Abstract
This article presents an unsupervised method for movement onset detection from electroencephalography (EEG) signals recorded during self-paced real hand movement. A Gaussian Mixture Model (GMM) is used to model the movement and idle-related EEG data. The GMM built along with appropriate classification and post processing methods are used to detect movement onsets using self-paced EEG signals recorded from five subjects, achieving True–False rate difference between 63 and 98%. The results show significant performance enhancement using the proposed unsupervised method, both in the sample-by-sample classification accuracy and the event-by-event performance, in comparison with the state-of-the-art supervised methods. The effectiveness of the proposed method suggests its potential application in self-paced Brain-Computer Interfaces (BCI).
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Brain–Computer Interface (BCI) is a new approach to communication between the human brain and the machine, which translates brain signals into commands for communication and control (i.e. computer cursor control, wheelchair control, robotic control, etc.) [1, 2]. A BCI system can be synchronous where the system timing is controlled by the machine [3] or self-paced (asynchronous) [4] where the system timing is controlled by the user. The user of a BCI system can perform several well-studied mental tasks to communicate and control. The machine must be able to recognize these tasks from brain signals accordingly within a suitable time window for control.
Motor imagery tasks are commonly used in BCI environment [5], due to their good separability and the understanding of their neurological mechanisms. Previous research on event-related desynchronization/synchronization (ERD/ERS) showed that during real movements relevant EEG activity can be found in both contralateral and ipsilateral hemispheres, but in the case of imagined movements only contralateral hemisphere gets activated [6]. This justifies the use of real movements to test new methods, because the experiments are easier to conduct and the labelling is much more accurate in the self-paced configuration.
One of the challenges in self-paced BCI is onset detection of a mental task, which is about detecting when the user shifts from the idle/non-control state (when he/she is not executing any of the predefined mental tasks) to execute a mental task. This is important for the following reasons:
-
Identifying the idle state: The identification of the idle state is very important for control applications. In this case, the false positive rate must be as low as possible, to increase the reliability of the system especially when safety is an issue.
-
on/off switch: For a full self-paced BCI system the user must be able to turn the system on and off when needed. A robust onset detection system can be used as an on/off switch for a self-paced BCI system.
In [7], the difference between both applications is discussed.
1.1 Motivation of using unsupervised methods
In motor imagery based self-paced BCIs true labels are hard to obtain online. As what the BCI system needs to know is the onset from baseline to movement, the recognition of the transition phase between the two states/classes is very important. The labels in these transition periods are hardly correct because the change happens gradually through preparation, execution and after-execution periods. When using real movement instead of imagery one, the use of electromyography (EMG) can give a very clear idea of the true labels, but it is not actually accurate enough during the transition phase.
Unsupervised learning provides a possible answer to handle mislabelled and lowly separable data. In theory, unsupervised methods are more able to handle uncertainty in data labelling and to separate the data according to their own structure without the need of labels to build the model. This particular property of unsupervised learning methods is very important to handle the onset detection problem as it would be more robust and natural in modelling the transition between the baseline and movement classes. This will eventually lead to higher recognition rate and consequently a faster and more reliable onset detection.
Classifying EEG data is usually challenging due to the overlapping between the data of the different classes/mental tasks. This is even a harder problem when working on onset detection as what is looked at by an onset detection algorithm is the overlapped area between the two classes. Unsupervised learning can cope well with this issue as the data modelling is independent from the labels and rather based on the data itself. Statistical/probabilistic unsupervised modelling is especially able to handle these overlapped areas by using prior knowledge and soft definition of boundaries between the classes.
In this article, we introduce for the first time a fully unsupervised system for EEG onset detection in self-paced BCIs. The system is based on feature extraction/selection methods using power spectral density (PSD) and Davis–Bouldin Index (DBI) and an unsupervised classification method based on a well-known statistical model, Gaussian Mixture Model (GMM). The experimental results show the effectiveness of the proposed method for EEG onset detection. Further study is required to study the potential of these unsupervised methods for onset detection in self-paced BCIs.
