Abstract
This article highlights the role of biophysical principles in biofilm growth and propagation in food environments, an area that is of increasing concern to food processors due to the high resistance of biofilms to conventional remediation methodologies. First, the general characteristics of biofilms are discussed including their structure and physiological characteristics. Transfer and propagation mechanisms consisting of attachment followed by growth and subsequent detachment are reviewed. General growth models that are currently used in laboratories focusing on biofilm research are compared and emerging characterization techniques are discussed. An overview over current practices and techniques to remediate biofilms in a variety of environments is given. Remediation techniques that are reviewed include application of sanitizers and detergents. Finally, future research needs are briefly summarized.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Surface growth of pathogens and spoilage organisms on food and food processing equipment leading to the formation of biofilms remains an extremely serious problem in the food industry. For example, outbreaks associated with Listeria monocytogenes, one of the five major organisms causing foodborne diseases, account for approximately 28% of the deaths resulting from foodborne illnesses [1]. These outbreaks have been linked to the inability to eradicate the organisms from contaminated processing equipment and environments. Problematic sites in food operations that have been identified include the surfaces of slicing, packaging, dicing, dosing, skinning, pumping and mixing equipment. The inability to inactivate pathogens is based on the fact that organisms have formed a highly structured community of cells, so-called biofilms, that have shown a remarkable ability to survive cleaning and disinfection procedures. Studies that compared planktonic with biofilm cultured pathogens reported greatly increased resistance against most growth-limiting factors including nutrient deficiency, low pH, low water activity, and presence of salts, antibiotics, and antimicrobials. In principle, pathogen contamination problems may also be the result of the pathogens growing on macroscopic accumulations of food residues that remain after inadequate cleaning, in which case expression of the biofilm phenotype may or may not be required. In this case, the focus of cleaning operations should be directed towards a proper removal of food residues. Nevertheless, removal of food residues is often more easily accomplished than removal of a firmly attached biofilm.
Because of this, it is becoming increasingly critical to gain a better understanding of the physicochemical, biochemical and genetic factors that govern the growth and maintenance of biofilms. Although the number of biofilm-related research projects in the biomedical field aimed at preventing the occurrence of infections on implanted medical devices and prosthetics is fairly large, the number of studies that focuses on biofilm growth in a food-related environment is much smaller. In the biomedical field, the resistance to antibiotics is amajor issue to the medical community. In a food processing environment, the inability of combinations of cleaners (surfactants) and disinfectants or antimicrobials to efficiently remove biofilms is one of the major problems that lead to contamination of food products and subsequent outbreaks of foodborne diseases. It is becoming increasingly clear that the foundation of this remarkable ability of biofilms to not only survive but flourish in hostile environments is due to fundamentally altered physical mechanisms such as cell–cell and cell–surface interactions and transport mechanisms. These changes in fundamental biophysical properties have enabled biofilms to attach to surfaces, resist superimposed flow profiles, and maintain a tightly controlled microenvironment that supports growth and propagation of biofilms. Although there are a respectable number of comprehensive reviews that cover biofilm characteristics and biofilm formation [2–6] as well as the role of biofilms in food processing [7–10], they generally place less emphasis on the role that biophysical principles play in biofilms, which is the purpose of this review.
This article is intended to familiarize the reader with the emerging field of biofilm research in food environments. Questions such as what is a biofilm, what is the structure of a biofilm, how do biofilms grow and propagate, how can biofilms be characterized, and what can be done to prevent biofilm formation in food environments are addressed. In particular, this article highlights the role that basic physical and chemical properties play in a complex biological system that consists of colonies of living cells that are attached to natural or artificial surfaces.
Biophysical characteristics of biofilms
Introduction
Researchers now recognize that in their natural environments, bacteria do not exist as isolated cells but grow and survive in organized communities known as biofilms [11]. Generally, biofilms have been characterized as matrix-enclosed microorganisms that adhere to a surface and/or to each other, producing a dynamic environment in which the component microbial cells appear to reach homeostasis, optimally organized to make use of all available nutrients [12–16]. Once they have colonized the surface, microorganisms form a monolayer or multilayer of cells at phase interfaces. Interfaces where biofilms may grow in food processing environments include solid/liquid, gas/liquid, or, in the case of solid foods, gas/solid interfaces [11,15]. Throughout natural ecosystems, biofilms can be found on almost any surface with a high enough level of moisture to support growth [17]. Biofilms are formed by almost every type of microorganism under suitable conditions, including spoilage microorganisms such as Pseudomonas and pathogens of great concern to the food industry including the genera of Bacillus, Vibrio, Listeria, Escherichia, and Salmonella [15, 18, 19].
Physiology of cells in biofilms
When growing in a biofilm, bacteria are known to have a different rate of growth, cellular morphology, and physiology than their planktonic counterparts and may exhibit varied physiological responses to nutrient conditions [17, 20–23]. Investigations of diffusional transport of gaseous and liquid components through the biofilm matrix have indicated that biofilm bacteria receive less oxygen and fewer nutrients than cells in suspension. Surprisingly, this allows the bacteria to survive and grow under a variety of different conditions, due to altered physiology that leads to increased resistance to toxic agents compared to their planktonic forms [16, 24, 25]. The fundamental alterations of diffusional mass transport processes and biophysical interactions with components present in the neighboring aqueous phase seem to allow commensal and mutual communities of organisms to survive low nutrient and decreased temperature conditions that are often found in food processing and storage environments. The ability to resist antimicrobial agents is of particular concern to both the medical and food processing communities, since once a biofilm has been established on a surface, it becomes exceedingly difficult to completely remediate the film [2, 26–34].
Cells growing in biofilm show differences in their metabolism and physiology when compared to planktonic cells. These cells may even express different phenotypes depending on their location within the biofilm. Werner et al. [35] showed that in a Pseudomonas aeruginosa biofilm, the cells located in the inner layers were in fact metabolically inactive due to the lack of oxygen. This might not be true for biofilms composed of facultative aerobic bacteria. In the case of Staphylococcus aureus, it has been shown that the production of acell–cell adhesin encoded by the ica gene is required in order to form a complex biofilm structure [36]. Cell-to-cell communication among Gram-negative bacteria seems to require N-acyl homoserine lactones, a pathway that is typically not followed in planktonic cultures [37]. Unusual interspecies metabolic interactions have also been observed in biofilms composed of different bacterial species [38].
Single species versus multispecies biofilms
The majority of biofilm research that is being discussed in this review is based upon single-species experiments performed in simplified laboratory systems. The two most studied biofilm systems use pure cultures of the clinical organisms P. aeruginosa or Streptococcus aureus as model Gram-negative and Gram-positive systems, respectively. In most foods and in food processing environments, the biofilms present will be significantly more complex, i.e., they may be composed of multiple species that form a community of microorganisms and that may or may not behave in a similar manner as pure laboratory systems. Similarly, studies of biofilms under laboratory conditions usually do not take into account that biofilms in food processing environments may contain proteins and fats derived from improperly cleaned or sanitized processing areas as integral part of their structure. Experimental evidence suggests that the formation of a multispecies biofilm is advantageous. For example, higher numbers of L. monocytogenes were counted in biofilms that were cocultured with Pseudomonas [39, 40]. In other cases, the adhesion of L. monocytogenes is limited in the presence of other bacteria [41, 42]. Chae and Scharft showed that the rate of biofilm formation for L. monocytogenes differs from their planktonic growth [43, 44]. Biofilms within the food processing environments are considered to be a major source of L. monocytogenes contamination, and quantifying transfer of L. monocytogenes pure culture will be useful in assessing the potential risk within food processing environments [45].
The structure of biofilms
Biofilms are composed of microbial cells that in the latter stages of the growth cycle are embedded in an exopolymer matrix [46]. However, it should be clearly stated that biofilms do not possess a uniform structure. For example, Wimpenny et al. recognized that biofilms exist that are porous and that are nonporous [47]. The structures that are formed depend on a large variety of intrinsic and extrinsic parameters such as species, temperature, flow conditions, pH, presence of salts, etc. In a short communication by Van Loosdrecht and coauthors, a strong argument for the development of heterogeneity of the porous structure in biofilms as a function of medium concentration was made [48]. The authors argued that similar to processes that occur in particle growth by crystallization or flocculation, the existence of diffusion gradients would inevitably lead to the development of a nonhomogeneous structure and that the process of structure development would be strongly influenced by the substrate concentration gradient at the biofilm‐liquid interface. Filamentous structures are consequently formed that protrude from the surface of the film into the liquid phase contributing to the complexity of the structure. These protuberances are an important part of the propagation mechanisms and have shown to have substantially different densities than the base biofilm. A more in-depth description of the development of biofilm structures can be found in the next section.
Biofilm formation and propagation
Attachment to surfaces
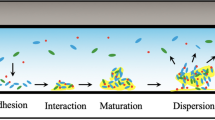
There are several steps in the formation of bacterial biofilms: (i) transport, (ii) initial adhesion, (iii) substrate attachment, and (iv) microcolony formation (cell–cell adhesion), leading to mature biofilms consisting of cells and a surrounding exopolymer matrix (Figure 1) [14, 46, 49]. The first step in biofilm formation consists of the transport of the organism to a solid surface. This can occur via motility of the organism, diffusion of the organism through the environment, or natural or forced convection in the system. Biofilm-forming bacteria may use all of these mechanisms at one time or another. It is well documented that flagella mutants often have lower biofilm production under static conditions, indicating that under these conditions flagella are involved in active cellular transport to surfaces. The role of the flagella in attachment was investigated by Vatanyoopaisarn [25]. Flagellin mutants of L. monocytogenes attached to stainless steel surfaces at levels that were 10-fold lower than wild-type cells with short incubation times (4 h). However, at longer incubation periods, the cell coverage by flagella negative mutants was similar to that of flagellated cells, suggesting that the flagella are important for initial attachment. The role of flow conditions on the attachment and growth of cells was investigated by various authors [40, 50, 51]. Contrary to expectations, greater deposition of bacteria under both laminar and turbulent flow conditions has been observed when compared to static conditions [52]. It has been speculated that turbulent flow may thrust bacterial cells onto the surface, thus enhancing probability of adhesion and biofilm formation [28].
Once the organisms approach the surface, physical interaction forces are thought to influence the initial adhesion of the organisms. Typical interactions that can take place include Van der Waals interactions (>50 nm from the surface), repulsive or attractive electrostatic interactions (2–10 nm from the surface), and hydrophobic interactions (0.5–2 nm from the surface) [53]. Van der Waals forces are due to dipole–dipole, induced dipole–dipole, and induced dipole–induced dipole interactions and are always attractive [54]. Electrostatic interactions arise because the cells, the developing exopolymer matrix, and the surface may carry a permanent positive or negative charge leading to the formation of a diffuse electrostatic double layer. Bacteria, as well as most natural solid surfaces, generally have an overall gross negative charge, but the origin of the overall charge is due to the combination of various charges from functional groups on the membrane constituent molecules, such as amino, carboxyl, phosphate, and, less commonly, sulfate groups and capsular macromolecules [55]. Ultimately, the magnitude of the electrostatic interactions is influenced by the nature of the environment, e.g., pH, ionic strength, valency of present counterions, and nature of the solvent [54].
