Abstract
Purpose
The aim of this study was to elucidate the impact of plant on the activity, abundance, and the community composition of the ammonia oxidizers, including both ammonia-oxidizing archaea (AOA) and bacteria (AOB). Moreover, the relationship between AOA and AOB in mangrove sediment was also analyzed.
Materials and methods
Sediment used for microcosm experiments was collected in the mangrove wetland. The native plant species Kandelia obovata and invasive species Spartina alterniflora were selected. Nitrifying activity was determined by assaying the potential nitrification rate (PNR). Abundances of gene and transcript were measured via real-time quantitative PCR (qPCR). Terminal restriction fragment length polymorphism (T-RFLP) was used to analyze the nitrifier community structures. Clone libraries were constructed for further phylogenetic analysis.
Results and discussion
PNR and abundances of both AOA and AOB were greatly enhanced in the vegetated sediments. S. alterniflora showed a greater promoting effect on nitrification activity, indicating the potential of exotic invasion on perturbing the nitrogen balance. Abundance of AOB transcript was hundreds of times higher than that of AOA. Moreover, ammonia-oxidizing communities were distinctly grouped responding to vegetation with two plant species. Along with direct impact of plants, these variations are as well related to the different sediment properties. Phylogenetic analysis revealed that both AOA and AOB communities formed apparent clusters. The latter contained two Nitrosomonas and Nitrosospira clusters. The Nitrosomonas genus is predominant over Nitrosospira genus, which might be due to the better adaption of Nitrosomonas to the flooded habitats in sediments.
Conclusions
Wetland plants were found to influence the activity, abundance, and community structure of ammonia oxidizers. Moreover, AOB is suggested to be more significant than AOA in nitrogen cycling in mangrove wetlands.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Mangrove ecosystems are natural intertidal wetlands widely distributed along estuaries in the tropical and subtropical regions, serving as breeding, growing, refuge, and feeding zones for many oceanic organisms (Holguin et al. 2001). Microorganisms, specifically bacteria, play dominant roles in all of the principal nitrogen transformation processes (Purvaja et al. 2008). Nitrification, as part of the nitrogen cycling, is a two-step aerobic process in which ammonia is stepwise oxidized to nitrate. The produced nitrate could be used by microorganisms through denitrification, a process reducing nitrate to gaseous products and returning nitrogen to the atmosphere. Nitrification–denitrification processes are responsible for nitrogen removal in wetlands, thus can prevent eutrophication (Angeloni et al. 2006).
Ammonia oxidation is the first and rate-limiting step of nitrification and is conducted by members of both bacteria and archaea. For a long time, only chemolithoautotrophic AOB were considered to be the driver for ammonia oxidization until the discovery and isolation of the ammonia-oxidizing Crenarchaeote Nitrosopumilus maritimus (Könneke et al. 2005). AOB are mainly composed of the β-proteobacteria genera Nitrosomonas and Nitrosospira (Head et al. 1993; Purkhold et al. 2003) and the γ-proteobacteria genus Nitrosococcus (Ward and O'Mullan 2002). AOA belongs to the phylum Thaumarchaeota (Alves et al. 2013). The catalytic subunit of ammonia monooxygenase (AMO) is encoded by the amoA gene which has been widely used to study the distribution and diversity of AOA and AOB (Francis et al. 2005; Schleper et al. 2005).
The relationship between AOA and AOB for ammonia oxidation remains unclear, and limited information is available on how these two groups respond and interact with each other in the nitrogen transformation processes. It is generally recognized that AOA predominate among ammonia-oxidizing prokaryotes in soil ecosystems (Leininger et al. 2006), particularly in acidic soils (Gubry-Rangin et al. 2010; Yao et al. 2011; Zhang et al. 2012). However, many recent studies have found that AOB are more abundant than AOA in agricultural soil (Jia and Conrad 2009), grassland soil (Di et al. 2009), and mangrove sediment (Li et al. 2011). These studies suggest that relationship between AOA and AOB varies in different environments. Nevertheless, the relationship between AOA and AOB is still limited in wetland ecosystems when compared with upland soils. It is therefore necessary to differentiate the roles of AOA and AOB in regulating biogeochemical nitrogen cycling in wetland ecosystems.
