermal Cycler (Bioer Technology ®).
Amplification products were resolved by electrophoresis in an agarose gel (2%), stained with ethidium bromide and visualized under UV illumination. The size of the amplification products was estimated with 1 kb ladder (Thermo Scientific©) as molecular weight marker. Unique amplification products and their respective primers were sequenced at Macrogen Inc. (Seoul, Korea) and edited using FinchTV software (Geospiza Inc.©). Once edited, the final sequences were analyzed using BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi) and the UniProt database (https://www.uniprot.org/). The sequences obtained were deposited to GenBank with accession numbers MZ130946-MZ13095.
Design of specific primers for qRT-PCR
Specific primers for qRT-PCR of target and reference genes were designed using Primer3Plus (Tables 3 and 4). More than one pair of primers for each gene of interest and reference gene was designed in order to obtain combinations with similar amplification efficiency for expression analysis. Target sequence size was confirmed on genomic DNA of A. sellowiana by PCR in 20-µl reactions containing: 100-ng genomic DNA, 1 × reaction buffer with 2-mM MgCl2, 1-mM dNTPs, 0.5 µM of each primer, and 0.5 U Taq DNA polymerase (Thermo Scientific©). The PCR program was as follows: 94 ºC for 1 min, followed by 35 cycles of 94 ºC for 30 s, 54 ºC for 30 s, and 72 ºC for 40 s, using a Gene Touch Thermal Cycler (Bioer Technology ®). Amplification products were resolved in agarose gels (2%) with 1 × TBE buffer solution and stained with ethidium bromide. The size of amplification products was estimated with 1 kb ladder (Thermo Scientific©) as molecular weight marker and visualized under UV illumination.
RNA isolation and cDNA synthesis
Samples were collected 12 and 36 h after rooting induction treatment with IBA. Total RNA was extracted from bottom Sects. (10 mm) of three micro-cuttings per replicate, further purified using Qiagen RNeasy Plant Mini Kit (Qiagen, Hilden, Germany) and treated with RNase-free DNase (Invitrogen, Carlsbad, USA) to eliminate residual DNA, following the manufacturer’s instructions. RNA concentration was determined using a NanoDrop spectrophotometer ND-1000 (Thermo Scientific©). Micro-cuttings used for RNA extraction were discarded, so different repetitions were used for the rooting experiment.
Synthesis of cDNA was performed from 1 µg total RNA to the final 12 µl reaction mixture using MMLV-RT/SS reverse transcriptase according to the manufacturer’s instructions (Invitrogen, Carlsbad, USA). We tested different cDNA dilutions (1/5, 1/10, 1/25, 1/50, 1/100) to determine the best concentration to be used. The optimum dilution was chosen with a quantification cycle (Cq) between 18 and 22 cycles for all samples. A standard curve was generated for each gene of interest using a fivefold dilution series, which was used to calculate primer efficiencies.
Gene expression analysis in micro-cuttings
The relative expression levels of TIR1, PIN1, and SHR at the base of micro-cuttings were determined by quantitative reverse transcription-PCR (RT-qPCR). The resulting data were analyzed by the Comparative Ct method (Livak and Schmittgen 2001). Results are presented as fold change in expression according to Eqs. 1 and 2.
First, the expression of these three genes in both genotypes without addition of exogenous auxin was compared to their expression in the R genotype at the beginning. We then compared the effect of exogenous IBA on the expression of the same genes, in both genotypes (R and NR) 12 and 36 h after the induction treatment.
PCR reactions were carried out using Maxima SYBR Green/ROX qPCR Master Mix (2x) (Thermo Scientific) in a Line-Gene K Fluorescence Quantitative PCR Detection System (Bioer Technology) as follows: 5 min pre-denaturing at 95 °C, followed by 40 cycles of 95 °C for 15 s, 60 °C for 20 s, and 72 °C for 15 s. Three biological and two technical replicates of each sample were done. To confirm the specificity of each PCR reaction, a heat dissociation curve (melting curve) was performed, from 60 °C to 90 °C, following the final PCR cycle. To study changes in gene expression at the onset of AR development, samples were harvested at two time points (12 and 36 h after excision).