The Derjaguin, Landau, Verwey, and Overbeek (DLVO) theory has been suggested as a first approximation to describe the interaction of bacteria with a solid surface as a function of the separation distance between the two systems. The DLVO theory assumes that the overall interaction is a sum of the attractive Van der Waals interaction and electrostatic repulsive interactions [56, 57]. Initially, Van der Waals attractions are thought to dominate the overall interaction leading to a reduction in the separation distance between bacteria and the substrate surface. However, as bacterial cells move closer to the substrate surface, repulsive electrostatic interactions may create an energy barrier that must be overcome before the two interacting bodies can come into close contact required for bacterial attachment [56]. Up to this point, the adhesion may be reversible, in particular if the minimum in the interaction potential is smaller than the thermal energy of the bacterial cells. The nature of the DLVO theory therefore does not explain the irreversible adhesion that actually occurs. Since hydrophobic interactions in water are much stronger than Van der Waals attraction at small separation distances [54], hydrophobic interactions between the cell surface and the solid substrate may be responsible for overcoming the repulsive electrostatic interactions. The origin of the hydrophobic interactions is thought to be enthalpically driven, that is, the exclusion of water molecules between the two surfaces and the direct interaction of the bacterial and substrate surface is believed to lower the overall free energy of the system. As a result, bacterial cells adhere irreversibly [58, 59]. This strict physicochemical approach, however, should not be overinterpreted. The bacterial surface is an extremely complex entity and contains a multitude of molecules that not only carry a variety of charges but are also more or less hydrophobic. In addition, the nature and composition of bacterial surfaces can vary greatly between different species. The fact that a single bacterial strain can adhere to a variety of surfaces with differing surface energies indicates that this simplified physicochemical interaction model is most likely not entirely correct. Strategies that attempted to prevent bacterial attachment by engineering the surface to be more or less hydrophobic have not led to the desired results. A large variety of bacterial cells have no difficulty attaching to both hydrophobic or hydrophilic surfaces [53]. Nevertheless, exceptions exists and, according to Assanta et al. [60] and Kaplan et al. [61], Aeromona hydrophila is able to attach in higher numbers to surfaces that have a low surface free energy as opposed to surfaces with a higher surface free energy such as stainless steel. What is clear, however, is that proteins are likely involved in the attachment [62]. For example, Smoot and Pierson observed a 99.9% reduction in bacterial attachment of L. monocytogenes to rubber and stainless steel in the presence of trypsin [63]. The role of bacterial capsules in bacterial attachment remains rather controversial. Hassan et al. showed that bacterial capsule production enhanced formation of Escherichia coli O157:H7 biofilms [39], whereas other researchers reported just the opposite [64].
The complexity of bacterial adhesion to surfaces and the difficulty in developing a fundamental biophysical attachment model has been noted by several researchers [63, 65, 66]. For example, the surface charge of L. monocytogenes cells is dependent upon growth temperature. As the growth temperature decreased to 8°C, the overall surface charge of cells became less negative [65]. Temperature dependencies with respect to hydrophobicity were found by Chavant et al. [67]; for example, L. monocytogenes LO28 became more hydrophobic at lower temperatures independent of the growth phase of cells. As a result, the organism could not attach to PTFE, a polymer with a strongly hydrophobic surface, at low temperatures [67]. In another study, the numbers of L. monocytogenes Scott A cells that adhered to a stainless steel surface increased with decreasing ionic strength in the surrounding media, which was attributed to electrostatic and Lewis acid–base interactions [65]. Clearly, a large number of questions remain to be answered before a comprehensive model explaining adhesion of cells can be developed.
Biofilm growth
After the initial adhesion occurs, bacteria begin to anchor themselves to the surface by synthesizing extracellular polymeric substances (EPS) that facilitate irreversible bacterial attachment to a surface and help maintain the microcolony and biofilm structure [13, 14, 16, 47, 68]. Azaredo and Oliveira found that the exopolymers produced by Sphingomonas paucimobilis possess surface-active properties that aided bacteria in their attachment to hydrophilic surfaces [69]. Interestingly, the presence of preadsorbed proteins on a surface prior to inoculation generally reduced the adhesion of L. monocytogenes regardless of the surface composition or free energy [70–73]. EPS have been shown to enhance nutrient capture and resistance to environmental stress and antimicrobial agents [11, 14–16, 74, 75]. When mature, biofilms exist as a structured matrix with a network of vertical and horizontal channels to allow liquid flow to guarantee supply of nutrients and disposal of waste products that are generated as part of the natural respiration activities of cells. The composition and structure of the extracellular polymeric matrix can vary greatly depending on the microorganism(s), their physiological status, the nutrients available, and the physical conditions present [16].
At sufficiently high concentrations of exopolymers, the biofilm begins to exhibit a gel-like character. Rheological characterization of the biofilm yielded a noticeable increase in the elastic modulus of the biofilm [76]. Overall, the rheological behavior of the film depends on superimposed flow conditions. In laboratory systems, for example, the structure of P. aeruginosa biofilm has shown to assume a denser and more streamlined configuration under turbulent flow with a semicircular appearance that offers little resistance to flow. Additional growth occurred primarily in the direction of flow, and ripple-like structures were occasionally formed. Under laminar flow, the same organism formed flat monolayers with rough surface topologies, and circular, hemispherical colonies only rarely formed [77].
Studies of growth of biofilms using pure cultures of L. monocytogenes have largely focused on the influence of environmental conditions such as growth temperature, pH, and media composition [9, 17, 63, 78–80], and the influence of absorbed food components also referred to as “preconditioning” of substrate surfaces [39, 70, 71]. Despite the number of studies, controversies remain. For example, L. monocytogenes was reported to propagate better when grown in minimal or diluted rich media [79]. For example, the use of MWB (minimum media) enhanced biofilm formation in six out of eight strains of L. monocytogenes [81]. Conversely, Stepanovic et al. [82] found that L. monocytogenes produced better biofilms on plastic surfaces in the presence of a rich medium as opposed to Salmonella spp. that produced better biofilms under low medium conditions. The increased production of flagella and extracellular matrix components was observed at lower incubation temperatures (20 or 4°C), although this may be due to increased incubation times required at lower temperatures [78, 83]. As in planktonic cultures, a neutral pH of 7.0 appears to be optimal for growth compared to basic or acidic media [63, 78].
Detachment and transfer of cells
Detachment in systems with superimposed flows
It is critical to keep in mind that while biofilms may be growing on surfaces they are in no way a stagnant system. Although not often included in biofilm developmental schemes, it is becoming clear that the final step of the biofilm growth cycle includes the detachment or dissemination of colonies from the biofilm (Figure 2). Some researchers have proposed that that physical constraint of the massive towers of cells and extracellular matrix in combination with a superimposed liquid flow causes a structural failure and clumps of bacterial cells leave via erosion. Others have hypothesized that single cell dispersal occurs related to a cell-to-cell quorum-signaling event. It is likely that both mechanisms occur simultaneously, but the exact mode and signals initiating biofilm dissemination will be dependent on the type of bacteria and the growth environment. When detachment rates and size distribution of cell clumps were measured from P. aeruginosa biofilms, both single cells and clumps were observed. Interestingly, the majority of the detachment events were due to the detachment of single cells. A closer look revealed, however, that the majority of detached cells (20–40%) were present in clumps of 300 or more cells [84]. In S. aureus biofilms the number of detached single-cell events was lower than the number of detachment events of clumps, but clumps contained only 10–100 cells [85]. Remarkably, the clumps of S. aureus retained their increased resistance to antibiotics. Obviously there are benefits to utilizing preformed minicolonies as mechanism for propagation.
Suggested mechanism of detachment of single cells and cell clusters from established biofilms. Initially, the biofilm is attached to the surface (black arrow). As shear forces are applied, the biofilm is detached. As it moves laterally with the flow, single cells and cell clusters (gray arrow) are being detached due to the flow-induced rotation. (Adapted from Rupp et al., 2005 [87]).
Recently, a report of organized biofilm single-cell dissemination of nonmucoid P. aeruginosa was published [86]. In this system, as microcolonies matured, cells were observed to differentiate into nonmoving cells and moving cells. Nonmoving cells were located in an outer layer or shell that surrounded a center core with highly motile, densely packed cells. Eventually, the moving cells left the microcolony in an organized swimming fashion termed “seeding dispersal,” leaving behind a hollowed mound of cells [23, 86]. The hollowed structures observed in biofilms were produced by rhamnolipid deficient strains (rhlI) but were not seen in a rhamnolipid, quorum-sensing double mutants (lasI, rhlI), indicating that quorum sensing is involved in this dispersal [86]. In addition, seeding dispersal was not observed in a mucoid P. aeruginosa.
Another mechanism of dissemination without complete physical detachment may be rolling of microcolonies or detached portions of biofilms within the flow. Rupp et al. [87] took time lapse images of S. aureus microcolonies slowly moving along with the liquid flow rolling across the surface of a glass capillary and a movie can be viewed on line [88]. As the microcolony rolled, multiple attachment events were observed in the direction of the flow. Localized detachment from the surface occurred behind the colony, as it tears away from flexible cellular tethers. Thus, in this pure S. aureus system, the biofilm was observed to move along with the fluid flow without detachment and the majority of cells remained in an organized state in order to retain the advantages of being located within a biofilm. It is not yet known if clumps and single cells have similar surface characteristics or adhesive qualities, but it is likely that the mode of dissemination may be species specific and may always consist of a combination of both single-cell and clumping events.
Although progress is being made to develop a mechanistic understanding of bacterial detachment in biofilms, many open questions remain because of the complexity of the process [89]. Results from a number of studies have shown that a large number of parameters may influence the detachment including the presence of matrix-degradation enzymes, gas bubbles generated by microbes, nutrient levels, availability of multivalent cross-linking cations, shear stress due to superimposed flow profiles, contact attrition, lytic bacteriophages, and, as previously mentioned, quorum-sensing signals (Figure 3). Several authors have hypothesized that among these factors, nutrient starvation plays a dominant role in biofilm detachment [89, 90]. Hunt et al. [89], for example, found starvation to be a trigger in P. aeruginosa biofilms. Hunt et al. speculated that under starvation conditions, the structure of the center cluster of a P. aeruginosa biofilm had a different motility as opposed to the outer layer of cells. Cells in the center cluster were motile, whereas the cells in the boundary layer were nonmotile. Similar results were also found by Kaplan et al. in biofilms produced by Actinobacillus actinomycetemcomitans where cell detachment occurred due to motility of cells inside the biofilm [61]. The site-specific action of a number of enzymes such as lyases was cited that aided in the dissolution of biofilm prior to detachment [91, 92]. Thormann et al. pointed out that in addition to the presence of nutrients, oxygen depletion might trigger detachment as well [93]. The researchers investigated the detachment of Shewanella oneidensis MR-1 and found that detachment occurred as soon as 5 min after a superimposed hydrodynamic flow was arrested. The authors concluded that sudden oxygen depletion served as a trigger factor for biofilm detachment and that it only occurred when the oxygen depletion was faster than the biofilm adaptation, a process that seems to be regulated by genes [93]. Typically, during operation of a food process, an abundance of nutrients and water is available inside the process equipment and nutrient and water starvation is therefore less likely to play a major role in the detachment [28, 73]. However, during cleanup and shutdown of the equipment or in case of floor drains that are only periodically exposed to nutrient-rich wastewater, genes may trigger detachment. Upon drying, aerosol formation from floor drains has also been reported and dry cells may slough off into air currents [94].
Shear-induced detachment of cells from P. aueroginosa biofilms under turbulent and laminar flow conditions. Biofilms grown under low shear force (black circle) showed no detachment until the average flow velocity was increased to 1 m/s. For biofilm grown under turbulent flow (white circle), the detachment did not occur until the average flow velocity was 2.5 m/s. Conversely, the rate of detachment was lower when the biofilm was grown under high shear compared to low shear. (Adapted from Stoodley et al. 2002100).
