Abstract
The presence of expansins was investigated in various developmental and ripening stages of cherry fruits by SDS-PAGE and immunoblotting. An expansin gene and three fragments (242, 607 and 929 bp) of its promoter region were cloned. The genomic clone of the expansin gene contained three introns, two exons spanning a 1.6 and a 1.0 kb upstream region. Semi-quantitative PCR analysis showed that this gene was ripening specific. Chimeric promoter—GUS constructs were made and truncated forms of the expansin promoter were introduced into tomatoes by agroinjection and fruits were analyzed for GUS expression by histochemical GUS staining and enzyme activity assays. The 0.60 kb expansin promoter efficiently induced GUS expression in transgenic tomatoes, whereas constructs with the 0.25 kb promoter did not display significant GUS staining. The highest GUS activity was detected in tomatoes containing the 1.0 kb promoter construct. Both large base pair promoter constructs drove the expression of the GUS gene at an equal or higher rate than the tomato E8 promoter.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
One of the major events of fruit ripening is the structural modification of the cell wall. Among the numerous enzymes and proteins found in plant cell walls endo-1,4-β-glucanases, xyloglucan endotransglycosylases and polygalacturonases drew attention in recent years because of their elevated expression and activity occurring concomitantly with fruit softening (Fry et al. 1992; Fry 1989; Nishitani and Tominaga 1992). However, the exact role of these enzymes in the softening process is not clear. Studies on transgenic tomato were not able to show a significant correlation between cell wall hydrolase activity and fruit softening (Giovannoni et al. 1989; Brummell and Harpster 2001). Isolation and characterization of expansins and their genes (McQueen-Mason et al. 1992; Scherban et al. 1995) led to the recognition that expansin accumulation and action is a prerequisite to cell wall hydrolase activity.
Expansins are small, cell wall localized proteins with a molecular weight range of 25–30 kDa (McQueen-Mason et al. 1992; Cosgrove 2000). It is thought that expansins may be involved in cell wall modification either by disrupting the hydrogen bonds between cellulose microfibrils or making carbohydrate polymers more accessible to the cell wall localized hydrolytic enzymes (Cosgrove 1998). Rose et al. (1997) showed the expression of a fruit and ripening-specific expansin gene (LeExp1) in tomatoes. Antisense suppression of this gene resulted in firmer tomato fruits and thus extended shelf life (Brummell et al. 1999). In contrast, expansin-overexpressed transgenic fruits were less firm than controls and softening occurred before commencement of the ripening reactions. Elevated expansin mRNA accumulation has been also detected in non-climacteric fruits such as grape and strawberry (Harrison et al. 2001) and expansin genes were cloned and sequenced from cherries by Gao et al. (2003). Cherries have been traditionally classified as non-climacteric fruits and their ripening do not seem to be under the control of ethylene. Therefore, suppression of expansin accumulation may represent a novel strategy to extend their shelf life.
Cauliflower mosaic virus promoter (35S CaMV) is presently used as the promoter of choice. 35S CaMV is a constitutive promoter that expresses the transgene in every tissue all the time. Because of this, it is not considered to be a viable promoter for up or down regulation of organ and development-specific events.
The goal of this investigation was to determine the expression pattern of expansin proteins in cherries and isolate a fruit and ripening-specific promoter to design an efficient and intelligent plant transformation vector as a first step to create higher quality fruits.
Materials and methods
Nucleic acid extraction
Genomic DNA was extracted from unexpanded young cherry leaves using the method of Lodhi et al. (1994). RNA used in this study was obtained from plant tissues by phenol/chloroform extraction according to method described by Lopez-Gomez and Gomez-Lim (1992). Total RNA quality and quantity was determined by denaturing formaldehyde gel and using a Spectronic® Genesys™ spectrophotometer, respectively.
Cell wall protein extraction
The extraction method described by Fils-Lycaon et al. (1996) was essentially used to obtain cherry cell wall proteins. Complete™ protease inhibitor cocktail tablets (Roche) were added to the extraction buffer to inhibit protease activity during extraction. Half volume of 20% trichloroacetic acid (TCA) was used to precipitate the cell wall proteins. The pellet was washed three times with acetone and air-dried to remove residual acetone. Protein was dissolved in SDS-PAGE sample buffer and stored at −80°C until used.
SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotting
Analytical SDS-PAGE was performed in Bio-Rad (Bio-Rad, Hercules, CA, USA) minigels (Laemmli 1970) using general laboratory techniques (Sambrook et al. 1989). Expansin proteins were detected using polyclonal tomato expansin (LeExp1) antibody diluted 1:1,500 in TBS buffer. The proteins cross-reacting with the expansin antibody were visualized with anti-rabbit antibody conjugated to alkaline phosphatase (Sigma). Nitroblue tetrazolium (NBT) and 5-bromo-4-chloro-3-indolyl phosphate (BCIP) were dissolved in alkaline phosphatase developing buffer and used as substrate for color development.
Inverse PCR and isolation of the expansin promoter
Expansin gene promoter isolation was accomplished using inverse PCR (IPCR) technique as described by Ochman et al. (1988). Fifteen microgram genomic cherry DNA was digested with TaqI restriction endonuclease; 4 µl of digested and purified genomic DNA (40 ng) was self-ligated to use as template for inverse PCR. PCR primers oriented in the reverse direction of the usual orientation were designed to amplify the flanking region (Table 1).
The PCR reaction profile was 94°C for 3 min for the initial denaturation step and 35 cycles of 94°C for 30 s, 62°C for 30 s, 70°C for 4 min and followed by one cycle of 70°C for 15 min. The PCR products were analyzed on 1.2% agarose gel. The correct size bands were excised from the gel, purified and subsequently sequenced. New primer pairs were designed targeting the putative promoter region and the coding sequence of PcExp2 gene to amplify the upstream region. PCR products were sequenced to confirm nucleotide sequence.