The reference genes (RGs) used as internal control were Histone H2B and Elongation factor EF2, reported as reference genes for qPCR during in vitro adventitious rooting of Eucalyptus globulus (de Almeida et al. 2010) and validated for A. sellowiana. To check that the expression of these RGs was not influenced by the conditions of the experiment, the effect of genotype, experimental treatment, and time of sampling on their expression was validated by one-way ANOVA (p = 0.01) using 2−ΔCt, where ΔCt \(\left(Ct sample-Ct cal\right)\) (Schmittgen and Zakrajsek 2000). Experimental data were analyzed by the Comparative Ct method (Livak and Schmittgen 2001), using the geometric mean of the RGs for normalization of the genes of interest expression (Vandesompele et al. 2002).
Experimental design and statistical analysis
The rooting experiment had a factorial design (2 × 2) with five replicates, where the factors were two genotypes (R and NR) and two levels of IBA (0 and 9.8 µM).
The gene expression experiment had a factorial design (2 × 2 × 2) with two genotypes (R and NR), two levels of IBA (0 and 9.8 µM) and two time points post treatment (12 and 36 h.). For each combination of factors, three biological and two technical replicates were performed.
Data were analyzed statistically by analysis of variance (ANOVA) and means were compared by Tukey’s test, with a confidence level of p ≤ 0.05, using Infostat® statistical software. Arcsine transformation was applied to response data before analysis. Data in figures are given as means ± SE.
Results
Adventitious root differentiation in response to exogenous auxin (IBA)
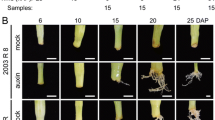
The effect of exogenous IBA on AR formation was observed after four weeks. In vitro rooting of NR micro-cuttings improved significantly (standard error 2.89, degrees of freedom 3) when exogenous IBA (9.8 µM) was added to the induction media (p < 0.0001). Rooting percentage increased 2.4-fold when compared to the control treatment in the absence of auxin (Fig. 1a) although the number of roots per explant was not significantly different (p = 0.4104) (standard error 0.25, degrees of freedom 3) (Fig. 1b). No differences were detected in rooting percentage or root number of the R genotype after the addition of exogenous IBA (Fig. 1a and b). New adventitious roots developed at the base of the micro-cuttings without callus formation in all cases (Fig. 2).
Amplification of target sequences in A. sellowiana
Unique amplification products of the expected size were obtained for the three genes of interest: TIR1, SHR, and PIN1. SHR and TIR1 amplified with only one pair of the specific primers that we designed (Fig. 3a and b). PIN1 amplified effectively with both pairs of specific primers designed. The resulting sequences (325 bp and 430 bp) partially overlapped and were assembled into a unique sequence (Fig. 3c). The three sequences obtained showed high identity with their homologous genes in E. grandis (> 90%) and A. thaliana (> 75%), according to NCBI and Uniprot databases respectively (Table 5).
The reference genes chosen for this study resulted in unique amplification products of the expected size in A. sellowiana: EF2, H2B, and UBI. The resulting sequences showed high identity with their homologous genes in E. grandis (> 90%) and A. thaliana (≥ 69%), according to NCBI and Uniprot databases respectively (Table 5, Fig. 4). The predicted amino acidic sequences also showed high identity with the corresponding proteins in Eucalyptus and A. thaliana (Table 6).
Primers for qPCR
Two pairs of specific primers for qRT-PCR for the genes of interest TIR1, SHR and the reference gene H2B were designed (shown in Fig. 2). However, for the gene of interest PIN1 and the reference genes EF2 and UBI, we were able to design only one forward and two reverse primers or two forward and one reverse primer for each (Tables 3 and 4). These primers were confirmed by conventional PCR using A. sellowiana genomic DNA, and amplicons of the expected size for each gene were obtained.