Finally, it should be noted that computer models have been developed to describe biofilm behavior if a fluid flow is superimposed. Unfortunately, most of these models are deficient because they are based on a single species of microorganisms. As food microbiologists well know, biofilms in food systems are typically composed of multiple species that might have a substantially more complex behavior. Furthermore, models describing the detachment behavior of biofilms in pipes have utilized Newtonian fluids, and the flow scenarios were mostly limited to laminar flows, but flow situation in food processing environments are often turbulent and fluids may be non-Newtonian.
Detachment due to direct surface-to-surface contact
A completely different mode of transfer involves direct contact between two different surfaces. For example moving parts may briefly touch a stationary surface during mechanical food process operations such as mixing, scraping, or cutting. Unfortunately, very few researchers in the food science community have investigated this process. The majority of studies that investigated surface-to-surface transfer focused on direct contact of human body parts (e.g., hands, feet) with other human body parts or with synthetic surfaces such as fabrics, food, and synthetic surfaces [95, 96]. Studies on hand washing have found that bacterial translocation occurs from dry unwashed hands at a level of 101–102 CFU, but increased to 104–103 CFU when water was present [96]. Other researchers have reported that in addition to the presence of water, the overall transfer of bacteria to fabrics was increased when friction was simultaneously applied [97]. When thetransfer of dried bacterial films from countertop laminated surfaces to stainless steel and cleaning cloths was investigated, it was found that even at low levels ofbacteria (E. coli, Salmonella spp., or S. aureus) on the original surface (200–300 CFU), almost 20% were transferred to the second surface at contact times of less than 30 s [62].
In a study more relevant to food science, the transfer of L. monocytogenes, Staphylococcus sciuri, Pseudomonas putida, and Comamonas sp. from three contaminated processing surfaces (stainless steel, polyvinyl chloride, and polyurethane) to beef was determined [98]. Surfaces were preconditioned with meat exudates to provide a substrate, inoculated with the test organisms, and allowed to form a biofilm over a 48-h period. The transfer efficiency was then tested by bringing the contaminated surface repeatedly (up to 12 times) in contact with the beef. A plot of the logarithm of the number of organisms transferred versus the number of contacts indicated that transfer occurred at higher levels if the surfaces were brought less than three times into contact and at significantly lower levels after the third contact, i.e., k n=1–3 > k n=4–12, where k is the transfer efficiency and n is the number of contact events. Interestingly, the initial transfer efficiency k n=1–3 varied depending on bacterial strains and nature of the surfaces, whereas k n=4–12 did not depend on bacterial strain or type of surface. The authors concluded that the biofilm was composed of two layers, a primary “soft” layer with a composition and structure that was less resistant to transfer and a secondary layer that was more resistant to transfer [98]. Regardless of the surface type or bacterial species, the potential for transfer between surfaces increased as the number of bacteria in the initial biofilm increased. Higher transfer efficiencies were found using stainless steel instead of polymers irrespective of bacterial strains tested. Of the tested organisms, L. monocytogenes had the lowest transfer efficiency of all bacteria, indicating that the biofilm was strongly attached to the surface [98]. It should be noted that a statistical estimate of initial numbers of adherent bacteria was used rather than an actual measurement, which reduces the accuracy of the determination of the transfer efficiency. Authors also did not systematically evaluate the influence of the hydration state of the biofilm on the transfer efficiency since only washed (hydrated) biofilms were used. Based on previously cited studies one would expect that a dried or partially dehydrated biofilm may show a substantially different transfer behavior. It is possible that during the subsequent contact events, more and more moisture was pressed out of the film. The increased dehydration may have contributed to the change in transfer efficiency.
In direct surface-to-surface transfer, the propagation of cells from one surface to the other likely depends on many factors. While several processes may be involved in the transfer, two key steps appear to be of paramount importance. First, cells must dislocate from the donor surface, which implies a failure of the internal structure of the biofilm; second, adhesion on the second surface must occur. The failure of the internal structure of the biofilm may depend on the composition of the film (e.g., type of microorganisms and nature of the exopolymers) and bacterial cell numbers [98]. In the process of bringing the two surfaces into contact, the film is compressed, resulting in the development of shear and normal stresses within the film. These forces may be sufficient to disrupt the integrity of the biofilm. Following the compression cycle, the stress in the biofilm relaxes as the distance between the two surfaces is initially increased during the expansion cycle. If the layer of the biofilm adheres strongly to the receptor surface, the biofilm may experience tensile stresses that may further aid in the dislocation of a part of the biofilm, which now adheres to the receptor surface. This transfer model, although rather simplistic, nevertheless clearly illustrates that the mechanical properties of the biofilm as well as the attractive and repulsive interaction forces between the film and the two surfaces play a key role. Unfortunately, because of the compositional complexity of biofilms, a complete rheological description of a biofilm is most likely impossible [99]. In addition to the compositional complexity, environmental conditions in a food processing environment such as concentration of nutrients, pH, and temperature are constantly changing, which further complicates the development of a predictive model. Nevertheless, a number of authors have begun to study the rheological behavior of biofilms [76, 100, 101]. Generally, the authors found that biofilms exhibit a viscoelastic behavior; that is, the film has solidlike characteristics under low shear stresses and a more viscous behavior at higher shear stresses. This behavior appears to be of particular importance when a flow profile is superimposed [87]. The shear thinning behavior seems to aid the detachment since the mechanical strength of the film is dramatically weakened as the flow speed is increased or the flow becomes turbulent. Again, nutrient starvation has been suggested to play a role as well [102].
Biofilm characterization techniques
Introduction
Researchers planning to investigate biofilms face many difficult choices, from the selection of a suitable model system to the techniques used to characterize the structure and behavior of the grown biofilm. Many different types of laboratory-based model systems for microbial biofilms can be found in the scientific literature. Unfortunately, none of them can be considered to be the one optimal model system that is universally applicable. On the contrary, the researcher must choose the particular model system that is able to give specific answers to questions that were formulated at the beginning of the study. The accurate simulation of conditions that are encountered in a processing environment is obviously an exceedingly difficult task due to variations in the nature of the food process operation, the natural microflora that may be present, and the food product that is being processed. For these reasons, the researchers, prior to designing an experiment, must make several key decisions. For example, what microorganisms should be used to grow the biofilm (single vs. multispecies), under what growth conditions is the biofilm to be incubated (this will have a substantial impact on the formation of exopolymers), what material is to be used to provide the substrate surface, and what are the basic surface characteristics (surface roughness, hydrophobicity, and charge)? The surface may be positioned vertically or horizontally, which will have important consequences since sedimentation may be involved if a horizontal surface is used. Rinsing and drying procedures are an important part of any bacterial adhesion study and are required to remove unattached (planktonic) or loosely attached bacteria. Finally, if the biofilm is to be prepared for characterization, the researcher faces a choice of methods to remove the film from the substrate surface, and high-intensity ultrasound, surfactants, or simple mechanical forces may be used. All of these methods may alter the structure of the biofilm in one way or another. In this part of the review, we will first review available biofilm growth techniques that are used by microbiologists and biophysicists to study the behavior of biofilm. We will then briefly review characterization techniques that may yield important information about the structure, mechanical behavior, and composition of biofilms.
Biofilm growth techniques
Most biofilms are cultured in simple batch systems. Usually a nutrient solution is inoculated with the desired organism, and a substrate surface is simultaneously supplied to support growth of the biofilm. However, actual setups may be quite complex and involve dripping of nutrients over the substrate surface or continuous pumping of the nutrients through a reactor vessel. An overview of the available growth systems together with a brief review of their advantages and limitations is shown in Table 1.
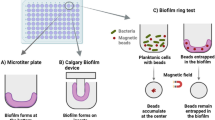
Colony biofilms grown in microtiter plates.
The microtiter plate assay is one of the most commonly used methods for estimation of growth of bacteria in situ [82, 103, 104]. The plates are composed of a polymer such as PVC and consist of 96 wells that can be filled with up to 0.2 ml of inoculated broth. For adhesion experiments, the wells are inoculated with bacteria and allowed to grow. After the incubation period, the liquid is removed and the wells containing the biofilm are washed with a buffer solution often up to four times. The biofilm may then be dried to fix it followed by staining with a fluorescent stain such as crystal violet, followed again by rinsing with water and drying. The dry plates are then read using a microtiter plate reader. Microtiter plate assays have been used to investigate biofilm formation by different bacteria such as Salmonella spp., L. monocytogenes, Helicobacter pylori [82, 103, 104], and fungus such as Candida albicans [105]. The assay has also been used to assess the effect of cleaning agents and disinfectants against P. aeruginosa biofilms using a fluorometric technique [106]. The microtiter plate method has shown to be useful in the genetic analysis of biofilm formers because of the high number of experiments that can be conducted simultaneously in the 96 wells of the plates [22]. This assay is also well suited to study the early stages of biofilm formation that involve colonization and initial biofilm structure development [107]. However, the choice of substrate surface materials is limited since the plates have to be optically transparent in order to read the UV or fluorescence spectra from which the cell numbers are estimated. In addition, the accuracy of determining cell numbers is low, in particular at higher cell concentrations due to multiple scattering effects. A variation of the standard microtiter assays is known as the Calgary biofilm device. It was designed for rapid and reproducible assays of P. aeruginosa, S. aureus, and E. coli biofilm susceptibility to antibiotics. The device produces 96 equivalent biofilms using the standard 96-well technology, with a plastic peg as substratum for the biofilm [108].
Batch and batch-fed growth system
Batch-fed growth systems have been successfully used to compare Staphylococcus epidermidis biofilm growth on acrylic surface over a period of 3 days. The acrylic surfaces were inserted in wells of a six-well tissue culture plate. In the batch-fed mode, every 12 h the medium containing the bacteria is replaced with fresh medium and biofilm dry weight is determined [109].
Colony biofilms grown on polycarbonate membranes
In this fairly simple experimental method, a planktonic culture of the target bacteria is first grown and a drop of the culture containing the desired initial inoculum level placed on a sterile, black, polycarbonate membrane filter that rests on an agar plate [35, 110, 111]. The membranes are sterilized by UV exposure for 15 min. The agar plate is inverted and incubated to allow for growth of the biofilm on the polycarbonate membrane. For viable bacteria enumeration, each membrane-supported biofilm is vortexed in tubes containing peptone water in order to detach the biofilm, and serial dilutions are plated onto appropriate agar plates [110, 111]. This technique is commonly followed by cryoembedding of the biofilm and freezing on dry ice in order to cut micrometer-sized sections of the embedded colonies in a microtome that may then be examined under the microscope [35]. There has been some critique of this technique stating that it does not represent conditions under which biofilms typically grow in nature. Wentland et al. grew a Klebsiella pneumoniae biofilm on polycarbonate filters and stained it with acridine orange [112]. They found different color intensities that were believed to be related to the different metabolic states of the cell, although the correlation was not very strong. Biofilms grown under these conditions are deemed to be less representative of natural biofilms. However, studies that investigated biofilm susceptibility to disinfectants and surfactants and that were conducted with the same organisms on both polycarbonate membranes and biofilm reactors have shown good agreement between the two methods [113]. Because of the ease of preparation, polycarbonate membranes are particularly useful in screening studies.