Tomato transformation
Constructs harboring different size of PcExp2 promoter fragments were electroporated into Agrobacterium LBA4404 strain and grown on media containing selective antibiotics. The agroinjection procedure described by Orzaez et al. (2006) was closely followed for the transient gene expression analysis.
Tomatoes stably transformed according to Boyce Thompson Institute (BTI) transformation protocol which is a modified version of the method described by Fillatti et al. (1987). Cotyledons were cut into smaller pieces (0.5 cm) and co-cultivated with the Agrobacterium LBA4404 culture harboring the constructs. Successful transformants were selected and rooted on media containing 100 mg l−1 kanamycin and then transferred to the greenhouse. The presence of the construct in the tomato plants was verified by PCR.
Reverse transcriptase PCR (semi-quantitative RT-PCR) and real time PCR (qRT-PCR)
Total RNA was extracted from different cherry tissues as described above (Lopez-Gomez and Gomez-Lim 1992) and treated with RQ1 RNase-Free DNase kit (Promega) to remove contaminating genomic DNA. SuperScript™ III One-Step RT-PCR system with Platinum® Taq DNA Polymerase kit was used for semi-quantitative RT-PCR assay according to manufacturer’s instructions (Invitrogen, Life technologies). Reactions were incubated at 55°C for 30 min for the cDNA synthesis and denatured at 94°C for 2 min prior to PCR amplification. Primers used for RT-PCR analysis are shown in Table 1. The PCR reaction profile was 34 cycles at 94°C for 15 s, 56°C for 30 s, 68°C for 1 min and followed by one cycle of 68 °C for 5 min. PCR products were separated on 1.2% agarose gel, visualized and the gels were photographed.
qRT-PCR analysis was carried out using iScript™ One-step RT-PCR Kit with SYBR® Green (Bio-Rad, Hercules, CA) in the iCycler instrument (Bio-Rad). Real Time PCR cycling conditions were 50°C for 10 min (cDNA synthesis), 95°C for 5 min (reverse transcriptase inactivation), 40 cycles of 95°C for 10 s and 56°C for 30 s (primer annealing and extension). Melting point analyses were performed after each run to confirm specificity of the primers and to detect primer–dimers and secondary products. The accumulation of uidA was normalized against the expression of the 26S-rRNA (Moon and Callahan 2004), primer sequences are shown in Table 1. The qRT-PCR data were analyzed using a comparative ∆∆C T method (Livak and Schmittgen 2001). The ∆C T was calculated according to the difference between C T of normalizer and target genes and expressed as relative mRNA levels.
GUS reporter gene constructions
The binary vector, pBI101.1, containing a GUS coding sequence, was used to make reporter gene constructs (Jefferson et al. 1987). Three different sizes of expansin gene promoter fragments (242, 607 and 929 bp) with respect to transcription start site were obtained by PCR amplification using primers ProF1, ProF2, ProF3 together with ProR (Table 1). The E8 promoter was obtained from tomato genomic DNA using primers E8F and E8R (Table 1). HindIII and XbaI restriction enzyme sites were introduced to all the promoter fragments via site-directed mutagenesis and cloned into the pBI101 vector double digested with HindIII and XbaI enzymes. Construct fidelity was assured by PCR and sequencing.
GUS histochemical staining and enzyme activity assay
GUS histochemical staining was carried out as described by Jefferson (1987). Methanol (20%) was added to the buffer to inhibit background β-glucuronidase activity (Kosugi et al. 1990) and staining was carried out at 37°C. To measure GUS enzyme activity, tomato fruit slices were homogenized in GUS extraction buffer and enzyme activity was determined spectrophotometrically (Jefferson 1987); 50 µl of extract was added to 950 µl of assay buffer containing methanol and mixed thoroughly by vortexing. Reaction tubes were incubated overnight at 37°C. The reaction was stopped by the addition of 0.4 ml 2.5 M 2-amino-2-methyl propanediol. Another reaction mixture was prepared and stopped immediately after addition of the GUS extract. Absorbance was measured against a stopped reaction mixture at 415 nm.
Results
Fractionation of cherry cell wall protein extracts and detection of the expansin proteins during cherry fruit development
Six major protein bands were detected in the unripe fruits and four of them comprised more than 90% of the total soluble cherry cell wall proteins with the molecular sizes ranging from 18 to 85 kDa (Fig. 1a). In addition to these six bands, new protein bands started to appear in the beginning of the ripening stage of fruits indicating the increasing metabolic activity in the fruits during ripening. Protein fractions obtained from ripe cherry fruit cell wall extracts displayed more uniform distribution and there were at least ten major protein bands detected. There was significant accumulation of soluble cherry cell wall proteins during fruit development at the molecular size of 60 kDa and in the range of 25 and 30 kDa (Fig. 1a). When the gels were blotted and the blots probed with a polyclonal tomato anti-expansin antibody (Rose et al. 2000), a 29 kDa cherry cell wall protein band cross reacted with the anti-expansin antibody (Fig. 1b). The blots also showed that the expansin protein band intensity increased in cherry fruits during the development and ripening stages.
SDS-PAGE and Immunoblot analysis of cell wall protein extracts from cherry fruit cv. ‘Montmorency’. a SDS-PAGE gel of proteins extracted with 1.5 M NaCl and 0.2 M Na-acetate from the cell walls of cherry fruits from different stages of ripening. b Immunoblot analysis of the cell wall proteins using the polyclonal LeExp1 antibody. c The expansin proteins detected by LeExp1 antibody as shown. M r markers are indicated
Even though cherry fruit maturation can be divided into three distinct stages (Tukey 1936), the presence of expansin activity may be most critical in Stage I (rapid cell division and expansion) and Stage III (final swell). Stage II is thought of as the “retarded growth phase” and obviously there is no exceptional cell wall modification in this period. Western blot data (Fig. 1b, c) revealed that expansin proteins started to accumulate in the early stage of cherry fruit development which is characterized by rapid cell division and expansion (Stage I) and their expression significantly increased in parallel with development of ripening. The highest amount of expansin proteins were found in the fully ripe and softest cherry fruits.