Analysis of gene expression in micro-cuttings
Relative expression at the onset of AR induction was measured in micro-cuttings of A. sellowiana by RT-qPCR using the geometric mean of two reference genes (H2B and EF2). AsTIR1, AsPIN1, and AsSHR transcripts were induced in response to exogenous auxin in the difficult-to-root genotype during the early steps of AR formation. First, we examined the expression of these genes in both genotypes (R and NR) without IBA, relative to their expression in the R genotype without IBA 12 h after the onset of the experiment. When no exogenous IBA was added to the medium, the expression of these three genes was lower in the NR genotype than in the R genotype (p < 0.0001) throughout the period under study (Fig. 5). Without the addition of exogenous IBA, the R genotype showed an increase in the expression of the three genes 36 h after the beginning of the experiment (p < 0.0001). In the NR genotype, on the other hand, AsTIR and AsPIN also increased their expression (p < 0.0001) but in a much lower magnitude. Data were analyzed by the Comparative Ct method (2−ΔΔCq) using the geometric mean of the reference genes (H2B and EF2). The coefficient of variation between technical replicates was < 1 for the three genes of interest. \(\Delta \Delta Ct=\left[\left(Ct \,gene \,of \,interest-Ct \,internal \,control\right)\,NR \,genotype-\left(Ct \,gene \,of \,interest-Ct \,internal \,control\right)Rgenotype\right]\)
Expression of AsTIR1, AsSHR and AsPIN1 without exogenous IBA in the R and NR genotypes of A. sellowiana micro-cuttings relative to their expression 12 h after the beginning of the experiment. R: easy-to-root genotype; NR: difficult-to-root genotype. Data were analyzed by the Comparative Ct method (2−ΔΔCq) using the geometric mean of the reference genes (H2B and EF2). Bars represent the mean relative expression ± SE of three independent biological replicates and two technical replicates
Next, we compared the expression of these genes between treated (0.98 µM IBA) and untreated (0 µM IBA) samples and found that the behavior of the R and NR genotypes was clearly different in response to the exogenous auxin. The relative expression of TIR1, SHR, and PIN1 increased in response to IBA, but a much greater response was observed in the NR genotype in the treated vs untreated samples. An increase in the expression of the auxin receptor TIR1 was induced 12 h after the treatment with exogenous IBA (p = 0.0111), while the expression of the auxin efflux carrier PIN1 and the transcription factor SHR increased in the NR genotype 36 h after treatment with exogenous IBA relative to their expression in the untreated sample (p = 0.0157 and 0.0189 respectively) (Fig. 6). The relative expression of TIR1 and SHR increased in the R genotype in response to IBA but to a lesser extent (p = 0.0455 and 0.0189 respectively), while changes in relative expression of PIN1 was not significant (p = 0.1216).
Relative expression of AsTIR1, AsSHR and AsPIN1 in treated (0.98 µM IBA) vs. untreated (0 µm IBA) samples of two genotypes of A. sellowiana micro-cuttings with contrasting rooting ability, 12 and 36 h after induction treatment with IBA (9.8 µM). R: easy-to-root genotype, NR: difficult-to-root genotype. Data were analyzed by the Comparative Ct method (2−ΔΔCq) using the geometric mean of the reference genes (H2B and EF2). Bars represent the mean relative expression ± SE of three independent biological replicates and two technical replicates. \(\Delta \Delta \,Ct=\left[\left(Ct \,gene \,of \,interest-Ct \,internal \,control\right)\,sample \,with \,IBA-(Ct \,gene \,of \,interest-Ct \,internal \,control)\,sample \,without \,IBA\right]\)
Discussion
When a well-curated genome of the species under study is not available, the genome of a related species can be used as reference; however, the exact sequence and the genomic location of the genes of interest are unknown. Amplification and sequence comparison can provide evidence to hypothesize that the transcripts that are being quantified are homologous to the genes reported in model species. In this study, we successfully amplified regions of genes which have been shown to be involved at different stages during the onset of AR formation based on published genomes of model species. We showed that the genomic sequences obtained from A. sellowiana are consistent with the target genes and observed a pattern of expression that can be interpreted in the light of the current general understanding of AR formation and previously published hypotheses about intraspecific variability in A. sellowiana for this trait.
Unique amplification products of the expected size were obtained for three genes of interest: TIR1, SHR, and PIN1. Sequence analysis of the obtained PCR fragments showed that they presented high sequence similarity with the candidate genes PIN1, SHR, and TIR1. For the three genes, identity with E. grandis and with A. thaliana was high (more than 90% and 70% respectively). Furthermore, the protein sequences resulting from the translation of the isolated gene fragments present, although partially, domains characteristic of the PIN1, SHR, and TIR1 proteins. AsPIN1 sequence includes part of a transmembrane domain, a domain present in PIN1 from Arabidopsis (Gälweiler et al., 1998), while AsSHR sequence contains a significant portion of the GRAS domain (Pysh et al. 1999; Helariutta et al. 2000). When analyzing the results in AsTIR1, the isolated fragment presents a partial AMN domain, which corresponds to 4 complete LRRs (Leucine-Rich Repeats). This AMN domain contains 16 LRRs in A. thaliana. Although the length of the isolated fragment did not include the FBox domain, which is the other descriptive feature for the TIR1 gene (Ruegger et al., 1998), the isolated fragment shows 95% and 84.6% identity with the homologous fragments of E. grandis and A. thaliana respectively. According to these results, TIR1, PIN1, and SHR genes are present in the A. sellowiana genome, and their sequences show a similarity of more than 90% with the respective E. grandis genes in the region of the gene that was isolated.