Capillary biofilm reactor
Capillary biofilm reactors consist of glass capillary tubes where biofilm may grow under continuous flow conditions. The glass tubes have a square cross section to allow for direct microscopic observation. The capillary cells are mounted in a flow cell holder to minimize the risk of breakage. The flow cell is connected to a vented feed carboy that contains the medium, a flow break, a filtered air entry, and a peristaltic pump. The system is also fitted with an inoculation port and a waste carboy. It has been reported that mixing of the fluid with air in the peristaltic pump mayaid in the development of some biofilms, e.g., P.aeruginosa [35]. For the inoculation, the flow is stopped and the downstream tubing is clamped. The culture is injected via the port to fill the glass capillary. The upstream tubing is then clamped and the system is allowed to stand without flow for a specific amount oftime. After attachment and initial growth, flow maybe initiated at varying flow rates. Biofilms may becounterstained by injecting a solution of rhodamine B into the capillary to allow for confocal scanning lasermicroscopy (CSLM) [114].
Flow cell reactor
Like most reactors, the external setup is similar to that of the capillary biofilm reactor and consists of a carboy holding the medium, a flow pump to regulate medium flow, and a waste carboy to collect the spent medium. The flow cell itself has a semicircular cross section and contains seven removable slides (stainless steel slides glued on rectangular pieces of Perspex that properly fit in the apertures of the flow cell) that allow sampling of biofilm at desired time intervals. This type of reactor has been used to observe biofilm growth of Pseudomonas fluorescens with a superimposed laminar or turbulent flow profile [51].
Robbins' device and modified Robbins' device
The Robbins device is a conventional method to establish a surface-associated biofilm. It is a multiport sampling device with evenly spaced sampling ports. The device is often used to examine colonization on engineered material surfaces [115]. The modified Robbins device is an extension of the original design where different surface materials are mounted in the can be used to test growth on several materials simultaneously under similar continuous flow conditions [116].
Rotating disk reactor
This reactor consists of one or more disks with several removable slides per disk that allow for sampling of biofilms. The discs are rotated by a connected motor. Rotational speeds may be adjusted to simulate different flow conditions. This system has been specifically used to grow Gram-negative bacteria biofilm, using Teflon coupons as substratum [117], and to test the activity of sulfate-reducing bacteria in aerobic wastewater biofilms [118].
CDC biofilm reactor
The CDC biofilm reactor is one of the most versatile reactor systems and was developed by Donlan and coauthors [119]. It incorporates 24 removable biofilm substrate surfaces, also known as coupons (eight independent rods with three chips per rod) that are grown inside a jacketed vessel with an effluent spout that is connected to the waste bottle [120]. The jacket is connected through two ports to an external water bath that can be used to regulate the temperature. A continuous mixing of the fluid may be ensured through a magnetically driven baffled stir bar. Each rod may be removed at a given time to access the coupon with the sample biofilm. CDC biofilm reactors have been used to continually monitor the formation of biofilms, characterize their structure, and assess the effect of antimicrobial substances [120, 121].
Rotating annular reactor
This type of reactor has been used to test bacterial biofilm growth from either a drinking water distribution system or a river water source. The reactor consists of a stationary outer cylinder and a rotating solid inner cylinder with 12 removable flush-mounted slides for biofilm sampling. One or several chemostats feed the reactor with the necessary solutions and substrate. Mixing is accomplished by the rotation of the inner cylinder as well as the presence of four draft tubes in the inner cylinder [122, 123].
Constant-depth film fermentor
This design closely mirrors that of a scraped surface heat exchanger. A stainless steel turntable houses 15 polytetrafluoroethylene (PTFE) pans, each containing six stainless steel plugs. Two PTFE scraper blades constantly move across the turntable surface to maintain the biofilms at a constant depth. This system was designed to assess the antimicrobial action of carvacrol on a dual-species biofilm, which reached a quasi-steady state after 12 days [124].
In vivo and animal models of biofilm infection
For in vivo studies of bacterial biofilms, implants containing adhered bacteria may be implanted into a subcutaneous tissue pouch, a peritoneal cavity, the biliary tract, or ureters. These implants may remain in the animals for long periods, e.g., days, weeks, or even months. Eventually the animals are sacrificed to remove the implant and evaluate the formed biofilm. Alternatively, biophotonic imaging has emerged as a method to investigate biofilm formation in vitro in animals. Biophotonic imaging uses bioluminescence to detect a bacterial biofilm. A modified complete lux operon is inserted in the bacterial chromosome and the total photon emissions from selected regions of the mouse are quantified [125].
Biophysical characterization techniques
After an appropriate growth model to produce biofilms has been selected, a need arises to reliably quantify the number of cells in the grown biofilm and to determine the structure and composition of the biofilm. Whereas a comprehensive review of available techniques is beyond the scope of this article, we will briefly discuss the most commonly used methods for characterization of biofilms.
Image-analyzed epifluorescence microscopy
This technique is based on the analysis of microscopy images of fluorescence stained biofilms and is feasible when the biofilm is fairly thin (less than 3–4 μm). Image-analyzed epifluorescence microscopy (IAEFM) is capable of determining the total number of adhered cells, the area coverage, and volume that is occupied by the film. Djordjevic et al. used this method to evaluate L. monocytogenes biofilm formation in microtiter plate assay in real time [104]. Due to the combination of the microscopy with image analysis, cell counts are rapidly obtained, allowing a large number of samples to be analyzed. More importantly, it allows determination of cell counts on opaque surfaces such as steel or rubber. Limitations of the technique include the inability to accurately count thicker films and possible problems in distinguishing single cells in dense clusters. In these cases, additional microscopy techniques may be needed to verify the counts such as scanning electron microscopy (SEM) and confocal laser scanning microscopy (CLSM) [28].
Transmission electron microscopy and SEM
Both techniques have been used to examine biofilms since both offer extremely high resolution, but because they operate in a high vacuum the samples need to be fixed and dehydrated by using graded solvents such as alcohol, acetone, and xylene [107]. The preparation of the samples (dehydration and staining) irrevocably changes the structure of the biofilm, which limits theapplicability of the technique [28]. In addition, the preparation is time-consuming and because of this, SEM or transmission electron microscopy (TEM) analysis is not conducted on a routine basis. Nevertheless, TEM has been used to characterize the structure of the extracellular polymer matrix in biofilms grown on medical devices using ruthenium red as a dye [126] and to enumerate the stratified growth in P. aeruginosa biofilms [35]. In a recent publication, Hunter and Beveridge used high-pressure freeze-substitution TEM as opposed to conventional TEM [127]. The authors were able to reveal remarkable structural details within the biofilm that had never before been imaged due to the limitations of other techniques. SEM has been used to study the spatial distributions ofL. monocytogenes cells attached to ready-to-eat meats [128], determine the effect of exposure to antibiotics or sanitizers on biofilm integrity [108, 129], and follow formation of biofilms from H. pylori [103] and C.albicans [105].
Environmental scanning electron and field emission scanning microscopy
Both of these techniques offer substantial improvements over SEM and TEM. In the case of environmental scanning electron microscopy (ESEM), sample analysis is conducted under reduced air pressure rather than a high vacuum, thereby allowing partially hydrated samples to be examined [130]. Using field emission scanning microscopy (FESEM), fully hydrated biofilms may be analyzed.
Confocal laser scanning microscopy
This technique was developed in the 1980s and allows examination of biofilms without the limitations imposed by SEM or TEM. Fully hydrated biofilms are analyzed by progressive laser scans at different focal planes within the sample. Computer analysis of the scanned images permits a recreation of the three-dimensional structure of the biofilm. The application of confocal laser scanning microscopy (CLSM) combined with a number of staining fluorescent techniques provides an important and effective tool to analyze the composition and structure of hydrated biofilms in situ, nondestructively and in real time [131]. Viability and distribution of cells within the biofilm may be analyzed as well. When using epifluorescence microscopy or CLSM, the choice of suitable fluorescent stains is critical in order to increase the contrast between the organisms and the exopolymers in the biofilms. Nucleic acid stains such as 4,6-diamino-2-phenylindole (DAPI) or acridin orange have been used to stain the DNA of cells regardless of their viability. Other dyes sensitive to viable cells such as propidium iodine or 5-cyano-2,3-ditolyl tetrazolium chloride [28] may be used to further resolve viable and dead cells.
Infrared spectroscopy
Fourier transform infrared spectroscopy (FTIR) and attenuated total internal reflectance spectroscopy (ATIR) have been used to study antibiotic penetration into biofilms [132], and to determine the composition of the exopolymeric matrix. The kinetics of Streptococcus pneumoniae biofilms formation has been studied in situ and real time [119], ATIR is a nondestructive technique, where infrared radiation is multiply reflected from the inner surface of an internal reflection element (IRE). At each reflection site, a longitudinal wave of radiation penetrates from the IRE into the adjacent biofilm sample to generate IR absorption bands that are characteristic of the chemical composition of the biofilm [133].
Analysis of biofilm morphology
Biofilms are composed of microbial cells and extracellular polymeric substances (EPS). EPS, primarily composed of polysaccharides, may comprise up to 50 to 90% of the total organic carbon found in biofilms [134]. As previously mentioned, biofilms are heterogeneous, containing clusters or microcolonies of bacterial cells that are encased in the EPS matrix. These clusters may be physically separated from other microcolonies by interstitial voids [135]. These interstitial voids are of key importance in the transport of nutrients and oxygen and have shown to play a role in the susceptibility to antimicrobial agents. Structural parameters that can easily be quantified include thickness, roughness and surface area coverage [51, 136, 137], density, porosity, and mean pore size of interstitial voids [138, 139]. Fractal dimensions of activated sludge or sulfate-reducing microorganism biofilms have also been calculated [139, 140]. The morphological properties of biofilms is important because it contributes to the development of internal pH gradients that may influence transport of nutrients, metabolic products, and oxygen throughout the biofilm [141, 142]. Hunter and Beveridge developed a novel pH fluoroprobe for the analysis of the pH microenvironment in P. aeruginosa biofilms [127]. The authors used seminaphthorhodafluor-4F 5-(and)-carboxylic acid (C-SNARF-4) as a quantitative indicator of pH microenvironments in microbial biofilms and discussed the results in terms of the biofilm morphology obtained from the analysis of CSLM images.
Increasingly two other techniques, atomic force microscopy (AFM) and X-ray photoelectron spectroscopy (XPS), are used in the compositional and structural analysis of biofilms. AFM is capable of imaging surfaces at nanometer or subnanometer resolutions [143]. AFM utilizes a small silicon nitride or silicon tip that is mounted on a cantilever that is then scanned across the surface of the sample. The use of AFM to visualize biofilms has been pioneered by Bremer et al. who sought to understand biofilm-induced deterioration of a variety of materials [144]. XPS is well established in materials science and has recently also been used to analyze microbial cell surfaces and biofilm surfaces [145]. XPS involves irradiation of the biofilm with an X-ray beam. As a result of the irradiation, electrons are emitted from the sample, each carrying a kinetic energy that is characteristic of the specific composition at the scanning coordinate. Unfortunately, similarly to SEM and TEM, samples have to be dehydrated before being introduced in the vacuum chamber of the spectrometer.
Image structural analysis
This analytical method calculates nine textural and dimensional parameters from two-dimensional biofilm images [146]. The parameters calculated include porosity, microcolony length and width, average diffusion distance, maximum diffusion distance, and fractal dimension. Image structural analysis (ISA) also calculates three textural parameters: textural entropy, angular second moment, and inverse difference moment. Because ISA was designed to analyze larger-scale biofilm patters in two-dimensional grayscale images, it is primarily used for the analysis of conventional or epifluorescence microscopy images [49].