Expression of the expansin genes during cherry maturation and ripening
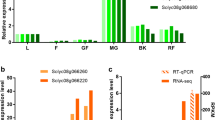
Cherry fruit development and ripening take 50–55 days which can be divided into three distinct stages and involve high level of cell wall metabolism (assembly–disassembly and degradation) (Tukey 1936). To evaluate the potential role of expansin genes in cherry growth and fruit development, mRNA accumulation of the cherry expansin genes has been investigated. Specific primers targeting the highly divergent 3′ untranslated region (UTR) were designed using cherry expansin sequence data published by Gao et al. (2003) and a semi-quantitative RT-PCR approach was employed to characterize the transcription of the specific expansin genes. Four expansin genes, including PcExp1, PcExp2, PcExp4 and PcExp5 were upregulated during different stages of cherry development (Fig. 2). Figure 2 shows the expression analysis of four cherry expansin genes on 1.0% (w/v) agarose gel stained with ethidium bromide. Each of the expansins found in cherries displayed different expression pattern and characteristic during fruit development and only two of them were expressed in the vegetative organs (Fig. 2).
Tissue-specific expression profile of expansin genes. Total RNA was extracted from leaves (L), buds (B), petals (Pt), petioles (Pl), and from green (G), pink (P), red (R) and fully ripe (FR) fruit. Specific primers targeting the divergent 3′ UTR region of the expansin genes were designed and used for semi-quantitative RT PCR analysis. 28S and 18S RNA were used for loading control. The PCR products were separated on 1.0% agarose gel and stained with ethidium bromide
The expression of the PcExp1 was very different than the other three expansin genes found in the cherries as shown in Fig. 2. mRNA accumulation of the PcExp1 was detected in leaves, petioles, petals and in fruits throughout growth and ripening, with the highest expression in the fully ripe fruits. PcExp1 transcription was detected first time at green fruit stage and increased gradually during maturation and ripening. The expression of this gene made the sharpest increase that paralleled the final swelling of the cherry fruit. PcExp1 mRNA was not detected in the buds. Its expression significantly increased after bud braking. The highest PcExp1 mRNA accumulation was detected in petals. There was no detectable PcExp2 mRNA accumulation in vegetative organs such as leaves, petioles, or buds. PcExp2 transcription displayed differential expression in the fruits: PcExp2 was upregulated at the pink fruit stage and high level of PcExp2 mRNA was detected at the later stages of ripening. In contrast to the PcExp1, PcExp2 mRNA was not present at the green fruit developmental stage. PcExp2 was the most abundant expansin gene in cherries displaying ripening specificity. PcExp4 transcription was also detected in fruits throughout fruit development and ripening. However, PcExp4 transcript accumulation peaked before the fruits have reached fully ripe stage and declined afterwards. There was no PcExp4 mRNA accumulation in the vegetative cherry organs such as leaves, petioles and buds. However, some PcExp4 transcripts were also detected in petals suggesting a specific role for this gene related to petal growth and organ abscission. PcExp5 gene transcription reached detectable levels only after the fruits turned red, and its expression was the highest in the fully ripe fruits. PcExp5 expression was only detected in red stage and ripe fruits and there was no detectable PcExp5 mRNA accumulation in other stages of fruits and vegetative organs. PcExp5 mRNA abundance was very low at the red fruit developmental stage and slightly increased in the final stages of fruit ripening.
Individual members of the cherry expansin family possesses distinct spatial and temporal expression pattern. This data may reflect their specific roles during plant growth and development. According to the expression data, PcExp1 and PcExp4 may be associated with cell expansion and thus plant growth; whereas, the cumulative accumulation of PcExp2 and PcExp5 mRNAs can be related to cell wall breakdown and softening. However, at this point, the role of PcExp1 and PcExp4 in fruit softening cannot be discounted.
Cloning of the ripening-specific expansin gene and its promoter fragments
Cherry expansin 2 gene (PcExp2) was identified as one of the expansin genes whose expression lighted up and upregulated during ripening in cherries. Substantial amount of PcExp2 mRNA accumulation was detected in cherries at later stages of fruit maturation. Gene-specific primers (PcExp2F and PcExp2R, Table 1) were designed to isolate the PcExp2 gene from cherry genome and amplified DNA fragments have been subsequently sequenced. Sequence data was used to obtain around 1,000 bp upstream region of the gene. To clone the promoter region of the fruit and ripening-specific PcExp2 gene, genomic DNA was isolated from young cherry leaves by the CTAB method (Lodhi et al. 1994) and digested by restriction endonucleases. Genomic DNA fragments were self-ligated using T4-DNA ligase (New England Biolabs) and upstream promoter region amplified using inverse PCR (IPCR) approach described by Ochman et al. (1988).
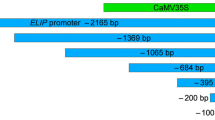
Eventually, a 2,596 bp sour cherry genomic DNA clone containing the expansin gene (PcExp2) and its putative promoter was obtained. This fragment contains 929 bp of the expansin gene upstream region and a 1,667 bp long open reading frame interspersed with two introns. Sequence comparison of the cherry genomic clone with its cDNA sequence confirmed that the 2.6 kb fragment encoded two introns, three exons and a 929-bp promoter region (Fig. 4). The genomic clone of the PcExp2 gene contains a 74 bp 5′ UTR region, one small (156 bp) and one large (440 bp) introns, and a 248 bp 3′ UTR region.