Our expression analysis indicates that AsTIR1, AsPIN1, and AsSHR have different levels of expression in R and NR genotypes when there is no hormonal treatment. The level of expression of these three genes in the difficult-to-root genotype 36 h after the beginning of the experiment was always lower than in the easy-to-root genotype. AsTIR1, AsPIN1, and AsSHR transcripts are induced in response to exogenous IBA in stem cuttings of difficult-to-root genotypes of A. sellowiana, during the early steps of AR formation. The R genotype already had a high expression without IBA, so the response to exogenous auxin is not as strong as the increase in relative expression of these genes in the NR genotype. This result is in agreement with the rooting behavior itself; the R genotype roots well without the addition of IBA while the NR genotype significantly improves rooting levels when the exogenous auxin is added to the culture media. This may be explained by the presence of an endogenous higher level of auxin in the R genotype. Among various conditions previously evaluated to induce rooting, exogenous IBA was the only treatment that improved the AR capacity of this genotype to levels that are similar to the easy-to-root genotype (Ross et al. 2017). However, the R genotype does not improve rooting in response to exogenous IBA, and the expression of these genes remains stable. Our data show that the expression of these genes was already much higher in the untreated R genotype, and the addition of exogenous IBA had a very small effect on this genotype. In the difficult-to-root genotype, on the other hand, the relative expression of AsTIR1 increases strongly in response to exogenous IBA, shortly after the induction treatment. In Populus, the expression of PagFBL1 (homolog of TIR1 in Arabidopsis) was similar to the distribution pattern of auxin during AR formation, with a high expression in the cambium and secondary phloem during the induction and initiation phases that decreased in the emerging primordia (Shu et al. 2019). Although our expression analysis was not focused on specific tissues, neo-formation of adventitious root meristems in A. sellowiana takes place outside the cambial ring of the stem, in the secondary phloem (Ross et al. 2021). Thus, it is possible that the increase in the expression of AsTIR1 that we found is concentrated in this tissue and it would be interesting to explore how the expression varies in different tissues of the stem cutting. The relative expression of AsPIN1 and AsSHR also increases, but 24 h later. The effect of IBA as a rooting agent for difficult-to-root materials of this species can thus be related to the increase in expression of at least these genes. These findings are congruent with our previous results and support our hypothesis of an earlier phase change from juvenile to mature of the NR genotype of A. sellowiana. We found that loss of competence to form AR was associated with an earlier phase change, evidenced by the differentiation of a periderm in the NR genotype (Ross et al. 2021).
As plants age, there is a lower expression of the main auxin receptor (TIR1) that explains the loss of sensitivity to auxin (Aumond et al. 2017). The improvement of rooting ability of the NR genotype of A. sellowiana when exogenous IBA was added may have resulted from the increased expression of the auxin receptor AsTIR1. This increase in the sensibility to auxin may have led to a further modification in the expression of other genes that act downstream auxin perception and that play essential roles in the rooting of cuttings. Among many other genes involved in AR differentiation, the increase in relative expression of AsPIN1 and AsSHR in NR genotype is congruent with the observed improvement of rooting ability after exogenous IBA treatment. As a consequence of the concerted action of these genes which showed higher expression in response to IBA, the NR genotype improved the rooting performance, reaching levels similar to the R genotype with or without exogenous IBA. Similar changes in gene expression in response to exogenous auxin have been reported in other species of Myrtaceae (Fett-Neto et al. 2011; de Almeida et al. 2015). The relative amount of mRNA of SHR and other transcription factors of the GRAS family has been observed to be significantly reduced in adult tissues that have lost the capacity to develop AR. The expression of SHR during AR formation is affected by age, auxin level and developmental stage of the cells (Abarca et al. 2014). In Arabidopsis, the same cells that are reactivated by auxin to differentiate ARs, are also able to initiate xylogenesis, with SHR among other transcription factors controlling the switch between the programs (Ricci et al. 2016).