COMSTAT
COMSTAT was developed to analyze high-resolution three-dimensional confocal image stacks [147]. The image stacks are analyzed pixel by pixel and digitized by assigning a value of one if biomass is present or zero if no biomass is present. The decision of whether to assign a zero or a one is based on a threshold gray or color value. COMSTAT has ten separate image-analysis features that yield the biovolume, the area occupied by bacteria in each layer in the biofilm, thickness and roughness, identification and distribution of microcolonies, microcolony volume, fractal dimension, average and maximum diffusion distance, and surface-to-volume ratio [147]. In most COMSTAT measurement, bacteria are tagged with green fluorescent protein (GFP) [148].
Other techniques
Attachment of bacterial cells to surfaces can be affected by many factors such as the cell surface charge and cell surface hydrophobicity, which influences electrostatic and hydrophobic interactions between cells, the exopolymers, and the substrate surface [149]. The cell surface net charge of L. monocytogenes may be determined by electrostatic interaction chromatography (ESIC). In another study where P. aeruginosa adhesion to PVC from endotracheal intubation tubes was studied, the electrophoretic mobility of cells was measured using a Doppler-electrophoretic light-scattering technique. Electrophoretic mobilities were converted to zeta potentials to estimate the bacterial surface charge using the Helmholtz–Smoluchowski equation [150]. Hydrophobic interaction chromatography (HIC) and contact angle measurement (CAM) may be used to examine cell surface hydrophobicity [128].
Biofilm remediation techniques
Introduction
One of the most remarkable properties of biofilms is their ability to survive remediation procedures that would be sufficient to completely inactivate planktonic cultures. Surprisingly few studies have been published that systematically evaluated the efficiency of remediation procedures to eliminate biofilms from food processing surfaces. While regular and rigorous sanitation programs have helped reduce the spread of microbial contamination due to cross contamination of infected food processing surfaces and food products, they have led to increased costs due to long downtimes and extensive water, detergent, and sanitizer usage. The disposal of waste detergents and sanitizers is also associated with substantial costs, and food companies must ensure that no toxic residues are introduced into the food due to the cleaning procedure. The correct selection of detergent and/or sanitizer type and concentration as well as the control of the water temperature is critically important to ensure the effectiveness of a remediation program [151]. Increasingly, consumers look to naturally occurring antimicrobials that are biodegradable and nontoxic and may even be part of the food system [151]. Compounds such as essential oils and chitosan are being evaluated to prevent infection of foods from contaminated food processing equipment. Factors such as food compatibility and partitioning of compounds may affect their ability to prevent the growth of biofilms. Food manufacturers that are battling infections of processing equipment need to identify the primary source of contamination. For example, is the repeated growth of biofilms due to an insufficient cleaning procedure or is it because a recontamination of the process surfaces occurs due to the raw material being contaminated? Additional considerations when developing sanitation protocols are the sequence in which sanitizers and detergents are added. Should the sanitizer be added before the detergent or vice versa? Interestingly, much more is known about biofilm killing than is known about biofilm removal [113], and more research is needed to develop a more comprehensive understanding.
Sanitizers and antimicrobials
Sanitizers
The distinction between sanitizers and anti-microbials is somewhat arbitrary, but sanitizers or disinfectants are typically low molecular weight compounds that are soluble in water and are highly reactive [152]. Sanitizers or disinfectants are chemical compounds capable of inactivating microorganisms, bacterial spores, and viruses. Chlorine-based compounds are the most commonly used sanitizers in food processing environments and include chlorine gas, hypochlorites, chloramines, and chlorine dioxide. Due to problems with corrosion and evaporation, they are mostly applied in cold water. Iodophors are a combination of iodine and a solubilizing agent that aid in the release of free iodine when the mixture is dispersed into water. Quaternary ammonium compounds (QAC) are odorless, colorless, and nontoxic and are therefore often used in food processing as part of the cleaning protocol. However, they are incompatible with chlorine based-sanitizers and because of their positive charge may not be combined with negatively charged detergents. Dilute acids (phosphoric acid, peracetic acid, acetic acid) and alkali reagents (NaOH and KOH) may also be used as sanitizing agents. These compounds are abundantly available at relatively low cost and therefore are widely used as part of cleaning protocols. Not only do theyaid in the removal of biofilms, but they are also highly efficient solubilizers of a variety of biopolymers such as proteins and carbohydrates. Finally, hydrogen peroxide or ozone have been used to inactivate planktonic and biofilm cultures as well. It should be noted that with most of these compounds, the system pH canhave a dramatic effect on their activity.
Interaction of sanitizers with biofilms
Organisms grown in biofilms may survive prolonged exposure to fairly high concentrations of sanitizers [26, 29, 32, 34, 153–158]. Schwach and Zottola [159] demonstrated as early as 1982 that treatment of Pseudomonas fragi, Salmonella montevideo, and Bacillus cereus with sodium hypochloride followed by rinsing with water was not effective in completely removing bacteria from food processing surfaces. Chen and Stewart, in an interesting study on a mixed P. aeruginosa and K. pneumoniae biofilm grown in a continuous flow annular reactor, remarked that killing and removal are two distinctly different phenomena [113]. They found that treatment of biofilms with a variety of sanitizers such as monochloramine and aminotri(methylene-phosphonic acid) pentasodium salt (Dequest 2006), did not simply result in killing but may or may not ease removal and that agents that promote removal may or may not kill the microorganisms. The question why sanitizing agents are sometimes not effective is therefore not easily answered. Although various models have been proposed, it is feasible that some compounds may lead to detachment of cultures from infected surfaces, but if cultures are not inactivated, they could reattach and regrow further down the processing line. Similarly, if biofilms are inactivated but not removed, they may provide a fertile ground for attachment of living bacteria that may originate upstream of the contaminated equipment after the sanitizing compounds have been removed. Similarly, a number of researchers reported that they were unable to achieve complete inactivation of L. monocytogenes using a combination of various sanitizers [156, 160]. Two models have been proposed to explain the increased resistance of organisms in biofilms to sanitizers. The first model proposes that the physiology of microorganisms within a biofilm changes due to adaptation of microorganisms to a microenvironment that has limitations in nutrient concentration, pH, and cell mobility [161]. Physiological factors such as biofilm age [158], nutrient deficiency [162], and growth rate [163], have been suggested to affect the susceptibility to disinfectants. A second, more recent model proposes that physical properties of the biofilm limit the rate of transport and activity of sanitizers [113, 164–168]. Investigators suggested that transport of active agents from the delivery phase (typically the solvent) through the biofilm to the adhering interface (the surface of the medical or processing equipment) might be reduced due to physicochemical interactions of disinfectants with organic material or microorganisms in the upper layer of the biofilm matrix. Most sanitizers are strong oxidizers that lose their activity once they have reacted with the target material. It is therefore feasible that due to the rapid reaction rate and aggressive nature of the compounds, the compounds would not be able to penetrate into the lower layer of the biofilm, leaving that part of the biofilm viable.
Antimicrobials
An antimicrobial is a substance that inactivates or inhibits the growth of microorganisms, fungi, or parasites. Antibiotics are a particular class of antibacterial and antifungal antimicrobials that may be used as medicinal drugs to treat infections because of their low toxicity for humans or animals [169, 170]. A significant amount of research has been conducted to determine the resistance of biofilms to the application of antibiotics and an excellent review has been published by Drenkard in 2003 [171]. Biofilm infection, in fact, is one ofthe principal problems that causes rejection of implanted medical devices. The list of antimicrobials and antibiotics is much larger than that of sanitizers. Antimicrobials may differ greatly in their molecular nature. Compounds may be hydrophobic, hydrophilic, or both; they may be high molecular weight compounds such as lysozyme (an enzyme) or chitosan (a polysaccharide) or fairly small molecular weight compounds such as nisin (a peptide) and carvacrol and eugenol (essential oil compounds). Their inhibitory or biocidal activity is not based on a direct chemical reaction with the microorganisms but instead is based on either insertion of the compound in the bacterial membrane, leading to leakage and loss of proton motive force or penetration of the compound into the inside of the cell followed by either a change of internal pH or damage to the reproductive system of the cell [169, 170].
Interaction of antimicrobials with biofilms
The increased resistance of biofilms to antimicrobials has been confirmed in numerous studies. For example, the concentration of a mixture of chlorofene, chlorocresol, and phenylphenol required to inactivate a biofilm composed of E. coli CIP 54 127 (ATCC 10536) was five times greater than the concentration required to inactivate the same strain of freely suspended E. coli [172]. The researchers hypothesized that since phenol-based antimicrobials are only effective against protein-synthesizing bacteria, the deeply embedded cells in the biofilm may not have exhibited an active metabolism. Indeed, when measuring intracellular ATP levels in biofilm bacteria, they found a reduced metabolic activity compared to bacteria in suspension. Recent studies using two naturally occurring essential oil compounds as model antimicrobials has led to some puzzling results. Knowles and Roller reported that carvacrol and eugenol had higher efficacies than common commercial disinfectants based on hydrogen peroxide and peroxyacetic acid and that their efficiency was in fact higher against biofilms than against planktonic bacteria [173]. Finally, retarded penetration of antibiotics such as ciprofloxacin, vancomycin [174], and gentamicin [175] has been reported by a variety of researchers, but a comparison of diffusion coefficients did not lead to a confirmation of a solely diffusion-driven mechanism [176].
Surfactants
Much less is known about the interactions of surfactants with biofilms. Chen and Stewart [113] found greatly varying reductions in biofilm viable cell area densities after addition of anionic (SDS) and nonionic (Triton X-100, Tween 20) surfactants. Whitekettle [177] found that addition of surfactants affected growth of biofilms on a variety of surfaces, but experiments were not conducted to evaluate the effect of addition of surfactants after the biofilm had been established. Of the few studies available, most simply compared the effect of selected surfactants on biofilms. For example, a comparison of anionic surfactants, chlorinated alkaline detergents, and enzyme blends concluded that anionic detergents were more efficient in the removal of biofilms [178]. The most efficient treatment reported was a two-step treatment with a cleaning agent (surfactant) being first applied followed by a subsequent treatment with a sanitizer [153]. Nevertheless, no mechanistic model has been introduced to explain the efficiency of such a sequential treatment or of a treatment with surfactants alone.
Kinetic aspects of the remediation process such as cell destruction and survival after repeated treatment with surfactant or surfactant followed by disinfectants have also not been explored. In order for surfactants to be effective in removing biofilms, they would have to penetrate into the interface between the substrate layer and the biofilm. If they in fact penetrate the biofilm matrix to reach the lower layer of the biofilm, they could adsorb at the interface due to their high surface activity and reduce the interfacial tension. Consequently, the attractive interactions between the bacterial surfaces and the substrate surface responsible for continued adhesion of the bacteria may be decreased, which would ease removal of the film. Interstitial voids that are used for nutrient and metabolic product transport may play an important role in the interaction of biofilms and surfactants. Clearly, an improved understanding of the penetration behavior of surfactants into biofilms is required to develop detergents with higher efficiencies.
Conclusions
Biofilms are a perfect case study to illustrate the importance of biophysical processes in biological systems. The mechanism involved in the formation and propagation of biofilms illustrates that biophysical processes such as cell-to-cell and cell-to-surface interactions play an important role in ensuring successful attachment to a variety of substrate surfaces. Evolution has led to the formation of a matrix structure that offers much better survival chances to superimposed physicochemical and biological stresses by deliberately limiting mass transport of nutrients and oxygen, which causes alterations in the physiology of cells. The molecular diversity of exopolymers that are synthesized to form the matrix of the biofilm ensures that potentially harmful compounds immediately interact with the top layer of the biofilm thereby quickly neutralizing them. As a result, biofilms are more resistant to sanitizers and detergents than planktonic cells. In most cases, this resistance arises from the fact that biofilms act as physical barrier limiting the penetration of the sanitizers into the biofilm. Much of the improved understanding of biofilms in recent years has come from an application of traditional physicochemical characterization techniques and a transfer of physicochemical principles such as mass transport phenomena and colloidal and molecular interaction theories. As the field of biophysics develops, we can expect to gain an even better understanding and appreciation of the underlying principles that govern biofilm formation and propagation. Ultimately, these insights may lead to better remediation strategies in situations where biofilms pose an inherent danger. Alternatively, we may see the beginning of a deliberate use of biofilms in a much more controlled fashion in fermentation and bioconversion processes.