The resulting putative promoter sequence was analyzed by Blast against a genome database (NCBI) containing all available plant DNA sequences. No significant similarity was found between the cherry expansin promoter and a number of promoters isolated from many other plant species, including tomato, cotton and Arabidopsis. Interestingly, the only similarity was between the promoter regions of Prunus cerasus expansin 2 (PcExp2) and Pyrus communis expansin 2 (PcoExp2) genes (data not shown).
It was reported that 30–40% of the promoter regions are conserved between the orthologous genes isolated from different genera and homology is higher if they are from the same the species . For instance, two peach 1-aminocyclopropane-1-carboxylic acid oxidase (ACO) promoters exhibited approximately 97% sequence similarity in their first 798-bp DNA sequence (Moon and Callahan 2004). There was only a 10–15% similarity between the promoter regions of cherry and pear expansin genes. However, particularly this region may be conserved due to its critical role for the temporal and spatial promotion of the expansin transcription.
This 929-nt sequence contains a TATA-box at −38 bp relative to the transcription start site (Fig. 3), which is considered as regular feature of eukaryotic promoters (Joshi 1987; Zhu et al. 1995) and consistent with the previously published reports. The upstream region exhibited consensus or highly conserved sequences of CCAAT-boxes, GATA-boxes, pyrimidine box, ethylene responsive element, gibberellin factors, anaerobiosis elements, transcriptional activators and cis-acting elements conferring tissue specificity.
A 350-bp region of the PcExp2 promoter is required for gene expression
The transcriptional activity of the portions of 5′ flanking region of the expansin promoter was investigated by GUS reporter gene analysis. Truncated series of the expansin promoter in the sizes of 242, 607, 929 bp and a 1.1 kb of core region of the tomato E8 promoter were amplified by PCR and ligated into the upstream region of the uidA (GUS-β-glucuronidase) gene in the pBI101.1 vector and named PcPro1, PcPro2, PcPro3, LeProE8. The primer pairs that had been used to amplify the expansin 5′ upstream region are shown in Table 1.
Transgenic tomato fruits agro-injected with constructs containing various size of truncated expansin gene promoters were subjected to histochemical GUS staining and enzyme activity assays. β-Glucuronidase activity was measured in the fruits at different developmental stages of ripening. The average level of β-glucuronidase activity in fruits harboring the constructs carrying expansin promoter fragments was determined spectrophotometrically and displayed in the graphs to show the large changes in GUS activity. According to the results obtained, the construct containing the smallest expansin promoter PcPro1 (242 bp) was not able to direct GUS expression (Fig. 4) in both ripe and unripe fruit.
GUS activity above the background level was detected with a PcPro2 promoter construct (607 bp) (Fig. 5a, b). Promoter strength and activity increased in parallel with the length of the 5′ flanking region of the expansin gene and reached the highest level in the PcPro3 promoter (Fig. 6a, b). This data suggested that cis-acting elements acting as positive regulator of expansin expression are most likely located in between the portions of the smallest and medium size promoters and in the distant fragment of the promoter.
Database searches revealed that several cis-acting-positive regulators of transcription were missing in the smallest expansin promoter, such as ethylene-responsive element, gibberellin-responsive factor, GATA-box, P-box, Myb-binding site. Therefore, an approximately 350-bp region between the smallest (242 bp) and medium size (607 bp) promoter established the first significant region conferring promoter activity—strength leading to the conclusion that an expansin promoter as long as around 600 bp is sufficient to drive the GUS expression. The transgenic tomatoes injected with the PcPro3 displayed the highest GUS activity in tomatoes from all ripening stages (Fig. 6a, b). These data indicated the presence of additional positive regulators of the expansin promoter located between −600 and −929 bp.
GUS enzyme activity measurement was carried out in parallel with histochemical analysis to quantitate the β-glucuronidase activity. In addition to the small green stage fruits, there was no measurable β-glucuronidase activity in the tomatoes injected with the shortest promoter fragment (Fig. 4) and empty transformation vectors in any of the developmental changes. There was significant activity in tomatoes injected with the medium size (607 bp) promoter. These data also confirmed that an expansin promoter around 0.6 kb size is capable of driving gene expression. Measurable β-glucuronidase activity increased in parallel with fruit ripening and reached the highest level in the fully ripe fruits.
Agro-injected tomato fruits were subjected to the GUS histochemical staining. Typically, GUS staining in fruits from various ripening stages initially was observed in vascular bundles. At the green fruit stage, GUS staining was confined to the vascular bundles and no significant staining was detected in the other parts of the fruit. However, staining was extended to the tomato pericarp and placental tissue in the tomatoes injected with the construct containing the 929 bp length promoter fragment (Fig. 6b).
Tomatoes injected with the longest promoter displayed more intense GUS staining especially in the pericarp, placental tissue and vascular bundles (Fig. 6b). The GUS staining intensity steadily increased in parallel with ripening in agro-injected tomatoes.
A ripening associated E8 promoter has been characterized in tomatoes (Deikman et al. 1992) and was widely used to drive transgene expression in heterologous plant transformation studies. A 1.1 kb fragment of the “late-expressing” E8 promoter has been used for vaccine expression in ripe fruits (He et al. 2008). The organ and developmental stage specificity of the E8 genes have been shown in tomatoes by GUS reporter gene assays (Montgomery et al. 1993). Therefore, a 1.1 kb core region of E8 promoter was fused to the upstream region of uidA gene open reading frame and introduced to the tomato fruits to investigate the expression pattern of another ripening and fruit-specific promoter. Fruits agro-injected with the constructs harboring the fragment of the E8 promoter were subjected to histochemical and enzyme activity assays to compare the promoter strength-activity of the E8 promoter and the cherry expansin promoters (Fig. 7a, b). E8 promoter injected unripe tomatoes did not display significant GUS staining (Fig. 7b). However, GUS staining increased to detectable levels after the fruits turned breaker stage and reached the highest level in the fully ripe fruit. The sharpest increase in GUS staining observed in the fully ripe fruits indicated the fruit and ripening specificity of the E8 promoter. GUS staining started to increase at the breaker stage fruits in tomatoes agro-injected with the PcExp-GUS constructs and reached the highest level in the ripe fruits. Both cherry expansin 5′ fragments (PcPro2 and PcPro3) displayed stronger promoter activity in comparison to the E8 promoter (Fig. 5a, 6a, 7a). This result makes both expansin promoters useful for transformation studies targeting the improvement of the cherry fruit quality.
