Abstract
Applications of large language models (LLMs) in the healthcare field have shown promising results in processing and summarizing multidisciplinary information. This study evaluated the ability of three publicly available LLMs (GPT-3.5, GPT-4, and Google Gemini—then called Bard) to answer 60 multiple-choice questions (29 sourced from public databases, 31 newly formulated by experienced breast radiologists) about different aspects of breast cancer care: treatment and prognosis, diagnostic and interventional techniques, imaging interpretation, and pathology. Overall, the rate of correct answers significantly differed among LLMs (p = 0.010): the best performance was achieved by GPT-4 (95%, 57/60) followed by GPT-3.5 (90%, 54/60) and Google Gemini (80%, 48/60). Across all LLMs, no significant differences were observed in the rates of correct replies to questions sourced from public databases and newly formulated ones (p ≥ 0.593). These results highlight the potential benefits of LLMs in breast cancer care, which will need to be further refined through in-context training.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Large language models (LLMs) are artificial intelligence tools able to process, summarize, and generate text, specifically trained on vast datasets comprising books, articles, websites, and other written material [1]. These models employ advanced deep neural network architectures: in particular, most of recently developed LLMs utilize the transformer architecture, which enables unsupervised learning from unlabeled datasets, leading to improved performance through more efficient text processing [2]. During inference, LLMs leverage their internalized knowledge to predict the probability distribution of the next word in a sequence. The self-attention mechanism within transformers allows LLMs to consider the importance of different words in a given context [3]. The quick improvements of these technologies have resulted in LLMs output being virtually indistinguishable from human replies to the same queries [4]. Promising results, for example, have come from LLMs applications in the healthcare field, on tasks ranging from responding to patients’ questions to the extraction of clinical information from medical reports [5]. Of note, LLMs could prove especially beneficial in areas where the amount of information needed to appropriately manage the different stages of a diagnostic and therapeutic pathway is seeing constant growth, such as several oncological topics—e.g., breast cancer, lung cancer, head and neck cancer [6,7,8]—where multidisciplinary approaches have long been established [9]. As LLMs applications continue to expand [10], their answers to questions dealing with these multidisciplinary scenarios can represent a benchmark to understand their potentials and pitfalls. Thus, focusing on breast cancer (i.e., one of the aforementioned multidisciplinary settings) the objective of this study is to assess the ability of three different LLMs (GPT-3.5, GPT-4, and Google Gemini—previously called Bard) to correctly answer questions—either drawn from public datasets or specifically generated for this study—involving breast cancer diagnosis (imaging interpretation and diagnostic interventions) and treatment (in the oncological, surgical, and radiation oncology domains).
Materials and methods
For the purposes of this study, three LLM-based chatbots (GPT-3.5, GPT-4, Google Gemini) were prompted to answer 60 questions divided into four groups of 15 questions each: breast cancer treatment and prognosis (Group I), breast cancer diagnostic and interventional techniques (Group II), breast cancer imaging interpretation (Group III), and breast cancer pathology (Group IV).
Of all 60 questions (detailed in the Supplementary Material), 29 were selected from publicly available repositories of questions developed by the following four sources: i) the training sample database of the European Diploma in Breast Imaging (European Society of Radiology); ii) the training samples from the 2020, 2021, 2022 Diagnostic Radiology In-Training Exam of the American College of Radiology; iii) the practice test database of the RadiologyKey website (queried for breast cancer); iv) the online database of Medscape (queried for breast cancer). The following criteria were used for the selection on all four sources: (i) questions not containing any reference to images or other multimedia file; (ii) questions with the multiple choice or true/false formats; (iii) questions with only one correct answer among those proposed. According to these criteria, we included 15 questions from the European Diploma in Breast Imaging training database, 9 from the Diagnostic Radiology In-Training Exam, 3 from the RadiologyKey website, 2 questions from Medscape.
To achieve the prespecified number of questions in each group, 31 other questions were formulated explicitly for this study by two board-certified breast radiologists (with 13 and 15 years of experience, respectively) and revised by a third board-certified breast radiologist with 17 years of experience, according to the following criteria: (i) no overlap with topics considered in the questions drawn from publicly available databases; (ii) subjects—related to the topics of the four groups—identified as clinically relevant by international guidelines and accompanying literature.
GPT-3.5 and GPT-4 (OpenAI, San Francisco, USA) and Google Gemini (Google LLC, Mountain View, USA) were accessed on March 2, 2024, using an account specifically created for this study. To reduce the influence of previous responses, each question was submitted in a new chat window, and the answers were recorded for subsequent evaluation.
After verifying the replies of each LLM as correct (scoring 1 point) or incorrect (scoring 0 points), the scores of each LLM (expressed as counts and percentages) were compared descriptively and then with the Cochran’s Q and McNemar tests for paired data. For overall comparisons with the Cochran’s Q test, p values < 0.05 were considered statistically significant, whereas for pairwise comparisons with the McNemar test, the Bonferroni correction was used, with an ensuing p value threshold of 0.017. Statistical analyses were conducted using SPSS v.26.0 (IBM SPSS Inc.).