In [8], the authors argued that Fisher linear discriminant analysis (FLDA) classifier along with bandpower features was the best classifiers for the current state-of-art BCI, but this study ignored the wide spectrum of unsupervised machine learning methods that are potentially very useful for EEG classification and hence better BCI performance.
2 Methods
Gaussian Mixture Model is a well-known statistical model for data clustering. It has been repeatedly used for BCI data [9, 10]. In this section, we briefly introduce the model with two different training approaches: an unsupervised approach where the data is modelled using one GMM, and a supervised approach where a separate model is built for each class. The supervised approach is presented here for comparison purposes only.
2.1 Gaussian Mixture Model
In a GMM configuration, the data are assumed to be generated from a finite number of Gaussian distributions. Gaussian mixture model then is simply a linear superposition of Gaussian components. The data is modelled by a probability density function as follows:
where K is the number of Gaussian components, \({{\mathcal{N}}}(x|\mu_k,\Upsigma_k)\) is a Gaussian distribution with mean μ k and variance \(\Upsigma_k,\) π1,…, π K are the mixing coefficients. π k should satisfy the following conditions
and
In [11], a formulation of Gaussian mixtures in terms of discrete latent variables was introduced and deeply discussed. Here, we use this formulation and extend it for an unsupervised classification scheme.
Let us introduce a K-dimensional binary random variable z having a 1-of-K representation, in which a particular element z k is equal to 1 and all other elements are equal to 0. The values of z k therefore satisfy z k ∈ {0, 1} and \(\sum_kz_k=1\). There are K possible states for the vector z according to which element is nonzero. We will define the joint distribution p(x, z) in terms of a marginal distribution p(z) and a conditional distribution p(x|z).
The marginal distribution over z is specified in terms of the mixing coefficients π k , with p(z k = 1) = π k . \(p(z)=\prod_{k=1}^K \pi_k^{z_k}.\) The conditional distribution of x given a particular value of z is \(p(x|z_k=1)={{\mathcal{N}}}(x|\mu_k,\Upsigma_k)\) and \(p(x|z)=\prod_{k=1}^K {\mathcal{N}}(x|\mu_k,\Upsigma_k)^{z_k}.\)
The joint distribution is then given by p(z)p(x|z) and
The introduction of z helps in calculating the responsibility of each component in the mixture: γ(z k ) = p(z k = 1|x). We will use this presentation in the unsupervised training method.
Expectation–Maximisation (EM) method [11] is used to estimate the model parameters \(\{\pi,\mu,\Upsigma\},\) where π = {π1,…, π K }, μ = {μ1,…, μ K } and \({\Upsigma=}\{\Upsigma_1,\ldots,\Upsigma_K\}.\) And C-means was used to initialize the model parameter values.
One issue to be considered when building GMMs is how many components are optimal for a particular data set. In this article, we used cross-validation method to select the best number of components for each data set. More details on how the hyper-parameters of the model were selected are presented in Sect. 3.5.
In this article, we divide the data into training and testing data, according to the cross-validation scheme. We assume that the labels are available only for some offline/training data. In the supervised training method, we use these labels to separate the data into two training sets and then build a separate model for each set, so the labels are directly used in building the classification model. In the unsupervised scheme, one model is built for the whole training data set regardless of its class. The available labels are used here only to calculate the probability of a certain component to belong to one of the two classes. This use of labels offline in the unsupervised scheme is justified as it is only used in calculating the priors not in the model building itself. This approach has been used in unsupervised neural networks, e.g. SOM for speech recognition builds up clusters first without using labels and then decides which class each cluster corresponds to by using labels for classification purpose [12]. The use of labels in this way suits the self-paced BCI in particular as the labels are not reliable.