Temporal and spatial activity of cherry expansin gene promoter
Tomato plants stably transformed with the construct containing the longest insert [929 bp (Fig. 8)], which was shown to be the strongest among the other promoter fragment. GUS staining and expression were assayed using histochemical staining and qRT-PCR (real time PCR) approach both in fruit and non-fruit tissues to investigate the temporal and spatial activity of the promoter. Despite the fact that there was no GUS staining in vegetative and floral tissues including stems, leaves and flowers, GUS histochemical staining significantly increased in tomato fruits during the development of ripening and reached the highest level in fully mature transgenic fruits (Fig. 9). Real-time PCR was used to determine accumulation of GUS transgene in leaf, flower, stems and fruit tissues and the data normalized against the amount of 26S-rRNA (Moon and Callahan 2004). In parallel with the results obtained from staining experiments there was no detectable transgene accumulation above background values in vegetative and floral organs however the uidA gene transcription significantly increased in the fruit tissues (Fig. 10). This result also confirmed the controlling ability of cherry expansin promoter on transgene expression tissue and developmental-specific manner. Findings obtained from the stable tomato transformation experiments indicated that this promoter is a valuable molecular tool to manipulate plant based food quality traits.
Histochemical staining of non-fruit and fruit tissues obtained from transgenic tomato plants transformed with PcPro3 construct. No GUS staining detected in stems, flowers and leaves. The longest promoter (PcPro3, 929 bp) displayed fruit and ripening specificity. The GUS staining was the highest in fully ripe transgenic tomatoes
Discussion
Tomato has been successfully used as a model system to unravel the underlying mechanisms of climacteric fleshy fruit ripening and thus much of the research has been focused on this fruit. Ethylene coordinates the ripening associated biochemical reactions in climacteric fruits. The majority of the tomato fruit research has been focused on ethylene biosynthesis and signal transduction pathways. There is limited amount of research on non-climacteric fruit ripening and the role of ethylene is not clear. However, there are several reports in the literature implicating ethylene in non-climacteric fruit ripening, such as pigmentation in citrus (Alonso et al. 1995) and gene expression in strawberries (Tesniere et al. 2004). One recent study reported that ethylene may be involved in the regulation of VvADH2 gene expression in grapes (Vitis vinifera, one of the non-climacteric type fruits) (Tesniere et al. 2004). The ethylene-responsive element (ERE) and the anaerobic-responsive element (ARE) are located in the promoter region of the VvADH2 gene. ERE and ARE elements are known as positive regulators of gene expression (Walker et al. 1987; Chen and Chase 1993). ARE elements were identified in the upstream regions of AtADH1 (Arabidopsis thaliana) and LeADH2 (Solanum lycopersicum) genes and their expression was upregulated under low-oxygen stress (Daraselia et al. 1996; Feldbrugge et al. 1994). The role of ERE cis-acting element on the induction of ADH expression was not established yet (Tesniere et al. 2004). However, exogenous ethylene application substantially induced VvADH2 expression. Ethylene data and presence of ERE and ARE elements suggested a role for ethylene in the non-climacteric fruit ripening.
An ERE and an ARE element were also found in the cherry expansin gene promoter region. Cherries have been classified as non-climacteric and there is no detectable ethylene burst at the onset of the ripening and ethylene synthesis rate remains very low during the time course of ripening (around 1–2 pmol kg h, data not shown). Ethylene production rate and expansin mRNA accumulation does not correlate well in cherries. Young green cherry fruits produce substantially higher amount of ethylene in comparison to the ripe fruits, while expansin mRNAs are most abundant in fully ripe fruits. Therefore, it does not seem feasible that expansin expression is positively regulated by ethylene in cherries.
Auxin and ethylene regulates the expression of AtExp7 (Arabidopsis thaliana expansin gene) at the root hair formation sites. Reporter gene analyses have been carried out to characterize the promoter region of the AtExp7. Database searches revealed that auxin (ARE) and ethylene-responsive elements (ERE) are located in the distal region of the AtExp7 promoter (Cho and Cosgrove 2002). However, removal of these elements did not alter the responsiveness of the promoter to the auxin and ethylene treatments. Deletion analysis indicated that DOF zinc finger-binding domains (AAAG) and a MYB-like (MYBSt1) protein-binding site (GGATA) located at the proximal region of the promoter is critical for inducibility of the AtExp7 expression. A second MYBSt1 core motif located at the distal region of the promoter was also found to be associated with AtExp7 expression (promoter activity). In addition to ethylene and gibberellin-responsive elements, a series of DOF zinc finger domain and MYB-like protein-binding motifs are located on the PcExp2 promoter in a similar pattern found in that of AtExp7 promoter. This region is composed of 371 nucleotides. An internal deletion covering this fragment significantly reduced expansin promoter activity. Interestingly, this fragment corresponds to the only conserved region in between promoter regions of PcoExp2 and PcExp2 genes.
Cis-acting elements found in the expansin promoter
Environmental and hormonal signals regulate differential expression of expansin genes and these are present in the upstream regions of expansin genes (Lee et al. 2001). It was found that expansin expression can be regulated by auxin (Catalá et al. 2000; Hutchison et al. 1999), gibberellin (Cho and Kende 1997; Lee and Kende 2001), cytokinin (Malley and Lynn 2000; Wrobel and Yoder 2001) and ethylene (Kim et al. 2000; Vriezen et al. 2000).