The kind of changes in gene expression that we observed in the different genotypes resembles those observed in different stages during AR development in other species of the Myrtaceae family. Studies in other species of the Myrtaceae with poor AR development also show lower expression of auxin receptors. Loss of AR ability in E. globulus micro-cuttings has been explained by a combination of lower expression of auxin receptors (TIR1, ABP1) and higher expression level for auxin repressors (IAA12, TPL, ARR1). Furthermore, the expression of genes related to auxin synthesis (TAA1, YUC3) and transport (PIN1, AUX1) was found to diverge between stages of development and auxin treatment (Vilasboa et al. 2018).
Although the regulatory gene network that controls AR formation in A. sellowiana or its specific differences from better known species is far from being completely understood, we found evidence that supports the hypothesis that earlier phase change underlies the reduced competence to differentiate new roots in the NR genotypes. Several techniques have been used to delay maturation of juvenile plants or reverse the physiological status of adult plants in other species (Wendling et al. 2014b; Benedini et al. 2015; Stuepp et al. 2016; Bisognin et al. 2017, 2018); among these, epicormic shoots induced by pruning, coppicing, or girdling of adult trees could be evaluated as a source of rejuvenated material for stem cuttings of the NR genotypes of A. sellowiana.
Conclusions
We identified and characterized three genes that are induced by IBA and are likely related to AR development in A. sellowiana micro-cuttings: AsPIN1, AsTIR1, and AsSHR. The results of the expression analysis showed that in the difficult-to-root genotype, AsTIR1 increases strongly in response to exogenous IBA, shortly after induction treatment improving sensibility to auxin of the cells. Relative expression of AsPIN1 and AsSHR also increases, but 24 h later.
These results indicate that AsTIR1, AsPIN1, and AsSHR transcripts are induced during the early steps of AR formation in response to exogenous IBA in stem cuttings of the difficult-to-root genotype of A. sellowiana, improving AR formation. This behavior is similar to that of mature tissues studied in other species.
Our results show that cloning of A. sellowiana by stem cuttings requires physiologically juvenile or rejuvenated material and that different genotypes may require different treatments to produce competent cuttings.
Data availability
The datasets generated and analyzed during the current study are available in the GenBank repository at https://www.ncbi.nlm.nih.gov/genbank/, reference number MZ130946-MZ13095.
References
Abarca D, Pizarro A, Hernández I et al (2014) The GRAS gene family in pine: transcript expression patterns associated with the maturation-related decline of competence to form adventitious roots. BMC Plant Biol. https://doi.org/10.1186/s12870-014-0354-8
Aumond ML, de Araujo AT, de Oliveira Junkes CF et al (2017) Events Associated with Early Age-Related Decline in Adventitious Rooting Competence of Eucalyptus globulus Labill. Front Plant Sci 8:1–10. https://doi.org/10.3389/fpls.2017.01734
Bellini C, Pacurar DI, Perrone I (2014) Adventitious roots and lateral roots: similarities and differences. Annu Rev Plant Biol 65:639–666. https://doi.org/10.1146/annurev-arplant-050213-035645
Benedini FJ, Brondani GE, de Almeida LV et al (2015) Vegetative rescue and cloning of Eucalyptus benthamii selected adult trees. New for. https://doi.org/10.1007/s11056-015-9472-x
Bisognin DA, Lencina KH, Kielse P et al (2017) Cuttings of post fire epicormic shoots of Ilex paraguariensis and Cabralea canjerana adult plants. Ciência Rural. https://doi.org/10.1590/0103-8478cr20151287
Bisognin DA, Lencina KH, da Luz LV et al (2018) Adventitious rooting competence and rescue of adult mate plants by cuttings. Rev Árvore 42:1–10. https://doi.org/10.1590/1806-90882018000300012