2.2 Unsupervised training
Here, we adopt the approach that we suggested in [13]. One GMM is built for all training/offline data and then the available labels are used to calculate the priors p(c = class i |z k ), i.e. the probability of the class to be class i when the data point is generated from component z k .
In order to calculate the probability of the data point x belonging to class i , p(c = class i |x) is calculated as follows:
where p(z k |x) (or p(z k = 1|x)) is the responsibility of component z k to generate the point x. Assuming p(c = class i |z k , x) is independent of x then
The probability p(c = class i |z k ) is calculated by the ratio:
where N i is the number of training samples that belong to class i , N ki is the size of the subset {x:p(z k = 1|x) = max(p(z j = 1|x)), j = 1 … K}, the training samples that belong to class i and were generated from z k . p(z k = 1|x) is calculated as follows:
In order to classify x in a two-class problem, p(c = class1|x) and p(c = class2|x) are calculated and then the output class is the one that equals
when the following condition holds:
where α is a rejection threshold. The introduction of the rejection criterion is important because of the overlapping between the two classes. In some cases, when the overlapping is so severe one would want the classifier to give an output only with high confidence. The rejection threshold α is the minimum confidence the classifier must have of one class to the other when classifying a new data point.
α takes a value in the range [0,1]. The selection of this value is done using cross-validation scheme, which means for different subjects and for different methods different values can be selected.
The problem with the rejection criterion is when the classifier is supposed to generate a continuous output. When classifying a point with a classification confidence less than α then usually one would not generate an output, this is not allowed when you have a continuous output. Here, we resolve this issue by simply replicating the output of the last “good” output (an output with a confidence level higher than α), the assumption here is that the classification outputs have temporal relation and the confidence level will drop in the area of moving from one class to another.
2.3 Supervised training
In this approach, the labelled offline data are separated into two sets, one for baseline and the other for movement. A separate GMM is built for each of these data sets. Each of these models is a probability density function (pdf) that shapes the probability distribution of each class. Now we can calculate the probability of any sample point to belong to one of these models.
In order to classify a new data point the log likelihood of each model given the data is calculated, log(p(x|M i )), where \(M_i=\{\pi_i,\mu_i,\Upsigma_i\}\) is the model built for class i . The model with the highest likelihood is the one that has most probably generated the data point.
In practice, we calculate the log likelihood value over a small window and average the results:
where N is the size of the window.
One may also use a Bayesian classifier, but for the data here the log likelihood comparison method gives better results.
The rejection method described in the previous section is used here as well. In this case, the comparison is not between the probabilities of the classes but rather between the normalized log likelihoods (which can be considered the probability/confidence of the classifier output):
3 Experiments
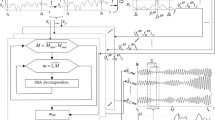
Figure 1 shows a block diagram of the system built for this study. Hereafter, we detail the motor tasks and the methods used for pre-processing and onset detection.
3.1 Subjects and motor tasks
Five right-handed subjects (three males and two females) were tested, who sit in an arm-chair with right arm resting on the arm rest. They were asked to perform the same real movements 40 times on their own pace in one session (The session lasted 534, 338, 400, 303 and 337 s for subjects 1, 2, 3, 4 and 5, respectively). The subjects were asked to leave at least 4 s between two movements. There was no prior training session, and the subjects had no experience of similar experiments. The designed movements were: extending right wrist, holding for about 1–2 s and relaxing.
3.2 EEG and EMG acquisition
EEG signals were recorded with 64 electrodes placed according to the International 10–20 Standard (ActiveTwo, Biosemi, The Netherlands). We used EMG to record muscle activities for establishing correct onset and offset time points of self-paced movements. This allows training data to be correctly labelled according to the real movement activities. EMG was recorded bipolar, from extensor carpi radialis muscle. Both EEG and EMG were sampled at 1,024 Hz, but were downsampled to 256 Hz for offline analysis. No artefact rejection or EOG correction was employed.