Transcription factors recognize and bind to short DNA sequences present at the upstream region of genes (Fickett and Hatzigeorgiou 1997). DNA-binding factors specifically bind to certain sequences and are significant elements of growth and differentiation. PLACE, PlantCare and MatInspector databases were used to identify the cis-acting and hormone responsive elements present at the upstream region of the PcExp2 gene. The elements were found to be critical for the activity and strength of the promoters isolated from the other species.
The cis-acting elements that confer tissue and developmental stage specificity to the expression of the plant genes, such as Myb protein-binding site (GGATA-box) (Baranowski et al. 1994), CCAAT-box (Rieping and Schoffl 1992; Hatamochi et al. 1998; Wenkel et al. 2006; Haralampidis et al. 2002), Telomere motif (Telo-box) (Liboz et al. 1991; Curie et al. 1993; Regad et al. 1995; Tremousayque et al. 1999), AC elements (Leyva et al. 1992; Kapoor and Sugiura 1999), GT-1 cis element (Park et al. 2004), and GAGA-box (Sangwan and O’Brian 2002) found to be conserved in the PcExp2 upstream region. These elements were shown to be associated with temporal and spatial regulation of the fruit and organ-specific gene expression. Further deletion analyses are necessary to elucidate the role of these elements in the activity and strength of the PcExp2 promoter.
Cloning of fruit and ripening-specific promoters
Early ripening related promoters were characterized from E4 and E8 genes (Deikman and Fischer 1988; Cordes et al. 1989) and followed by isolation of polygalacturonase (Bird et al. 1988) and the 2A11 (Van Haaren and Houck 1993) promoters in tomatoes. Transient transformation approaches were employed for the investigation of activity of homologous and heterologous promoters in strawberries. Similar to results we have obtained in this study, 1.2 kb of the promoter region of strawberry GalUR gene was shown to be containing the required cis-acting elements conferring specific transcription in fruits (Agius et al. 2004). Agius et al. (2004) found that a 397 bp core region of GalUR gene promoter is necessary for its function. Truncated promoter fragments with an equal size of PcExp2 promoter (around 1.0 kb) were shown to be involved in specific expression of Cucumisin gene in fruit tissues (Yamagata et al. 2002). One of the few promoter studies in tree fruits were carried out in peaches. Moon and Callahan (2004) showed that 403 bp fragment of 1-aminocyclopropane-1-carboxylic acid oxidase (ACO) promoter is related to its tissue-specific expression.
Cherry is a true (ovary-derived) fruit and represent a valuable model to study the underlying mechanisms of softening in non-climacteric fruits (Gao et al. 2003) including the isolation of tissue-ripening-specific promoters for the genetic engineering of a variety of fruits. The expansin promoter which is cloned in this study is a potentially valuable molecular tool to manipulate fruit quality factors in both climacteric and non-climacteric fruits including commercially important tomato. The availability of the tissue- and development-specific promoter can provide an advantage to develop precise molecular tools to interfere with gene expression related to fruit softening. Promoter research holds the promise to precise manipulation of gene expression and thus biological pathways without affecting other metabolic events. Expansin promoter can be utilized to genetically engineer cherries and other crops to produce transgenic fruits that have superior quality and extended shelf life.
References
Agius F, Amaya I, Botella MA, Victoriano V (2004) Functional analysis of homologous and heterologous promoters in strawberry fruits using transient expression. J Exp Bot 56:37–46
Alonso JM, Chamaro J, Granell A (1995) Evidence for the involvement of ethylene in the expression of specific RNAs during maturation of the orange, a non-climacteric fruit. Plant Mol Biol 29:385–390
Baranowski N, Frohberg C, Prat S, Willmitzer L (1994) A novel DNA binding protein with homology to Myb oncoproteins containing only one repeat can function as a transcriptional activator. EMBO J 13:5383–5392
Bird CR, Smith CJS, Ray JA, Moureau P, Bevan MW, Bird AS, Hughes S, Morris PC, Grierson D, Schuch W (1988) The tomato polygalacturonase gene and ripening-specific expression in transgenic plants. Plant Mol Biol 11:651–662
Brummell DA, Harpster MH (2001) Cell wall metabolism in fruit softening and quality and its manipulation in transgenic plants. Plant Mol Biol 47:311–340
Brummell DA, Harpster MH, Civello PM, Palys JM, Bennett AB, Dunsmuir P (1999) Modification of expansin protein abundance in tomato fruit alters softening and cell wall polymer metabolism during ripening. Plant Cell 11:2203–2216
Catalá C, Rose JKC, Bennett AB (2000) Auxin-regulated genes encoding cell wall-modifying proteins are expressed during early tomato fruit growth. Plant Physiol 122:527–534
Chen ARS, Chase T Jr (1993) Alcohol dehydrogenase 2 and pyruvate decarboxylase induction in ripening and hypoxic tomato fruit. Plant Physiol Biochem 31:875–885
Cho H-T, Cosgrove DJ (2002) Regulation of root hair initiation and expansin gene expression in Arabidopsis. Plant Cell 14:3237–3253
Cho HT, Kende H (1997) Expression of expansin genes is correlated with growth in deepwater rice. Plant Cell 9:1661–1671
Cordes S, Deikman J, Margossian LJ, Fischer RL (1989) Interaction of a developmentally regulated DNA binding factor with sites flanking two different fruit-ripening genes from tomato. Plant Cell 1:1025–1034