Blakesley D (1994) Auxin Metabolism and Adventitious Root Initiation. In: Davis TD, Haissig BE (eds) Biology of Adventitious Root Formation. Plenum Press, New York, pp 143–154
Blilou I, Xu J, Wildwater M et al (2005) The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433:39–44. https://doi.org/10.1038/nature03184
Bontempo P, Mita L, Miceli M et al (2007) Feijoa sellowiana derived natural Flavone exerts anti-cancer action displaying HDAC inhibitory activities. Int J Biochem Cell Biol 39:1902–1914. https://doi.org/10.1016/j.biocel.2007.05.010
Davies PJ (2010) Plant Hormones: Biosynthesis, Signal, Transduction, Action!, Revised 3r. Springer, New York
de Almeida MR, Ruedell CM, Ricachenevsky FK et al (2010) Reference gene selection for quantitative reverse transcription-polymerase chain reaction normalization during in vitro adventitious rooting in Eucalyptus globulus Labill. BMC Mol Biol 11:73. https://doi.org/10.1186/1471-2199-11-73
de Almeida M, de Almeida CV, Graner EM et al (2012) Pre-procambial cells are niches for pluripotent and totipotent stem-like cells for organogenesis and somatic embryogenesis in the peach palm: A histological study. Plant Cell Rep 31:1495–1515. https://doi.org/10.1007/s00299-012-1264-6
de Almeida MR, de Bastiani D, Gaeta ML et al (2015) Comparative transcriptional analysis provides new insights into the molecular basis of adventitious rooting recalcitrance in Eucalyptus. Plant Sci 239:155–165. https://doi.org/10.1016/j.plantsci.2015.07.022
de Almeida MR, Schwambach J, Silveira V et al (2020) Proteomic profiles during adventitious rooting of Eucalyptus species relevant to the cellulose industry. New for 51:213–241. https://doi.org/10.1007/s11056-019-09728-7
De Lucas M, Brady SM (2013) Gene regulatory networks in the Arabidopsis root. Curr Opin Plant Biol 16:50–55. https://doi.org/10.1016/j.pbi.2012.10.007
Della Rovere F, Fattorini L, D’Angeli S et al (2013) Auxin and cytokinin control formation of the quiescent centre in the adventitious root apex of Arabidopsis. Ann Bot 112:1395–1407. https://doi.org/10.1093/aob/mct215
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19(1):11–15
Druege U, Franken P, Hajirezaei MR (2016) Plant Hormone Homeostasis, Signaling, and Function during Adventitious Root Formation in Cuttings. Front Plant Sci 7:381. https://doi.org/10.3389/fpls.2016.00381
Druege U, Hilo A, Pérez-Pérez JM et al (2019) Molecular and physiological control of adventitious rooting in cuttings: phytohormone action meets resource allocation. Ann Bot 123:929–949. https://doi.org/10.1093/aob/mcy234
Fett-Neto AG, De AM, Ruedell C (2011) Expression of auxin carrier genes during adventitious rooting in Eucalyptus globulus. BMC Proc 5:P64. https://doi.org/10.1186/1753-6561-5-S7-P64
Ford Y, Bonham EC, Cameron RWF et al (2001) Adventitious rooting: examining the role of auxin in an easy- and a difficult-to-root plant. Plant Growth Regul 36:149–159
Franzon RC, Antunes LEC, Raseira M (2004) Efeito do AIB e de diferentes tipos de estaca na propagação vegetativa da Goiabeira-serrana (Acca sellowiana Berg). Rev Bras Agrociência 10:515–518
Gonin B, Nguyen, et al (2019) What Makes Adventitious Roots? Plants 8:240. https://doi.org/10.3390/plants8070240
Guan L, Murphy AS, Peer WA et al (2015) Physiological and Molecular Regulation of Adventitious Root Formation. CRC Crit Rev Plant Sci 34:506–521. https://doi.org/10.1080/07352689.2015.1090831
Guerra MP, Cangahuala-Inocente GC, Vesco LLD, et al (2012) Micropropagation Systems of Feijoa (Acca sellowiana (O. Berg) Burret). In: Lambardi M, Ozudogru E, Jain S (eds) Protocols for Micropropagation of Selected Economically Important Horticultural Plants. Methods in Molecular Biology (Methods and Protocols). Humana Press, Totowa, NJ, pp 45–62
Hall TA (1999) BioEdit_ user friendly biological sequence alignment editor. Nucleic Acids Symp Ser 41:95–98
Han H, Zhang S, Sun X (2009) A review on the molecular mechanism of plants rooting modulated by auxin. African J Biotechnol 8:348–353. https://doi.org/10.5897/AJB2009.000-9062