3.3 EEG data labelling
The continuous EEG data were labelled into two classes: baseline and movement. The movement data is further divided into three subclasses. Samples of 1.5 s prior to EMG onset were labelled as “preparation”, samples between an EMG onset and offset of one movement as “execution”, and samples of 1.5 s after an EMG offset as “after-execution”. The rest of the samples were labelled as “baseline”, because these samples should indicate no EEG activity with respect to the right wrist movement. The motivation of this division is the nature of the EEG signal itself during movement. Figure 2 shows an example of labelling 27.5 s of data.
3.4 Feature extraction and selection
In [14], we have proposed a novel onset detection method based on narrow band spectral analysis. The idea is to divide the mu, beta and lower gamma bands into even finer bands, so that feature selection method can be applied more efficiently. Here, we used the same proposed method for feature extraction and selection but we changed the classification scheme to use an unsupervised method. Following is a detailed description of the feature extraction and selection methods.
First, EEG data were filtered with common average reference method. To extract features for narrow band spectral analysis, the Thomson Multitaper Method was used to estimate the PSD of each EEG channel over a 1 s moving window with an overlap of 7/8 s (i.e. the moving window is shifted 1/8 of a second each time). The PSD over 8–45 Hz was sampled. Over 8–27 Hz it was sampled and averaged every 2 Hz, and over 28–45 Hz it was sampled and averaged every 3 Hz, resulting in a vector of 16 features. For 64 channels, there are 1,024 features in total.
Davis–Bouldin Index (DBI) [15] was used to select a subset of the best features. DBI measures the similarity between two clusters (classes) i and j by measuring the within-class scatters (S k ) and inter-class distances (M ij ) as follows:
where
where a i = [a 1i ,…, a ni ] is the centroid of cluster i, n is the dimensionality of the data, and T i is the number of data points in cluster i. In this study p = 2 and q = 2. DBI is then calculated as follows:
where
N features (with smallest DBI values) that maximise the separability of “preparation” against other subclasses were selected, and another N features that maximise the validity of “execution” against other classes were also selected. Therefore, 2N features were used for classification and evaluation. These features were selected as they are the most important for the onset/offset detection. In practice, N may be set to 50. This is still a high-dimensional problem. Directly using all the available features would affect severely the performance and will cause overfitting problems, which can be explained by the difficultly in building accurate probabilistic models in such high-dimensional space. To avoid these problems, we tested the methods using different number of features (in the range 2–100) and the number of features that gave the best result was used.
3.5 Data modelling and hyper-parameters
GMMs were built to model the data using both supervised and unsupervised methods discussed in Sect. 2. In order to build these models the number of components involved and the initial parameters are important factors in evaluating the model.
As discussed earlier, C-means was used to build the initial model and then EM was used to train the model. C-means itself was initialized using random vectors in the feature space. This initialization can affect severely the built model. For this reason, 10 models were built and a 10-fold cross validation test was done to select the model with the highest average classification accuracy.
The problem of using a suitable number of components to model the data is solved in a similar way. From our previous experience with BCI data, the number of components is usually in the range 4–20. Cross validation test was used to select the best number.
For the case of building two models (supervised training) a grid search like method was used, where the number of components for each model ranges from 2 to 20. For each number of components, 10 initialization values were tested and the one with the highest training accuracy was used.
Algorithm 1 shows the steps followed to select the hyper-parameters required for the unsupervised training method described in Sect. 2.2, where selectedFeat are the features selected via DBI, SAMPLECROSSVALID is a 10-fold cross validation method where the result is the average sample-by-sample classification accuracy, and TASKCROSSVALID is a 10-fold cross validation method where the result is the average TF (Sect. 3.8). In SAMPLECROSSVALID 10 different initializations of each model were tried to insure better modelling of the data.