Cosgrove DJ (1998) Cell wall loosening by expansins. Plant Physiol 118:333–339
Cosgrove DJ (2000) Loosening of plant cell walls by expansins. Nature 407:321–326
Curie C, Axelos M, Bardet C, Atanassova R, Chaubet N, Lescure B (1993) Modular organization and developmental activity of an Arabidopsis thaliana eEF1A gene promoter. Mol Gen Genet 238:428–436
Daraselia ND, Tarchevskaya S, Narita JO (1996) The promoter for tomato 3-hydroxy-3-methylglutaryl coenzyme A reductase gene has 2 unusual regulatory elements that direct high-level expression. Plant Physiol 112:727–733
Deikman J, Fischer RL (1988) Interaction of a DNA binding factor with the 5’ flanking region of an ethylene-responsive fruit ripening gene from tomato. EMBO J 7:3315–3320
Deikman J, Kline R, Fischer RL (1992) Organization of ripening and ethylene regulatory regions in fruit-specific promoter from tomato. Plant Physiol 100:2013–2017
Feldbrugge M, Sprenger M, Dinkelbach M, Yazaki K, Harter K, Weisshaar B (1994) Functional analysis of a light-responsive plant bZIP transcriptional regulator. Plant Cell 6:1607–1621
Fickett JW, Hatzigeorgiou EG (1997) Eukaryotic promoter recognition. Genome Res 7:861–878
Fillatti JJ, Kiser J, Rose R, Comai L (1987) Efficient transfer of a glyphosate tolerance gene into tomato using a binary Agrobacterium tumefaciens vector. Biotechnology 5:726–730
Fils-Lycaon BR, Wiersma PA, Eastwell KC, Sautiere P (1996) A cherry protein and its gene abundantly expressed in ripening fruit, have been identified as thaumatin-like. Plant Physiol 111:269–273
Fry SC (1989) Cellulases, hemicelluloses and auxin-stimulated growth: a possible relationship. Physiol Plant 75:532–536
Fry SC, Smith RC, Renwick KF, Martin DJ, Hodge SK, Matthews KJ (1992) Xyloglucan endotransglycosylase, a new wall-loosening enzyme activity from plants. Biochem J 282:821–828
Gao Z, Maurousset L, Lemoine R, Yoo S-D, van Nocker S, Loescher W (2003) Cloning, expression, and characterization of sorbitol transporters from developing sour cherry fruit and leaf sink tissues. Plant Physiol 131:1566–1575
Giovannoni JJ, DellaPenna D, Bennett AB, Fischer RL (1989) Expression of a chimeric polygalacturonase gene in transgenic rin (ripening inhibitor) tomato fruit results in polyuronide degradation but not fruit softening. Plant Cell 1:53–63
Haralampidis K, Milioni D, Rigas S, Hatzopoulos P (2002) Combinatorial interaction of cis elements specifies the expression of the Arabidopsis AtHsp90-1 gene. Plant Physiol 129:1138–1149
Harrison PE, McQueen-Mason SJ, Manning K (2001) Expression of six expansin genes in relation to extension activity in developing strawberry fruit. J Exp Bot 52:1437–1446
Hatamochi A, Golumbek PT, Vanschaftingen E, Decrombrugghe B (1998) A CCAAT DNA-binding factor consisting of 2 different components that are both required for DNA-binding. J Biol Chem 263:5940–5947
He Z-M, Jiang X-L, Qi Y, Luo D-Q (2008) Assessment of the utility of the tomato fruit-specific E8 promoter for driving vaccine antigen expression. Genetica 133:207–214
Hutchison KW, Singer PB, McInnis S, Diaz-Sala C, Greenwood MS (1999) Expansins are conserved in conifers and expressed in hypocotyls in response to exogenous auxin. Plant Physiol 120:827–831
Jefferson RA (1987) Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol Biol Rep 5:387–405
Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: glucuronidase as a sensitive and versatile gene marker in higher plants. EMBO J 6:3901–3907
Joshi CP (1987) An inspection of the domain between putative TATA box and translation start site in 79 plant genes. Nucleic Acid Res 15:6644–6652
Kapoor S, Sugiura M (1999) Identification of two essential sequence elements in the nonconsensus type II PatpB-290 plastid promoter by using plastid transcription extracts from cultured tobacco BY-2 Cells. Plant Cell 11:1799–1810
Kim JH, Cho HT, Kende H (2000) α-Expansins in the semiaquatic ferns Marsilea quadrifolia and Regnellidium diphyllum: evolutionary aspects and physiological role in rachis elongation. Planta 212:85–92
Kosugi S, Ohashi Y, Nakajima K, Arai Y (1990) An improved assay for β-glucuronidase in transformed cells: methanol almost completely suppresses a putative endogenous β-glucuronidase activity. Plant Sci 70:133–140
Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685
Lee Y, Kende H (2001) Expression of β-expansins is correlated with elongation of internodes in deepwater rice. Plant Physiol 2001(127):645–654
Lee Y, Choi D, Kende H (2001) Expansins ever-expanding numbers and functions. Curr Opin Plant Biol 4:527–532
Leyva A, Liang X, Pintor-Toro JA, Dixon RA, Lamb CJ (1992) cis-Element combinations determine phenylalanine ammonia-lyase gene tissue-specific expression patterns. Plant Cell 4:263–271
Liboz T, Bardet C, Le Van Thai A, Axelos M, Lescure B (1991) The four members of the gene family encoding the Arabidopsis thaliana translation elongation factor eEF1A ara actively transcribed. Plant Mol Biol 14:107–110
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real time quantitative PCR and the 2−∆∆CT method. Methods 25:402–408
Lodhi MA, Guang-Ning Ye, Weeden NF, Reisch BI (1994) A simple and efficient method for DNA extraction from grapevine cultivars, Vitis species and Ampelopsis. Plant Mol Biol Rep 12:6–13
Lopez-Gomez R, Gomez-Lim MA (1992) A method for extracting intact RNA from fruits rich in polysaccharides using ripe mango mesocarp. Hortiscience 27:440–442