Kelen M, Ozkan G (2003) Relationships between rooting ability and changes of endogenous IAA and ABA during the rooting of hardwood cuttings of some grapevine rootstocks. Eur J Hortic Sci 68:8–13
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2^(-ΔΔCT) method. Methods 25:402–408. https://doi.org/10.1006/meth.2001.1262
Lloyd G, McCown B (1980) Commercially feasible micropropagation of Mountain Laurel, Kalmia latifolia, by use of shoot-tip culture. In: Proc. Int. Plant Propagator’s Soc. http://www.pubhort.org/ipps/30/99.htm. Accessed 28 Dec 2015
Ludwig-Müller J (2000) Indole-3-butyric acid in plant growth and development. Plant Growth Regul 32:219–230
Mosbah H, Louati H, Boujbiha MA et al (2018) Phytochemical characterization, antioxidant, antimicrobial and pharmacological activities of Feijoa sellowiana leaves growing in Tunisia. Ind Crops Prod 112:521–531. https://doi.org/10.1016/j.indcrop.2017.12.051
Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15:473–497
Negishi N, Oishi M, Kawaoka A (2011) Chemical screening for promotion of adventitious root formation in Eucalyptus globulus. BMC Proc 5:P139. https://doi.org/10.1186/1753-6561-5-S7-P139
Negishi N, Nakahama K, Urata N et al (2014) Hormone level analysis on adventitious root formation in Eucalyptus globulus. New for 45:577–587. https://doi.org/10.1007/s11056-014-9420-1
Niella F, Rocha P, Thalmayr P, Duarte E (2018) Propagación vegetativa de dos frutales nativos de interés para productores de Misiones Argentina. In: III Congresso Paranaense de Agroecologia. Foz do Iguaçu, Brazil
Pacurar DI, Perrone I, Bellini C (2014) Auxin is a central player in the hormone cross-talks that control adventitious rooting. Physiol Plant 151:83–96. https://doi.org/10.1111/ppl.12171
Pierre-Jerome E, Moss BL, Nemhauser JL (2013) Tuning the auxin transcriptional response. J Exp Bot 64:2557–2563. https://doi.org/10.1093/jxb/ert100
Pizarro A, Díaz-Sala C (2019) Cellular dynamics during maturation-related decline of adventitious root formation in forest tree species. Physiol Plant 165:73–80. https://doi.org/10.1111/ppl.12768
Rasmussen A, Hosseini SA, Hajirezaei MR et al (2014) Adventitious rooting declines with the vegetative to reproductive switch and involves a changed auxin homeostasis. J Exp Bot 66:1437–1452. https://doi.org/10.1093/jxb/eru499
Ricci A, Rolli E, Brunoni F et al (2016) 1,3-Di(Benzo[D]Oxazol-5-Yl)Urea Acts As Either Adventitious Rooting Adjuvant or Xylogenesis Enhancer in Carob and Pine Microcuttings Depending on the Presence/Absence of Exogenous Indole-3-Butyric Acid. Plant Cell Tissue Organ Cult 126:411–427. https://doi.org/10.1007/s11240-016-1010-9
Ross S, Speroni G, Souza-Pérez M et al (2021) Stem - cutting anatomy and biochemical responses associated with competence for adventitious root differentiation in Acca sellowiana ( Myrtaceae ). Trees 35:1221–1232. https://doi.org/10.1007/s00468-021-02110-1
Ross S, Pechi E, Speroni G, et al (2017) In vitro rooting of Acca sellowiana microshoots. Acta Hortic https://doi.org/10.17660/ActaHortic.2017.1155.79
Ruedell CM, de Almeida MR, Fett-Neto AG (2015) Concerted transcription of auxin and carbohydrate homeostasis-related genes underlies improved adventitious rooting of microcuttings derived from far-red treated Eucalyptus globulus Labill mother plants. Plant Physiol Biochem 97:11–19. https://doi.org/10.1016/j.plaphy.2015.09.005
Schmittgen TD, Zakrajsek BA (2000) Effect of experimental treatment on housekeeping gene expression: Validation by real-time, quantitative RT-PCR. J Biochem Biophys Methods 46:69–81. https://doi.org/10.1016/S0165-022X(00)00129-9
Shu W, Zhou H, Jiang C et al (2019) The auxin receptor TIR1 homolog (PagFBL 1) regulates adventitious rooting through interactions with Aux/IAA28 in Populus. Plant Biotechnol J 17:338–349. https://doi.org/10.1111/pbi.12980
Stevens ME, Woeste KE, Pijut PM (2018) Localized gene expression changes during adventitious root formation in black walnut (Juglans nigra L.). Tree Physiol. https://doi.org/10.1093/treephys/tpx175