3.6 Sample-by-sample classification
After the GMM models are built in a supervised or unsupervised method as described in Sect. 2, classification is carried out on a sample by sample basis on the test data. The probability of each class is calculated and then the rejection criterion discussed in Sect. 2.2 is applied. If the output of the classifier is rejected, the predicted class is assigned the value of the last known output with high confidence. This is justified for EEG data due to its temporal nature. In practice α value can be selected depending on the subject and the training method. For unsupervised training, its value ranges between 0.1 and 0.3 and for supervised training its value ranges between 0 and 0.3. For every subject and method the α value ( ∈ [0, 1]) was selected manually by checking the sample-by-sample cross validation result.
3.7 Onset detection
To find an EEG onset, a 1.375 s long (11 samples in feature space) moving decision window was applied on the classification results. In the moving window, if there were four predicted baseline samples, followed by four predicted movement samples, then the current position of this window was recognised to be an EEG onset. A Debounce window was applied after each recognition to reduce false negative effect. In performance evaluation this predicted EEG onset is considered correct, if there is a real movement onset that occurs either 2 s before or after this predicted point. Figure 3 shows an example of the onset detection method at work using the unsupervised training GMM, in one fold of the cross-validation test. The first line from top is the true labels, the second line is the predicted labels, the third line is |p(class1|x) − p(class2|x)| and the last line shows the detected onsets.
3.8 Evaluation
The evaluation was conducted by 10-fold cross-validation. Each fold had 4 trials for testing and 36 trials for training. The number of true-positive (TP) detections and the number of false-positive (FP) detections from all the folds were combined to produce true-false difference (TF), which is an event-by-event measurement. Given that E is the total number of movements or events, TF is defined by
This measure was defined for practical asynchronous BCI evaluation in [16].
3.9 Results
The results of onset detection using the proposed method are presented in Table 1, where Ave refers to the sample by sample 10-fold cross-validation result, Max refers to the maximum sample by sample accuracy achieved, and Dev refers to the average time deviation in seconds of the detected onset time. The table also contains the average and standard deviation of the TF results over all the subjects. Table 2 shows the different parameters used to achieve these results on different subjects, where K is the number of components in the built GMM model, Features is the number of features selected for this subject, α is the rejection threshold, and Debounce is the size of the Debounce window in seconds. Tables 3 and 4 contain the results and parameters for supervised trained GMM models. Here, K contains the number of components selected for each class, where the first value is the number of components for baseline and the second is the number of components for movement.
The results clearly show that the unsupervised approach outperforms the supervised one, with higher TP and lower FP within smaller Debounce window for subjects 3 and 4.
In general unsupervised GMM requires lower dimensions to give a better performance than the supervised one. Subjects 1–3 require as low as 5, 5 and 22 features for unsupervised GMM while the supervised method requires 16, 10 and 28, respectively. Subjects 4 and 5 require the same number of features for both methods.
The sample-by-sample accuracy is similar between the two methods, and it is even higher for the supervised method for subjects 1, 3 and 5, but that clearly does not mean a better event-by-event (TF) accuracy. This is because of the overlapping problem between the two classes and the fluctuations in the classifier output as a result. It is clear that the unsupervised model is able to handle this issue much better than the supervised one. More discussion about sample-by-sample and event-by-event evaluation can be found in [16].
4 Discussion and conclusion
Onset detection is an important step in building real-life self-paced BCI systems. The switch design named low-frequency asynchronous switch design (LF-ASD) [7, 17] is one of the first few self-paced BCI systems. This 2-state self-paced system was later extended to 3-state system in [18]. This system uses supervised methods for classification (LDA, KNN). In [19] a 4-class BCI system is built, which uses Common Spatial Patterns (CSP) for feature extraction and LDA, SVM and MDA classifiers for combination purposes. In [20] onset detection based on the changes in average PSD features is used to enhance classification in continuous EEG problem. In [21] separate SVM classifiers were built for ERD and ERS to detect onset in cued BCI data of foot movement, an onset is detected when ERS is found after ERD. Receiver operating characteristics (ROC) was used to balance TP and FP, dwell time and refractory(debounce) window were also used for post-processing. The authors in [22] proposed a new BCI method, where users perform either sustaining or stopping a motor task with time locking to a predefined time window, they however worked on synchronous protocol and used supervised methods for classification. EEG onset detection was also studied in the field of seizure detection, in [23] SVM was used for this purpose.