References
T. Moretro and S. Langsrud, Listeria monocytogenes: biofilm formation and persistence in food-processing environments. Biofilms 1, 107 (2004).
R.M. Donlan, Biofilms: microbial life on surfaces. Emerg Infect Dis 8, 881 (2002).
C.A. Fux, J.W. Costerton and P.S. Stewart et al., Survival strategies of infectious biofilms. Trends Microbiol 13, 34 (2005).
P. Stoodley, S. Wilson and L. Hall-Stoodley et al., Growth and detachment of cell clusters from mature mixed-species biofilms. Appl Environ Microbiol 67, 5608 (2001).
K. Kierek-Pearson and E. Karatan, Biofilm development in bacteria. Adv Appl Microbiol 57, 79 (2005).
R. Van Houdt and C.W. Michiels, Role of bacterial cell surface structures in Escherichia coli biofilm formation. Res Microbiol 156, 626 (2005).
J.F. Frank, Microbial attachment to food and food contact surfaces. Adv Food Nutr Res 43, 319 (2001).
C. Ganesh Kumar and S.K. Anand, Significance of microbial biofilms in food industry: a review. Intl J Food Microbiol 42, 9 (1998).
S.K. Hood and E.A. Zottola, Adherence to stainless steel by foodborne microorganisms during growth in model food systems. Intl J Food Microbiol 37, 145 (1997).
S. Wong, D. Street and S.I. Delgado et al., Recalls of foods and cosmetics due to microbial contamination reported to the U.S. Food and Drug Administration. J Food Prot 63, 1113 (2000).
H.F. Jenkinson and H.M. Lappin-Scott, Biofilms adhere to stay. Trends Microbiol 9, 9 (2001).
Y.H. An and R.J. Friedman, Laboratory methods for studies of bacterial adhesion. J Microbiol Methods 30, 141 (1997).
R.J. Doyle, Microbial Growth in Biofilms—Part A: Developmental and Molecular Biological Aspects (Academic Press, San Diego, CA 2001).
G.A. O'Toole, H.B. Kaplan and R. Kolter, Biofilm formation as microbial development. Annu Rev Microbiol 54, 49 (2002).
L.V. Poulsen, Microbial biofilm in food processing. Lebensm-Wiss Technol 32, 321 (1999).
I.W. Sutherland, The biofilm matrix—an immobilized but dynamic microbial environment. Trends Microbiol 9, 222 (2001).
K.Y. Kim and J.F. Frank, Effect of nutrients on biofilm formation by Listeria monocytogenes on stainless steel. J Food Prot 58, 24 (1995).
N.G. Marriott, Principles of Food Sanitation (Aspen Publishers, Gaithersburg, MD, 1999).
P. Sommer, C. Martin-Rouas and E. Mettler, Influence of the adherent population level on biofilm population, structure and resistance to chlorination. Food Microbiol 16, 503 (1999).
A.E. Hodgson, S.M. Nelson and M.R.W. Brown et al., A simple in vitro model for growth control of bacterial biofilms. J Appl Bacteriol 79, 87 (1995).
S.L. Kuchma and G.A. O'Toole, Surface-induced and biofilm-induced changes in gene expression. Curr Opin Biotechnol 11, 429 (2000).
G.A. O'Toole and R. Kolter, Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol Microbiol 28, 449 (1998).
S. Sauer, A.K. Camper, G.D. Ehrlich, et al., Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J Bacteriol 184, 1140 (2002).
J.F. Frank and R.A.N. Chmielewski, Effectiveness of sanitation with quaternary ammonium compoind or chlorine on stainless steel and other domestic food-preparation surfaces. J Food Prot 60, 43 (1997).
S. Vatanyoopaisarn, A. Nazli and C.E.R. Dodd et al., Effect of flagella on initial attachment of Listeria monocytogenes to stainless steel. Appl Environ Microbiol 66, 860 (2000).
K.J. Bolton, C.E.R. Dodd and G.C. Mead et al., Chlorine resistance of strains fo Staphylococcus aureus isolated from poultry processing plants. Lett Appl Microbiol 6, 31 (1988).
C.K. Bower and M.A. Daeschel, Resistance reponses of microorganisms in food environments. Int J Food Microbiol 50, 33 (1999).
R.M. Donlan and J.W. Costerton, Biofilms: Survival mechanisms of clinically relevant microorganisms. Clin Microbiol Rev 15, 167 (2002).
W.M. Dunne, Bacterial Adhesion: Seen any good biofilms lately? Clin Microbiol Rev 15, 155 (2002).
J.F. Frank, R.A.N. Gillett and G.O. Ware, Association of Listeria spp. contamination in the dairy processing plant environment with the presence of staphylococci. J Food Prot 53, 928 (1990).
J.F. Frank and R.A. Koffi, Surface-adherent growth of Listeria monocytogenes is associated with increased resistance to surfactant sanitizers and heat. J Food Prot 53, 550 (1990).
M.W. LeChevallier, C.D. Cawthon and R.G. Lee, Inactivation of biofilm bacteria. Appl Environ Microbiol 54, 2492 (1988).
J.C. Nickel and J.W. Costerton, Bacterial biofilms and catheters: A key to understanding bacterial straegies in catheter-associated urinary tract infection. Can J Infect Dis 3, 619 (1992).
G. Reid, C. Tieszer and R. Foerch et al., Adsoption of ciprofloxacin to urinary catheters and effect on subsequent bacterial adheion and survival. Colloid Surf B Biointerfaces 1, 9 (1993).
E. Werner, F. Roe and A. Bugnicourt et al., Stratified growth in Pseudomonas areuginosa biofilms. Appl Environ Microbiol 70, 6188 (2004).
S.E. Crampton, C. Gerke and F. Gotz, In: Methods in Enzymology, Vol. 336 Microbial Growth in Biofilms Part A. Developmental and Molecular Biological Aspects, edited by R.J. Doyle (San Diego, CA 2001), p. 239.
G.M. Dunny and S.C. Winans, Cell–cell Signaling in Bacteria (ASM Press, Washington, DC 1999).
S. Moller, C. Sternberg and J.B. Andersen et al., In situ gene expression in mixed-cultures biofilms: evidence of metabolic interactions between community members. Appl Environ Microbiol 64, 721 (1998).
A.N. Hassan, D.M. Birt and J.F. Frank, Behavior of Listeria monocytogenes in a Pseudomona putida biofilm on a condensate-forming surface. J Food Prot 67, 322 (2004).
K.C. Sasahara and E.A. Zottola, Biofilm formation by Listeria monocytogenes utilizes a primary colonizing microorganism in flowing systems. J Food Prot 56, 1022 (1993).
V. Leriche and B. Carpentier, Limitation of adhesion and growth of Listeria monocytogenes on stainless steel surfaces by Staphylococcus sciuri biofilms. J Appl Microbiol 88, 594 (2000).
D.E. Norwood and A. Gilmour, The differential adherence capabilities of two Listeria monocytogenes strains in monoculture and multispecies biofilms as function of temperature. Lett Appl Microbiol 33, 320 (2001).
M.S. Chae and H. Schraft, Cell viability of Listeria monocytogenes biofilms. Food Microbiol 18, 103 (2001).
M.S. Chae and H. Schraft, Comparative evaluation of adhesion and biofilm formation of different Listeria monocytogenes strains. Int J Food Microbiol 62, 103 (2000).
R.B. Tompkin, V.N. Scott and D.T. Bernard et al., Guidelines to prevent post-processing contamination from Listeria monocytogenes. Dairy Food Environ Sanit 19, 551 (1999).
M.C.M. van Loosdrecht, D. Eikelbook and A. Gjaltema et al., Biofilm structures. Water Sci Technol 32, 35 (1995).
J. Wimpenny, W. Manz and U. Szewzyk, Heterogeneity in Biofilms. FEMS Microbiol Rev 24, 661 (2000).
M.C.M. van Loosdrecht, C. Picioreanu and J.J. Heijnen, A more unifying hypthesis for biofilm structures. FEMS Microbiol Lett 24, 181 (1997).
L.B. Purevdorj-Gage and P. Stoodley, Biofilm structure, behaviour, and hydrodynamics. In: Microbial biofilms, edited by M. Ghannoum and G.A. O'Toole (ASM Press, Washington, DC 2004), p. 160.
K.W. Millsap, G. Reid and H.C. van der Mei et al., Adhesion of Lactobacillus species in urine and phosphate buffer to silicone rubber and glass under flow. Biomaterials 18, 87 (1996).
M.A. Pereira, M. Kuehn and S. Wuertz et al., Effect of flow regime on the architecture of a Pseudomonas fluorescens biofilm. Biotechnol Bioeng 78, 164 (2002).
H.H.M. Rijnaarts, W. Norde and E.J. Bouwer et al., Bacterial adhesion under static and dynamic conditions. Appl Environ Microbiol 59, 3255 (1993).
M. Fletcher, Bacterial attachement in aquatic environments: a diversity of surfaces and adhesion strategies. In: Bacterial Adhesion: Molecular and Ecological Diversity, edited by M. Fletcher (Wiley-Liss, New York 1996), p. 1.
J.N. Israelachvili, Intermolecular and Surface Forces (Academic Press, San Diego, CA 1992).
A.M. James, Charge properties of microbial cell surfaces. In: Microbial Cell Surface Analysis—Structural and Physicochemical Methods, edited by N. Mozes, P.S. Handley, H.J. Busscher and P.G. Rouxhet (VCH Publishers, New York 1991), p. 221.
D.J. McClements, Food Emulsions: Principles, Practice and Techniques (CRC Press, Boca Raton, FL 1999).
D.R. Olivera, Physio-Chemical Aspects of Adhesion. In: Biofilms-Science and Technology, edited by L.F. Melo, T.R. Bott, M. Fletcher and B. Capdeville (Kluwer Academic Publishers, Dordecht, the Netherlands 1992).
H.J. Busscher, J. Sjollema and H.C. v.d. Mei, Relatie importance of surface free energy as a measure of hydrophobicity in bacterial adhesion to solid surfaces. In: Microbial Cell Surface Hydrophobicity, edited by R.J. Doyle and M. Rosenberg (ASM, Washington, DC 1990).
G. Reid, C. Tieszer and R. Foerch et al., Adsoption of ciprofloxacin to urinary catheters and effect on subsequent bacterial adheion and survival. Colloids Surf, B Biointerfaces 1, 9 (1993).
M.A. Assanta, D. Roy and D. Montpetit, Adhesion of Aeromonas hydrophila to water distribution systems pipes after different contact times. J Food Prot 61, 1321 (1998).
J.B. Kaplan, M.F. Meyenhofer and D.H. Fine, Biofilm growth and detachment of Actinobacillus actinomycetemcomintans. J Bacteriol 185, 1399 (2003).
E. Scott and S.F. Bloomfield, The survival and transfer of microbiological contamination via cloths, hands and utensils. J Appl Bacteriol 68, 271 (1990).