In wetlands, nitrification is mainly related to the sediment and other submersed surfaces (Purvaja et al. 2008; Ruiz-Rueda et al. 2009) and as well in the micro-oxic zones created by the diffusion of oxygen via aerenchyma to the plant roots and surrounding sediments (Philippot et al. 2009). Plant roots can also exude low molecular weight compounds serving as carbon sources for microorganisms (Philippot et al. 2013a). However, wetland plants also compete with microbes for nutrients since most of them are able to assimilate ammonium, nitrate, urea, and amino acids as nitrogen sources (Skiba et al. 2011), which indicates the potential of plants in regulating the nitrification process. A case study in mangrove sediments revealed that the effect of mangrove plants on abundances and community structures of AOA and AOB is species dependent (Li et al. 2011). Moreover, Laanbroek et al. (2012) showed that soil chemical properties associated with vegetation densities affected community structures of AOB, while an earlier study suggested that variations of nitrification potential between different soils were related to vegetation type rather than the soil physiochemical properties (Wedin and Tilman 1990). These studies emphasize the importance of plant and plant species in driving the microbial nitrification process, resulting in variations in the abundances and community structures of corresponding nitrifiers in the vicinity of plant roots. Additionally, only a research group from the University of Hong Kong has comprehensively assessed the nitrification process with respect to both AOA and AOB in mangrove wetland (Cao et al. 2011; Li et al. 2011; Li and Gu 2013). However, these studies have not fully answered how the activities, abundances, and community structures of AOA and AOB respond to wetland plants. Hence, investigations on the relationship between ammonia oxidizers and mangrove plants are required.
Over the last few decades, the exotic plant Spartina alterniflora has largely invaded eastern China since their first introduction from North Carolina to China in 1979 (An et al. 2007). S. alterniflora has formed single habitat in the Jiulong River estuary in Fujian Province, which has threatened to the native mangrove ecosystems (Zhang et al. 2011). In this Mangrove Nature Reserve, several field studies have been conducted to investigate the response of biogenic elements and microorganisms to the invasion of S. alterniflora with comparison to the native mangrove species Kandelia obovata (Zhang et al. 2011; Yu et al. 2014). However, it is somehow difficult to intuitively understand the role of single environmental factor in an open field study. Hence, in the present study, we performed microcosm experiments to investigate the shifts of the activities, abundances of both functional genes and their transcripts, and community structures of AOA and AOB in bulk and vegetated mangrove sediments. The invasive species S. alterniflora and native species K. obovata were selected for better understanding the invasive disturbance on the sediment nitrogen cycling. This study further aims to elucidate the impact of plant and plant species on the nitrification process in the mangrove wetland, thus providing new insights into the relationship between plants and ammonia-oxidizing microorganisms.
2 Materials and methods
2.1 Experiment setup
Sediment was sampled in the mangrove wetland located in the mud flat of Jiulong River estuary in Fujian Province, China (24° 23′ to 24° 30′ N; 117° 45′ to 118° 05′ E). The sediment used for microcosm was collected from 0 to 50 cm in the unvegetated area and homogenized evenly after collection. The seedlings (embryonic axis) of K. obovata hanging on the tree were also collected for cultivation, and S. alterniflora seedlings were from seeds germinated in the lab. An aliquot of 4 kg fresh sediment was added to each PVC container (height 25 cm; volume 5 L), and then, four similar seedlings of K. obovata (KO treatment) and S. alterniflora (SA treatment) were planted in the corresponding container, respectively. The containers without plants were considered as the control (bulk treatment). Each treatment contained at least three replicates, and all the treatments were cultivated for 2 months under the same conditions in the greenhouse. The air temperature in the greenhouse was kept around 30 °C in the day time and around 20 °C in the night.