In a previous study [14] we showed the potential of the usage of narrow band spectral analysis for onset detection in self-paced ongoing EEG. Tsui [24] showed for this data the highest separability is achieved between the preparation and execution tasks. In this article, we used the same approach for feature extraction and selection but different classification and postprocessing schemes. The unsupervised method presented here shows promising results for onset detection. Compared to the supervised methods with GMM, the unsupervised GMM is significantly better (more than 7% enhancement on average).
Tables 5 and 6 show the results of using LDA and naive Bayesian classifiers (which are standard methods to use with BCI) on the same data sets. The results show clearly the superiority of the proposed unsupervised method in sample-by-sample classification rate and the TF measure. It is clear that supervised LDA and naive Bayesian are unable to handle the last two subjects’ data very well. Figure 4 provides a box plot view of the results for a comparison among the methods.
The GMM model is able to better present the shifts between the idle and movement states, as it is able to model the structure of the data when the labels are not reliable. It is clear that even when using a supervised training method for GMM it is still able to give better results than the ones with LDA or naive Bayesian, which means that the GMM model is able to capture better the statistics of EEG data.
Although the supervised and unsupervised GMMs give similar sample-by-sample results, the True–False difference using unsupervised trained models is significantly better than the supervised trained ones. This is because the unsupervised trained classifier is able to generate more stable results, while the supervised trained classifier suffers from fluctuations in the output. These fluctuations can be explained by the overlapping between the two classes. Unsupervised trained model is clearly able to model better these areas in the feature space.
Another advantage of using unsupervised training can be seen in the number of features required to get better results. The results suggest that unsupervised trained GMMs need lower dimensions than the supervised trained models. This is important for building online system as this will require less processing time online.
Using this approach the time deviation is larger than the one presented in [14], but it is still in an acceptable window (mostly within 1 s). This is acceptable in BCI applications as the response time is not expected to be very fast.
The study here focused on the onset detection, but the same approach can be taken for offset detection as well.
References
Birbaumer N, Hinterberger T, Kubler A, Neumann N (2003) The thought-translation device (ttd): neurobehavioral mechanisms and clinical outcome. IEEE Trans Neural Syst Rehabil Eng 11(2):120–123
Tsui CSL, Gan JQ, Roberts SJ (2009) A self-paced brain–computer interface for controlling a robot simulator: an online event labelling paradigm and an extended Kalman filter based algorithm for online training. Med Biol Eng Comput 47(3):257–265
Pfurtscheller G, Neuper C, Müller GR, Obermaier B, Krausz G, Schlögl A, Scherer R, Graimann B, Keinrath C, Skliris D, Wörtz M, Supp G, Schrank C (2003) Graz-BCI: state of the art and clinical applications. IEEE Trans Neural Syst Rehabil Eng 11(2):177–180
Borisoff JF, Mason SG, Birch GE (2006) Brain interface research for asynchronous control applications. IEEE Trans Neural Syst Rehabil Eng 14(2):160–164
Pfurtscheller G, Linortner P, Winkler R, Korisek G, Müller-Putz G(2009) Discrimination of motor imagery-induced EEG patterns in patients with complete spinal cord injury. Comput Intell Neurosci article ID 104180, 6 pp