L.M. Smoot and M.D. Pierson, Influence of environmnetal stress on the kinetics and strength of attachement of Listeria monocytogenes Scott A to Buna-N rubber and stainless steel. J Food Prot 61, 1286 (1998).
J. Josephsen and F.K. Vogensen, Identificaiton of three different plasmid-encoded restriciton/modification systems in Streptococcus lactis subsp. cremoris W56. FEMS Microbiol Lett 59, 161 (1989).
R. Braindet, V. Leriche and B. Carpentier et al., Effects of the growth procedure on the surface hydrophbicity and Listeria monocytogenes cells and their adhesion to stainless steel. J Food Prot 62, 994 (1999).
A.A. Mafu, D. Roy and J. Goulet et al., Characterization of physiochemical forces involved in adhesion of Listeria monocytogenes to surfaces. Appl Environ Microbiol 57, 1969 (1991).
P. Chavant, B. Martinie and T. Meylheuc et al., Listeria monocytogenes L028: Surface physiochemical properties and ability to form biofilms at different temperatures and growth phases. Appl Environ Microbiol 68, 728 (2002).
G. Wirtanen and T. Mattila-Sandholm, Epifluorescence image analysis and cultivation of foodborn biofilm bacteria grown on stainless steel surfaces. J Food Prot 56, 678 (1993).
J. Azaredo and R. Oliveira, The role of exopolymers in the attachment of Sphingomonas paucimobilis. Biofouling 16, 59 (2000).
H. Al-Makhlafi, J. McGuire and M. Daeschel, Influence of preabsorbed milk proteins on adhesion of Listeria monocytogenes to hydrophobic and hydrophilic silica surfases. Appl Environ Microbiol 60, 3560 (1994).
H. Al-Makhlafi, A. Nasir and J. McGuire et al., Adhesion of Listeria monocytogenes to silica surfaces after suqential and competitive adsoption of bovine serum albumin and B-lactoglobulin Appl Environ Microbiol 61, 2013 (1995).
L.-M. Barnes, M.F. Lo and M.R. Adams et al., Effect of milk proteins on adhesion of bacteria to stainless steel surfaces. Appl Environ Microbiol 65, 4543 (1999).
D. Cunliffe, C.A. Smart and C. Alexander et al., Bacterial adesion at synthetic surfaces. Appl Environ Microbiol 65, 4995 (1999).
J.W. Costerton, Overview of microbial biofilms. J Ind Microbiol 15, 137 (1995).
J.W. Costerton, K.J. Cheng and G.G. Geesey et al., Bacterial biofilms in nature and disease. Annu Rev Microbiol 41, 435 (1987).
I. Kapper, C.J. Rupp and R. Cargo et al., Viscoelastic fluid description of bacterial biofilm material properties. Biotechnol Bioeng 80, 289 (2002).
B. Purevdorj, J.W. Costerton and P. Stoodley, Influence of hydrodynamics and cell signaling on the structure and behavior of Pseudomonas aeruginosa biofilms. Appl Environ Microbiol 68, 4457 (2002).
P.J. Herald and E.A. Zottola, Attachment of Listeria monocytogenes to stainless steel surfaces at various temperature and pH values. J Food Sci 53, 1549 (1988).
K.Y. Kim and J.F. Frank, Effect of growth nutrients on attachement of Listeria monocytogenes to stainless steel. J Food Prot 57, 720 (1994).
L.M. Smoot and M.D. Pierson, Effect of environmental stress on the ability of Listeria monocytogenes Scott A to attach to food contact surfaces. J Food Prot 61, 1293 (1998).
A.G. Moltz and S.E. Martin, Formation of biofilms by Listeria monocytogenes under various growth conditions. J Food Prot 68, 92 (2005).
S. Stepanovic, I. Cirkovic and L. Ranin et al., Biofilm formation by Salmonella spp. and Listeria monocytogenes on plastic surface. Lett Appl Microbiol 38, 428 (2004).
A.A. Mafu, D. Roy and J. Goulet et al., Attachment of Listeria monocytogenes to stainless steel, glass, polypropylene, and rubber surfaces after short contact times. J Food Prot 53, 742 (1990).
S. Wilson, M.A. Hamilton and G.C. Hamilton et al., Statistical quantification of detachment rates and size distributions of cell clumps form wild-type (PAO1) and cell signaling mutant (JP1) Pseudomonas aeuginosa biofilms. Appl Environ Microbiol 70, 5847 (2004).
C. Fux, S. Wilson and P. Stoodley, Detachment characteristics and oxacillin resitance of Staphylococcus aureus biofilm emboli in an in vitro catheter infection model. J Bacteriol 186, 4486 (2004).
B. Purevdorj-Gage, W.J. Costerton and P. Stoodley, Phenotypic differentiation and seeding dispersal in non-mucoid and mucoid Pseudomonas aeruginosa biofilms. Microbiology 151, 1569 (2005).
C.J. Rupp, C.A. Fux and P. Stoodley, Viscoelasticity of Staphylococcus aureus biofilms in responce to fluid shear allows resistance to detachment and facilitates rolling microation. Appl Environ Microbiol 71, 2175 (2005).
C. Rupp, S. Wilson and P. Stoodley, Staphylococcus aureus Biofilm Rolling Along the Lumen of a Glass Tube. (ASM MicrobeLibrary.org, 2002).
S.M. Hunt, E.M. Werner and B. Huang et al., Hypothesis for the role of nutrient starvation in biofilm detachment. Appl Environ Microbiol 70, 7418 (2004).
L.K. Sawyer and S.W. Hermanowicz, Detachment of Aeromonas hydrophila and Pseudomonas aeruginosa due to variations in nutrient supply. Water Sci Technol 41, 139 (2000).
D.G. Allison, B. Ruiz and C. SanJose et al., Extracellular products as mediators of the formation and detachment of Pseudomonas fluorescens biofilms. FEMS Microbiol Lett 167, 179 (1998).
A. Boyd and A.M. Chakrabarty, Role of alginate lyase in cell detachment of Pseudomona aeruginosa. Appl Environ Microbiol 60, 2355 (1994).
K.M. Thormann, R.M. Savilee and S. Shukla et al., Induction of rapid detachment in Shewanella oneidensis MR-1 biofilms. J Bacteriol 187, 1014 (2005).
A.C. Lee Wong, Biofilms in food processing environments. J Dairy Sci 81, 2765 (1998).
A.F. Merry, T.E. Miller and G. Findon et al., Touch contamination levels during anaesthetic procedures and their relationship to hand hygiene procedures: a clinical audit. Br J Anaesth 87, 291 (2001).
D.R. Patrick, G. Findon and T.E. Miller, Residual moisture determines the level of touch-contact-associated bacterial transfer following hand washing. Epidemiol Infect 119, 319 (1997).
S.A. Sattar, S. Springthorpe and S. Mani et al., Transfer of bacteria from fabrics to hands and other fabrics: development and application of a quantitative method using Staphylococcus aureus as a model. J Appl Bacteriol 90, 962 (2001).
G. Midelet and B. Carpentier, Transfer of microorganisms, including Listeria monocytogenes from various materials to beef. Appl Environ Microbiol 68, 4015 (2001).
K. Dastorri and B. Makin, Adhesion measurements for electrostatic powder coating using drop test rig and virtual oscilloscope. J Electrost 51, 509 (2001).
P. Stoodley, R. Cargo and C.J. Rupp et al., Biofilm material properties as related to shear induced deformation and detachment phenomena. J Ind Microbiol Biotechnol 29, 361 (2002).
P. Stoodley, D. deBeer and Z. Lewandowski, Liquid flow in biofilm systems. Appl Environ Microbiol 60, 2711 (1994).
C. Picioreanu, M.C.M. van Loodstrecht and J.J. Heijnen, Two-dimensional model of biofilm detachment caused by internal stress from liquid flow. Biotechnol Bioeng 72, 205 (2001).
S.P. Cole, J. Harwood and R. Lee et al., Characterization of monospecies biofilm formation by Helicobacter pylori. J Bacteriol 186, 3124 (2004).
D. Djordjevic, M. Wiedmann and L.A. McLandsborough, Microtiter plate assay for assessment of Listeria monocytogenes biofilm formation. Appl Environ Microbiol 68, 2950 (2002).
G. Ramage, K. VandeWalle and B.L. Wickes, et al., Characteristics of biofilm formation by Candida albicans. Rev Iber Micol 18, 163 (2001).
M. Augustin and T. Ali-Vehmas, Assessment of enzymatic cleaning agents and disinfectants against bacterial biofilms. J Pharmacol Pharm Sci 7, 55 (2004).
R.J.C. McLean, C.L. Bates and M.B. Barnes et al., Methods of studying biofilms. In: Microbial Biofilms, edited by M. Ghannoum and G.A. O'Toole (ASM Press, Washington, DC 2004), p. 379.
H. Ceri, M.E. Olson and C. Stremick et al., The Calgary Biofilm Device: new technology for rapid determination of antibiotic susceptibilities of bacterial biofilms. J Clin Microbiol 37, 1771 (1999).
N. Cerca, G.B. Pier and M. Vilanova et al., Influence of batch or fed-batch growth on Staphylococcus epidermidis biofilm formation. Lett Appl Microbiol 39, 420 (2004).
J.N. Anderl, M.J. Franklin and P.S. Stewart, Role of antibiotic penetration limitation in Klebsiella pneumoniae biofilm resistance to ampicilin and ciprofloxacin. Antimicrob Agents Chemother 44, 1818 (2000).
G. Borrielo, E. Werner and F. Roe et al., Oxygen limitation contributes to antibiotic tolerance of Pseudomonas aeruginosa in biofilms. Antimicrob Agents Chemother 48, 2659 (2004).
E.J. Wentland, P.S. Stewart C.-T. Huang et al., Spatial variations in growth rate within Klebsiella pneumoniae colonies and biofilm. Biotechnol Prog 12, 316 (1996).
X. Chen and P.S. Stewart, Biofilm removal caused by chemical treatments. Water Res 34, 4229 (2000).
B.B. Christensen, C. Sternberg and J.B. Andersen et al., Molecular tools for the study of biofilm physiology. Methods Enzymol 310, 20 (1999).
W.F. McCoy, J.D. Bryers and J. Robbins et al., Observations of fouling biofilm formation. Can J Microbiol 27, 910 (1981).
J.C. Nickel, I. Ruseska and J.B. Wright et al., Tobramycin resistance of Pseudomonas aeruginosa cells growing as a biofilm on urinary catheter material. Antimicrob Agents Chemother 27, 619 (1985).
R.M. Donlan, R. Murga and L. Carson, Growing biofilms in intravenous fluids. In: Biofilms: The Good, the Bad, and the Ugly, edited by J. Wimpenny, P. Gilbert, J. Walker, M. Brading, and R. Bayston (Bioline, Cardiff, Wales, 1999), p. 23.
S. Okabe, T. Itoh and H. Satoh et al., Analyses of spatial distributions of sulfate-reducing bacteria and their activity in aerobic wastewater biofilms. Appl Environ Microbiol 65, 5107 (1999).
R.M. Donlan, J.A. Piede and C.D. Heyes et al., Model system for growing and quantifying Streptococcus pneumonieae biofilms in situ and in real time. Appl Environ Microbiol 70, 4980 (2004).
R.M. Donlan, R. Murga and J. Carpenter et al., Monochloramine disinfection of biofilm-associated Legionella pneumophila in a potable water model system. In: Legionella, edited by R. Marre, Y.A. Kwaik, C. Bartlett, N.P. Cianciotto, B.S. Fiekds, M. Frosch, J. Hacker, and P.C. Lück (American Society for Microbiology, Washington, DC 2002).