Malley RC, Lynn DG (2000) Expansin message regulation in parasitic angiosperms: marking time in development. Plant Cell 12:1455–1465
McQueen-Mason SJ, Durachko DM, Cosgrove DJ (1992) Two endogenous proteins that induce cell wall extension in plants. Plant Cell 4:1425–1433
Montgomery J, Pollard V, Deikman J, Fischer RL (1993) Positive and negative regulatory regions control the spatial distribution of polygalacturonase transcription in tomato fruit pericarp. Plant Cell 5:1049–1062
Moon H, Callahan AM (2004) Developmental regulation of peach ACC oxidase promoter–GUS fusions in transgenic tomato fruits. J Exp Bot 55:1519–1528
Nishitani K, Tominaga R (1992) Endo-xyloglucan transferase, a novel class of glycosyltransferase that catalyzes transfer of a segment of xyloglucan molecule to another xyloglucan molecule. J Biol Chem 267:21058–21064
Ochman H, Gerber AS, Hartl DL (1988) Genetic applications of an inverse polymerase chain reaction. Genetics 120:621–623
Orzaez D, Mirabel S, Wieland WH, Crannell A (2006) Agroinjection in tomato fruits. A tool for rapid functional analysis of transgenes directly in fruits. Plant Physiol 140:3–11
Park HC, Kim ML, Kang YH, Jeon JM, Yoo JH, Kim MC, Park CY, Jeong JC, Moon BC, Lee JH, Yoon HW, Lee SH, Chung WS, Lim CO, Lee SY, Hong JC, Cho MJ (2004) Pathogen- and NaCl-induced expression of the SCaM-4 promoter is mediated in part by a GT-1 box that interacts with a GT-1-like transcription factor. Plant Physiol 135:2150–2161
Regad F, Herve ÂC, Marinx O, Bergounioux C, Tremousaygue D, Lescure B (1995) The tef1 box, a ubiquitous cis-acting element involved in the activation of plant genes that are highly expressed in cycling cells. Mol Gen Genet 248:703–711
Rieping M, Schoffl F (1992) Synergistic effect of upstream sequences, and HSE sequences for enhanced expression of chimaeric heat shock genes in transgenic tobacco. Mol Gen Genet 231:226–232
Rose JKC, Lee HH, Bennett AB (1997) Expression of a divergent expansin gene is fruit-specific and ripening regulated. Proc Natl Acad Sci USA 94:5955–5960
Rose JKC, Cosgrove DJ, Albersheim P, Darvill AG, Bennett AB (2000) Detection of expansin proteins and activity during tomato fruit ontogeny. Plant Physiol 123:1583–1592
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning. A laboratory manual, second edition. Cold Spring Harbor Laboratory Press
Sangwan I, O’Brian MR (2002) Identification of a soybean protein that interacts with GAGA elements dinucleotide repeat DNA. Plant Physiol 129:1788–1794
Scherban TY, Shi J, Durachko DM, Guiltina MJ, McQueen-Mason SJ, Shieh M, Cosgrove DJ (1995) Molecular cloning and sequence analysis of expansins, a highly conserved multigene family of proteins that mediate cell wall extension in plants. Proc Natl Acad Sci USA 92:9245–9249
Tesniere C, Pradal M, El-Kereamy A, Torregrosa L, Chatelet P (2004) Involvement of ethylene signaling in a non-climacteric fruit: new elements regarding the regulation of ADH expression in grapevine. J Exp Bot 55:2235–2240
Tremousayque D, Manevski A, Bardet C, Lescure N, Lescure B (1999) Plant interstitial telomere motifs participate in the control of gene expression in root meristems. Plant J 20:553–561
Tukey HB (1936) Development of cherry and peach fruits as affected by destruction of the embryo. Botanical Gazette 98:1–24
Van Haaren MJJ, Houck CM (1993) A functional map of the fruit-specific promoter of the tomato 2A11 gene. Plant Mol Biol 21:625–640
Vriezen WH, De Graaf B, Mariani C, Voesenek LACJ (2000) Submergence induces expansin gene expression in flooding-tolerant Rumex palustris and not in flooding-intolerant R. acetosa. Planta 210:956–963
Walker JC, Howard EA, Dennis ES, Peacock WJ (1987) DNA sequences required for anaerobic expression of the maize alcohol dehydrogenase 1 gene. Proc Natl Acad Sci USA 84:6624–6628
Wenkel S, Turck F, Singer K, Gissot L, Le Gourrierec J, Samach A, Couplans G (2006) CONSTANS and the CCAAT box binding complex share a functionally important domain and interact to regulate flowering of Arabidopsis. Plant Cell 18:2971–2984
Wrobel RL, Yoder JI (2001) Differential RNA expression of α-expansin gene family members in the parasitic angiosperm Triphysaria versicolor (Scrophulariaceae). Gene 266:85–93
Yamagata H, Yonesu K, Hirata A, Aizono A (2002) TGTCACA motif is a novel cis-regulatory enhancer element involved in fruit specific expression of the Cucumisin gene. J Biol Chem 273:11582–11590
Zhu Q, Dabi T, Lamb C (1995) TATA box and initiator functions in the accurate transcription of a plant minimal promoter in vitro. Plant Cell 1:1681–1689
Acknowledgments
The authors would like to thank Dr. Jim Giovannoni for providing transformation vectors and tomato seeds, Dr. Susan Brown for cherry plant materials, Dr. Jocelyn Rose for tomato expansin antibody, Drs. Minou Hemmat and Desen Zheng for technical assistance, Jay Freer for assistance in the field. MK thanks Harran University for a scholarship, the Arthur Boller Endowment and Cornell University Department of Food Science and Technology for partial financial support.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by S. Abe.
Rights and permissions
About this article
Cite this article
Karaaslan, M., Hrazdina, G. Characterization of an expansin gene and its ripening-specific promoter fragments from sour cherry (Prunus cerasus L.) cultivars. Acta Physiol Plant 32, 1073–1084 (2010). https://doi.org/10.1007/s11738-010-0499-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11738-010-0499-5