Stuepp CA, de Bitencourt J, Wendling I et al (2016) Indução de brotações epicórmicas por meio de anelamento e decepa em erva-mate. Cienc Florest 26:1009–1022
Teale WD, Paponov I, Palme K (2006) Auxin in action: signalling, transport and the control of plant growth and development. Nat Rev Mol Cell Biol 7:847–859. https://doi.org/10.1038/nrm2020
Tortora F, Notariale R, Maresca V et al (2019) Phenol-Rich Feijoa sellowiana (Pineapple Guava) Extracts Protect Human Red Blood Cells from Mercury-Induced Cellular Toxicity. Antioxidants 8:220. https://doi.org/10.3390/antiox8070220
Vandesompele J, De Preter K, Pattyn I et al (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3:34–41. https://doi.org/10.1186/gb-2002-3-7-research0034
Vanneste S, Friml J (2009) Auxin: a trigger for change in plant development. Cell 136:1005–1016. https://doi.org/10.1016/j.cell.2009.03.001
Vielba JM, Varas E, Rico S et al (2016) Auxin-mediated expression of a GH3 gene in relation to ontogenic state in Chestnut. Trees Struct Funct 30:2237–2252. https://doi.org/10.1007/s00468-016-1449-7
Vilasboa J, Da Costa CT, Fett-Neto AG (2018) Rooting of eucalypt cuttings as a problem-solving oriented model in plant biology. Prog Biophys Mol Biol. https://doi.org/10.1016/j.pbiomolbio.2018.12.007
Vuotto ML, Basile A, Moscatiello V et al (2000) Antimicrobial and antioxidant activities of Feijoa sellowiana fruit. Int J Antimicrob Agents 13:197–201. https://doi.org/10.1016/S0924-8579(99)00122-3
Wachsman G, Sparks EE, Benfey PN (2015) Genes and networks regulating root anatomy and architecture. New Phytol 208:26–38. https://doi.org/10.1111/nph.13469
Wendling I, Trueman SJ, Xavier A (2014a) Maturation and related aspects in clonal forestry-Part I: Concepts, regulation and consequences of phase change. New for 45:449–471. https://doi.org/10.1007/s11056-014-9421-0
Wendling I, Trueman SJ, Xavier A (2014b) Maturation and related aspects in clonal forestry-part II: Reinvigoration, rejuvenation and juvenility maintenance. New for 45:473–486. https://doi.org/10.1007/s11056-014-9415-y
Wendling I, Brooks PR, Trueman SJ (2015) Topophysis in Corymbia torelliana x C. citriodora seedlings: adventitious rooting capacity, stem anatomy and auxin and abscisic acid concentrations. New Forests 46:107–120. https://doi.org/10.1007/s11056-014-9451-7
Xuan L, Xu M, Chen C et al (2014) Identification and characterization of three PeSHRs and one PeSCR involved in adventitious root development of Populus. Plant Cell Tissue Organ Cult. https://doi.org/10.1007/s11240-014-0437-0
Acknowledgements
This research was funded by AGENCIA NACIONAL DE INVESTIGACIÓN E INNOVACIÓN; grant number FMV-2-2011-1-6320. Mother plants from the breeding program of Acca sellowiana used in this study were kindly provided by Ing. Agr. (M.Sc.) Danilo Cabrera (Instituto Nacional de Investigación Agropecuaria, Canelones, Uruguay) and Ing. Agr. Beatriz Vignale (Facultad de Agronomía, Salto, Uruguay).
Funding
The research leading to these results received funding from AGENCIA NACIONAL DE INVESTIGACIÓN E INNOVACIÓN under Grand Agreement FMV-2–2011-1–6320.
Author information
Authors and Affiliations
Contributions
Conceptualization: SR and PS; Methodology: SR, SR-D; JPS and GP; Formal analysis and investigation: SR; Writing—original draft preparation: SR; Writing—review and editing: SR, PS and OB; Funding acquisition: SR and PS; Project administration: SR; Supervision: PS.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare they have no financial interests. The authors have no competing interests to declare that are relevant to the content of this article.
Additional information
Communicated by V. P. Singh.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ross, S., Rodríguez-Decuadro, S., Pérez, G. et al. Validation and expression analysis of candidate genes for adventitious rooting, in micro-cuttings of Acca sellowiana (Myrtaceae). Acta Physiol Plant 46, 53 (2024). https://doi.org/10.1007/s11738-024-03682-4
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11738-024-03682-4