D.M. Goeres, L.R. Loetterle and M.A. Hamilton et al., Statistical assessment of a laboratory method for growing biofilms. Microbiology 151, 757 (2005).
A.K. Camper, W.L. Jones and J.T. Hayes, Effect of growth conditions and substratum composition on the persistence of coliforms in mixed-population biofilms. Appl Environ Microbiol 62, 4014 (1996).
L.M. Laurence and A. Gilmore, Characterization of Listeria monocytognes isolated form poultry products and from the poulty-processing envronment by random amplification of polymorphic DNA and multilocus enzyme electrophorsis. Appl Environ Microbiol 61, 2139 (1995).
J.R. Knowles, S. Roller and D.B. Murray et al., Antimicrobial action of carvacrol at different stages of dual-species biofilm development by Staphylococcus aureus and Salmonella enterica Serovar Typhymurium. Appl Environ Microbiol 71, 797 (2005).
J.L. Kadurugamuwa, L.V. Sin and J. Yu et al., Noninvasive optical imaging method to evaluate postantibiotic effects on biofilm infection in vivo. Antimicrob Agents Chemother 48, 2283 (2004).
I. Raad, W. Costerton and U. Sabharwal et al., Ultrastructural analysis of indwelling vascular catheters: a quantitative relationship between luminal colonization and duration of placement. J Infect Dis 168, 400 (1993).
R.C. Hunter and T.J. Beveridge, Application of a pH-sensitive fluoroprobe (C-SNARF-4) for pH microenvironment analysis in Pseudomonas aeruginosa biofilms. Appl Environ Microbiol 71, 2501 (2005).
S.C.C. Foong and J.S. Dickson, Attachment of Listeria monocytogenes on ready-to-eat meats. J Food Prot 67, 456 (2004).
P. Chavant, B. Gaillard-Martinie and M. Hébraud, Antimicrobial effects of sanitizers against planktonic and sessile Listeria monocytogenes cells according to the growth phase. FEMS Microbiol. Lett. 236, 241 (2004).
B. Little and P. Wagner, An overview of microbiologically influenced corrosion of metals and alloys. Can J Microbiol 42, 367 (1996).
J.R. Lawrence and T.R. Neu, Confocal laser scanning microscopy for analysis of microbial biofilms. Methods Enzymol 310, 131 (1999).
P.A. Suci, M.W. Mittelman and F.P. Yu et al., Investigation of ciprofloxacin penetration into Pseudomonas aeruginosa biofilms. Antimicrob Agents Chemother 38, 2125 (1994).
D.E. Nivens, J.Q. Chambers and T.R. Anderson et al., Monitoring microbial adhesion and biolfilm formation by attenuated total reflection/Fourier transform infrared spectroscopy. J Microbiol Methods 17, 199 (1993).
H.C. Flemming, J. Wingender and T. Griegbe et al., Physico-chemical properties of biofilms. In: Biofilms: Recent Advances in their Study and Control, edited by L.V. Evans (Harwood Academic Publishers, Amsterdam 2000), p. 19.
Z. Lewandowski, Structure and function of biofilms. In: Biofilms: Recent Advances in their Study and Control, edited by L. V. Evans (Harwood Academic Publishers, Amsterdam, 2000), Vol. 1–17.
R. Murga, J.M. Miller and R.M. Donlan, Biofilm formation by gram-negative bacteria on central venous catheter connectors: effect of conditioning films in a laboratory model. J Clin Microbiol 39, 2294 (2001).
P.S. Stewart, B.M. Peyton and W.J. Drury et al., Quantitative observations of heterogenesis in Pseudomonas aeruginosa biofilms. Appl Environ Microbiol 59, 327 (1993).
T.C. Zhang and P.L. Bishop, Density, porosity, and pore structure of Biofilms. Water Res 28 (1994).
H. Beyenal, C. Donovan and Z. Lewandowski et al., Three-dimensional biofilm structure quantification. J Microbiol Methods 59, 395 (2004).
S.W. Hermanowicz, U. Schindler and P.A. Wilderer, Fractal structure of biofilms: new tools for investigation of morphology. Water Sci Technol 32, 99 (1995).
V.J.M. Allan, L.E. Macaskie and M.E. Callow, Development of a pH gradient within a biofilm is dependent upon the limiting nutrient. Biotechnol Lett 21, 407 (1999).
J.W. Costerton, Z. Lewandowski and D. DeBeer et al., Biofilms, the customized microniche. J Bacteriol 176, 2137 (1994).
I.B. Beech, J.R. Smith and A.A. Steele et al., The use of atomic force microscopy for studying interactions of bacterial biofilms with surfaces. Colloids Surf, B Biointerfaces 23, 231 (2002).
P.J. Bremer, G.G. Geesey and B. Drake, Atomic force microscopy examination of the topography of hydrated bacterial biofilm on a copper surface. Curr Microbiol 24, 223 (1992).
P.G. Rouxhet, N. Mozes and P.B. Dengis et al., Application of X-ray photoelectron spectroscopy to microorganisms. Colloids Surf, B Biointerfaces 2, 347 (1994).
X. Yang, H. Beyenal and G. Harkin et al., Quantifying biofilm structure using image analysis. J Microbiol Methods 39, 109 (2000).
A. Heydorn, A.T. Nielsen and M. Hentzer et al., Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 146, 2395 (2000).
C. Niu and E.S. Gilbert, Colorimetric method for identifying plant essential oil components that affect biofilm formation and structure. Appl Environ Microbiol 70, 6951 (2004).
C.J. Van Oss, R.J. Good and M.K. Chaudry, The role of van der Waals forces and hydrogen bonds in ‘hydrophobic interactions’ between biopolymers and low energy surfaces. J Colloid Interface Sci 111, 378 (1986).
K. Triandafillu, D.J. Balazs and B.O. Aronsson et al., Adhesion of Pseudomonas aeruginosa strains to untreated and oxygen-plasma treated poly(vinyl chloride) (PVC) from endotracheal intubation devices. Biomaterials 24, 1507 (2003).
L.S. Stone and E.A. Zottala, Effect of cleaning and sanitizing on the attachment of Pseudomonas fragi to stainless steel. J Food Sci 50, 957 (1985).
S.E. Lentsch, Sanitizers for an effective cleaning program. In: Sanitation Notebook for the Seafood Industry, edited by G.J. Flick, C.L. Kassem, F. Huang, D.R. Ward, M.J. Thompson and C. Fletcher (Virginia Polytechnic Institute and State University Blacksburg, VA 1978), p. 11.
C.K. Bower, J. McGuire and M.A. Daeschel, The adhesion and detachment of bacteria and spores on food-contact surfaces. Trends Food Sci Technol 7, 152 (1996).
R.M. Donlan and J.W. Costerton, Biofilms: Survival mechanisms of clinically relevant microorganisms. Clin Microbiol Rev 15, 167 (2002).
R.M. Donlan, Biofilms: microbial life on surfaces. Emerg Infect Dis 8, 881 (2002).
J.F. Frank and R.A. Koffi, Surface-adherent growth of Listeria monocytogenes is associated with increased resistance to surfactant sanitizers and heat. J Food Prot 53, 550 (1990).
J.C. Nickel and J.W. Costerton, Bacterial biofilms and catheters: A key to understanding bacterial strategies in catheter-associated urinary tract infection. Can J Infect Dis 3, 619 (1992).
H. Anwar, J.L. Strap and J.W. Costeron, Establishment of aging biofilms: possible mechanism of bacterial resistance to antimicrobial therapy. Antimicrob Agents Chemother 36, 1347 (1992).
T.S. Schwach and E.A. Zottola, Use of scanning eletron microscopy to demonstrate microbial attachment to beef and beef contact surfaces. J Food Sci 47, 1401 (1982).
S.H. Lee and J.F. Frank, Inactivation of surface-adherent Listeria monocytogenes hypochlorite and heat. J Food Prot 54, 4 (1991).
M.R.W. Brown and P. Gilbert, Sensitivity of biofilms to antimicrobial agents. J Appl Bacteriol 74, S87 (1993).
D.E. Jenkins, J.E. Schultz and A. Matin, Stravation-induced cross protection against heat or hydrogen peroxide challeneg of Escherichia coli. J Bacteriol 170, 3909 (1988).
D.J. Evans, D.G. Allison and M.R.W. Brown et al., Susceptibility of Pseudomonas aeruginos and Escherichia coli biofilms towards ciprofloxacin: Effect of specific growth rate. J Antimicrob Chemother 27, 177 (1991).
P.S. Stewart, Biofilm accumulation model that predicts antibiotic resistance of Pseudomonas aeruginosa biofilms. Antimicrob Agents Chemother 38, 1052 (1994).
P.S. Stewart, Theoretical aspects of antibiotic diffusion into microbial biofilms. Antimicrob Agents Chemother 40, 2517 (1996).
C.M. Stewart, M.B. Cole and J.D. Legan et al., Modeling the growth boundary of Staphylococcus aureus for risk assessment purposes. J Food Prot 64, 51 (2001).
P.S. Stewart, J. Rayner and F. Roe et al., Biofilm penetration and disinfection efficiancy of alkaline hypochlorite and chlorsulfamates. J Appl Microbiol 91, 525 (2001).
C.M. Stewart, M.B. Cole and J.D. Legan et al., Staphylococcus aureus growth boundries: moving towards mechanistic predictive models based on soute-specific effects. Appl Environ Microbiol 68, 1864 (2002).
P.M. Davidson, Chemical Preservatives and Natural Antimicrobial Compounds. In: Food Microbiology: Fundamentals and Frontiers, edited by M.P. Doyle, L.R. Beuchat and T.J. Montville (American Society for Microbiology, Washington, DC 2001), p. 593.
P.M. Davidson and A.S. Naidu, Phytochemicals. In: Natural Food Antimicrobial Systems, edited by A.S. Naidu (CRC Press, Boca Raton, FL 2000).
E. Drenkard, Antimicrobial resistance of Pseudomonas aeruginosa biofilms. Microbes Infect 5, 1213 (2003).
E. Le Magrex-Debar, J. Lemoine and M.-P. Gelle et al., Evaluation of biohazards in dehydrated biofilms on foodstuff packaging. Int J Food Microbiol 55, 239 (2000).
J. Knowles and S. Roller, Efficacy of Chitosan, Carvacrol, and a Hydrogen Peroxide-Based Biocide against Foodborne Microorganisms in Suspension and Adhered to Stainless Steel. J Food Prot 64, 1542 (2001).
W.M. Dunne, E.O. Mason and S.L. Kaplan, Diffusion of rifampin and vancomycin through a Staphylococcus epidermis biofilm. Antimicrob Agents Chemother 37, 2522 (1993).
H. Kumon, K. Tomochika and T. Matunaga et al., A sandwich cup method for the penetration assay of antimicrobial agents through Pseudomonas exopolysaccharide. Microbiol Immunol 38, 615 (1994).
P.S. Stewart, Diffusion in Biofilms. J Bacteriol 185, 1485 (2004).
W.K. Whitekettle, Effects of surface-active chemicals on microbial adhesion. J Ind Microbiol 7, 105 (1991).
E.P. Krysinski, L.J. Brown and T.J. Marchisello, Effect of cleaners and sanitizers on Listeria monocytogenes attached to product contact surfaces. J Food Prot 55, 246 (1992).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
McLandsborough, L., Rodriguez, A., Pérez-Conesa, D. et al. Biofilms: At the Interface between Biophysics and Microbiology. Food Biophysics 1, 94–114 (2006). https://doi.org/10.1007/s11483-005-9004-x
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11483-005-9004-x