Results
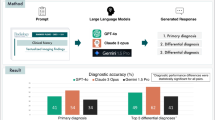
Overall, the rate of correct answers significantly differed among LLMs (Cochran’s Q statistic 9.294, p = 0.010). The rate of correct answers provided by GPT-4 (95%, 57/60) did not differ from that of GPT-3.5 (90%, 54/60, adjusted p = 1.000) but was significantly higher than that of Google Gemini (80%, 48/60, adjusted p = 0.009). Across all LLMs, no significant differences were observed in the rates of correct replies according to the questions’ origin, i.e., those selected from publicly available repositories and those formulated explicitly for this study (GPT-3.5: 89.7%, 26/29, vs. 90.3%, 28/31, p = 0.931; GPT-4, 96.6%, 28/29, vs. 93.6%, 29/31, p = 0.593; Gemini 79.3%, 23/29 vs. 77.4%, 24/31, p = 0.859). Table 1 shows four examples of questions and answers by the LLMs, while Fig. 1 details the rates of correct answers provided by the LLMs in the four groups of questions.
Rates of correct answers of the three different LLMs (GPT-3.4, ChatGPT-4, and Google Gemini) in the four groups of questions: breast cancer treatment and prognosis (Group I), breast cancer diagnostic and interventional techniques (Group II), breast cancer imaging interpretation (Group III), and breast cancer pathology (Group IV)
No significant difference in the rates of correct answers by the different LLMs was found among the 15 questions about breast cancer treatment and prognosis (Cochran’s Q statistic 3.500, p = 0.174), GPT-4 still having the highest rate (93.3%, 14/15), followed by GPT-3.5 (86.7%, 13/15) and Google Gemini (73.3%, 11/15).
The rate of correct answers among the 15 questions concerning interventional and diagnostic procedures differed significantly among LLMs (Cochran’s Q statistic 6.333, p = 0.042): the 100% rate of correct answers achieved by GPT-4 was significantly higher than that of Google Gemini (66.7%, 10/15, adjusted p = 0.037), while no other significant difference (adjusted p values ≥ 0.401) was observed between these rates and that of GPT-3.5 (86.7%, 13/15).
GPT-3.5 and GPT-4 had the same rate of correct answers (93.3%, 14/15) among the 15 questions related to image interpretation: Google Gemini displayed a lower rate (80.0%, 12/15), without any significant difference (Cochran’s Q statistic 2.000, p = 0.368).
Finally, all three LLMs had the same rate of correct answers (93.3%, 14/15, p = 1.000) for the 15 questions regarding breast cancer pathology.
Discussion
LLMs can reply quickly with suitable responses to user queries across various domains, providing immediate and contextually appropriate answers. This makes LLMs effective for applications requiring real-time interaction: for example, in the healthcare field, they could be employed to answer questions from patients or to extract clinical data from medical records [3, 4].
Findings from this study show that three major publicly available LLMs correctly reply to questions about different aspects of breast cancer care, achieving a rate of correct answers beyond 80%. Overall, there was a statistically significant difference in the rate of correct answers among LLMs (p = 0.010), the best performance being achieved by GPT-4 (95%, 57/60). The different rates of correct answers among the four groups—with the lowest rates in Group I (breast cancer treatment and prognosis) and Group II (breast cancer diagnostic and interventional techniques)—may be partially explained by the influence of clinical experience: this aspect is very difficult to incorporate into the training data of LLMs, as these models are primarily trained on text-based datasets [1], which may lack the nuanced knowledge coming from hands-on clinical practice.
The results of this study suggest that LLMs have the potential to be ultimately integrated into the breast cancer care pathway, at first focusing on tasks like providing evidence-based recommendations and streamlining the diagnostic and treatment planning processes, particularly when clinicians face uncertainties or multiple decision-making options. Additionally, LLMs could then serve as educational tools for medical professionals.
Our findings—obtained on a mixed set of questions drawn from public databases and specifically formulated for this study—are in line with results from other studies that included only questions from public datasets or newly generated ones. For example, as in our study, Brin et al. [11] showed how GPT-4 had the highest rate of correct answers on United States Medical Licensing Examination questions; likewise, in a study by Holmes et al. [12], GPT-4 had the highest rate of correct answers when confronted with newly-generated questions about radiation oncology physics, where it outperformed all other LLMs and medical physicists.
This study is one of the first exploring the potential roles of LLMs in breast cancer care [13,14,15], as discussed by Sorin et al. [6] in a recent review identifying three macro-domains of LLMs application: as decision-support systems in the multidisciplinary tumor board, as question-answering tools for patients and physicians, and as tools to extract information from imaging and pathology reports. Although the clinical impact of LLMs has been evaluated—at least preliminarily—in these studies, there is still a knowledge gap regarding patient perceptions and the economic aspects of implementing these tools in healthcare settings. These aspects are also reflected in the main limitations of this study, such as its restriction to 60 questions—none of which had an open answer format—that were all related to a single oncological field, the exclusive use of three publicly-available LLMs without any specific in-context training, and the uneven distribution of pre-existing and new questions among the different groups.