Neuper C, Pfurtsheller G (1999) Motor imagery and ERD. In: Pfurtscheller G, Lopes da Silva FH (eds) Handbook of electroencephalography and clinical neurophysiology—event-related desynchronization. Elsevier, Amsterdam
Mason SG, Birch GE (2000) A brain-controlled switch for asynchronous control applications. IEEE Trans Biomed Eng 47(10):1297–1307
Boostani R, Graimann B, Moradi MH, Pfurtscheller G (2007) A comparison approach toward finding the best feature and classifier in cue-based BCI. Med Biol Eng Comput 45(4):403–415
Awwad Shiekh Hasan B, Gan JQ (2009) Unsupervised adaptive GMM for BCI. In: International IEEE EMBS conference on neural engineering, Antalya, Turkey
Buttfield A, Millan J del R (2006) Online classifier adaptation in brain–computer interfaces. Tech. Report, IDIAP 06-16
Bishop CM (2006) Pattern recognition and machine learning. Springer, Berlin
Haykin S (2008) Neural networks and learning machines. Prentice Hall, Upper Saddle River
Awwad Shiekh Hasan B, Gan JQ (2009) Sequential EM for unsupervised adaptive Gaussian mixture model based classifier. In: International conference on machine learning and data mining, Leipzig, Germany
Tsui CSL, Vuckovic A, Palaniappan R, Sepulveda F, Gan JQ (2006) Narrow band spectral analysis for movement onset detection in asynchronous BCI. In: The 3rd international workshop on brain–computer interfaces, Graz, Austria, pp 30–31
Bezdek JC, Pal NR (1998) Some new indexes of cluster validity. IEEE Trans Syst Man Cybern 28(3):301–315
Townsend G, Graimann B, Pfurtscheller G (2004) Continuous EEG classification during motor imagery—simulation of an asynchronous BCI. IEEE Trans Neural Syst Rehabil Eng 12(2):258–265
Birch GE, Mason SG, Borisoff JF (2003) Current trends in brain-computer interface research at the Neil Squire Foundation. IEEE Trans Neural Syst Rehabil Eng 11(2):123–126
Bashashati A, Ward RK, Birch GE (2007) Towards development of a 3-state self-paced brain-computer interface. Comput Intell Neurosci article ID 84386, 8 pp
Sadeghian EB, Moradi MH (2007) Continuous detection of motor imagery in a four-class asynchronous BCI. In: Proceedings of the 29th annual international conference of the IEEE EMBS, Lyon, France
Galán F, Oliva F, Guàrdia J (2007) Using mental tasks transitions detection to improve spontaneous mental activity classification. Med Biol Eng Comput 45(6):603–612
Solis-Escalante T, Müller-Putz G, Pfurtscheller G (2008) Overt foot movement detection in one single Laplacian EEG derivation. J Neurosci Methods 175:148–153
Bai O, Lin P, Vorbach S, Floeter MK, Hattori N, Hallett M (2008) A high performance sensorimotor beta rhythm-based brain–computer interface associated with human natural motor behavior. J Neural Eng 5(1):24–35
Shoeb A, Edwards H, Connolly J, Bourgeois B, Treves T, Guttag J (2004) Patient-specific seizure onset detection. In: Proceedings of the 26th annual international conference of the IEEE EMBS, San Francisco, USA
Tsui CSL (2009) Adaptive self-paced brain-actuated control of mobility devices. PhD Thesis, School of Computer Science and Electronic Engineering, University of Essex
Acknowledgements
The authors would like to thank C.S.L. Tsui for providing the data sets and the source code for LDA and naive Bayesian classifiers. This work is part of the project “Adaptive Asynchronous Brain Actuated Control” funded by UK EPSRC. The first author’s study is funded by Aga Khan Foundation (AKF).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Awwad Shiekh Hasan, B., Gan, J.Q. Unsupervised movement onset detection from EEG recorded during self-paced real hand movement. Med Biol Eng Comput 48, 245–253 (2010). https://doi.org/10.1007/s11517-009-0550-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11517-009-0550-0