In conclusion, three publicly available LLMs achieved high—albeit significantly different—rates of correct answers to questions regarding breast cancer care, ranging from 80% (Google Gemini) to 95% (GPT-4). Further applications of LLMs in this field must take into account performance augmentation through in-context training and the generalizability of these results over a larger number of questions.
Abbreviations
- LLM:
-
Large language model
References
Singhal K, Azizi S, Tu T et al (2023) Large language models encode clinical knowledge. Nature 620:172–180. https://doi.org/10.1038/s41586-023-06291-2
Moor M, Banerjee O, Abad ZSH et al (2023) Foundation models for generalist medical artificial intelligence. Nature 616:259–265. https://doi.org/10.1038/s41586-023-05881-4
Nerella S, Bandyopadhyay S, Zhang J et al (2024) Transformers and large language models in healthcare: a review. Artif Intell Med 154:102900. https://doi.org/10.1016/j.artmed.2024.102900
Clusmann J, Kolbinger FR, Muti HS et al (2023) The future landscape of large language models in medicine. Commun Med 3:141. https://doi.org/10.1038/s43856-023-00370-1
Thirunavukarasu AJ, Ting DSJ, Elangovan K, Gutierrez L, Tan TF, Ting DSW (2023) Large language models in medicine. Nat Med 29:1930–1940. https://doi.org/10.1038/s41591-023-02448-8
Sorin V, Glicksberg BS, Artsi Y et al (2024) Utilizing large language models in breast cancer management: systematic review. J Cancer Res Clin Oncol 150:140. https://doi.org/10.1007/s00432-024-05678-6
Rahsepar AA, Tavakoli N, Kim GHJ, Hassani C, Abtin F, Bedayat A (2023) How AI responds to common lung cancer questions: ChatGPT versus Google Bard. Radiology 307:e230922. https://doi.org/10.1148/radiol.230922
Kuşcu O, Pamuk AE, SütaySüslü N, Hosal S (2023) Is ChatGPT accurate and reliable in answering questions regarding head and neck cancer? Front Oncol 13:1256459. https://doi.org/10.3389/fonc.2023.1256459
Shao J, Rodrigues M, Corter AL, Baxter NN (2019) Multidisciplinary care of breast cancer patients: a scoping review of multidisciplinary styles, processes, and outcomes. Curr Oncol 26:385–397. https://doi.org/10.3747/co.26.4713
Omiye JA, Gui H, Rezaei SJ, Zou J, Daneshjou R (2024) Large Language models in medicine: the potentials and pitfalls. Ann Intern Med 177:210–220. https://doi.org/10.7326/M23-2772
Brin D, Sorin V, Vaid A et al (2023) Comparing ChatGPT and GPT-4 performance in USMLE soft skill assessments. Sci Rep 13:16492. https://doi.org/10.1038/s41598-023-43436-9
Holmes J, Liu Z, Zhang L et al (2023) Evaluating large language models on a highly-specialized topic, radiation oncology physics. Front Oncol 13:1219326. https://doi.org/10.3389/fonc.2023.1219326
Griewing S, Knitza J, Boekhoff J et al (2024) Evolution of publicly available large language models for complex decision-making in breast cancer care. Arch Gynecol Obstet 310:537–550. https://doi.org/10.1007/s00404-024-07565-4
Cozzi A, Pinker K, Hidber A et al (2024) BI-RADS category assignments by GPT-3.5, GPT-4, and Google Bard: a multilanguage study. Radiology 311:e232133. https://doi.org/10.1148/radiol.232133
Wu Q, Wu Q, Li H et al (2024) Evaluating large language models for automated reporting and data systems categorization: cross-sectional study. JMIR Med Informatics 12:e55799. https://doi.org/10.2196/55799
Funding
None.
Author information
Authors and Affiliations
Contributions
Conception and design of the study: G. I., A. C.; Manuscript drafting: G. I., A. C., G. D. P, C. D. B., E. D., M. Cè, M. Cellina; Critical revision of the manuscript: A. C., C. D., G. S.; Final approval of the version to be published: All authors.
Corresponding author
Ethics declarations
Confict of interest
The authors have no relevant financial or non-financial interests to disclose.
Ethics approval
This study did not involve any human or animal subject and did not require any specific ethical approval.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Irmici, G., Cozzi, A., Della Pepa, G. et al. How do large language models answer breast cancer quiz questions? A comparative study of GPT-3.5, GPT-4 and Google Gemini. Radiol med (2024). https://doi.org/10.1007/s11547-024-01872-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11547-024-01872-1