2.2 Sediment sampling and analysis of physicochemical properties
Three containers in which the plant growths were in a similar condition for each treatment were selected, and surface sediment (0–10 cm) for each container was sampled. For the vegetated treatments, the sediments were collected in the vicinity of the plant roots. Roots and gravel were removed using sterile tweezers. Samples for DNA extraction were frozen immediately in liquid nitrogen and kept at −80 °C for further study. However, RNA was extracted immediately from fresh sediment and kept at −80 °C until analysis. RNA analysis was finished within 1 month. Sediment moisture was determined gravimetrically by drying the soil at 105 °C for 12 h. The salinity in pore water was measured by the MASTER-S/MillM salinity refractometer (ATAGO, Japan). For the pH, sediments were freeze-dried and sieved (<2 mm) and then measured with a XL60 pH meter (Fisher Scientific, USA) in a dry sediment to H2O proportion of 1/2.5 (g/mL). Sediment was further sieved (<0.15 mm) for the determination of total carbon (TC) and total nitrogen (TN) by using a Vario MAX CNS elemental analyzer (ELEMENTAR, Germany). Ammonium (N-NH4 +), nitrate, and nitrite (N-NO x −) concentrations were analyzed using 2 M KCl extracts with a fresh sediment to KCl solution ratio of 1:10 (g/mL), which were further measured by a FIA QC8500 continuous flow injection analyzer (LACHAT, USA).
2.3 Potential nitrification rate (PNR)
The PNR was determined as accumulated nitrate in a short incubation according to Hart et al. (1994). Briefly, 100 mL of phosphate buffer (1.0 mM) with ammonium sulfate (1.5 mM) at pH 7.2 was added to 15 g fresh sediment followed by incubation at 25 °C for 24 h on a rotatory shaker at 180 rpm. Aliquots of 10-mL subsample were collected at 2, 4, 22, and 24 h after the start of the incubation. The samples were filtered (0.22 μm), and the nitrate concentration was measured by an ion chromatography (Dionex ICS-3000, USA). The nitrification rates were calculated by linear regression of the accumulated NO3 −-N g−1 dry weight sediment versus time.
2.4 Nucleic acid extraction and quantification of amoA gene and transcript
Total DNA was extracted from 0.5 g fresh sediment using the FastDNA SPIN Kit for Soil (MP Biomedicals, Santa Ana, CA, USA) according to the manufacturer’s instructions. Total RNA was extracted from 5-g fresh sediment with the RNA PowerSoil Total RNA Isolation Kit (MO BIO, Carlsbad, USA) according to the manufacturer’s instructions. Concentration and purity of the extracted DNA and RNA were determined by a UV–vis spectrophotometer ND-1000 (NanoDrop, USA). The extracted RNA was treated with DNase I (RNase free) (Thermo Scientific, MA, USA) to remove the residual DNA. Then, cDNA was synthesized by reverse transcription with RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific) using the same amount of RNA from each sample. Transcription system without transcriptase was conducted as a control to ensure that DNA was completely removed in the extracted RNA. Both of the synthesized cDNA and controls were stored at −80 °C and used for qPCR.
The primer pairs of Arch-amoAF/Arch-amoAR (Francis et al. 2005) and amoA-1F/amoA-2R (Rotthauwe et al. 1997) were used for amplifying the archaeal amoA gene and the β-proteobacteria amoA gene, respectively. A 20-μL of PCR mixture contained 10 μL of 2× SYBR Premix Ex Taq II, 0.4 μL of 50× ROX Reference Dye II (Takara, Japan), 0.2 μM of each primer, 2 μL of DNA or cDNA (20 ng), and 0.2 μL of 0.1 % bovine serum albumin (BSA) (Takara). Reactions were conducted on an Applied Biosystems 7500 Real-Time PCR System (USA). For AOA, the condition was 1 cycle of 94 °C for 8 min and then 40 cycles of 94 °C for 45 s, 61.5 °C for 1 min, and 72 °C for 1 min. For AOB, the condition was 1 cycle of 94 °C for 3 min and then 40 cycles of 94 °C for 30 s, 55 °C for 30 s, and 72 °C for 45 s. The fluorescence was recorded during each cycle at 72 °C. Standard curves were obtained using serial dilutions of standard plasmids containing archaeal amoA gene and β-proteobacterial amoA gene with known copy numbers. Standard plasmids were extracted from positive clones obtained by ligating the gel-purified PCR products into the pMD19-T Vector (TAKARA, Japan) and using ligation products to transform Escherichia coli DH5α competent cells. Negative controls without DNA template were included for each amplification. Each sample was quantified in three parallel qPCR reactions to ensure the correct amplification. Inhibition was eliminated by highly diluting the DNA extracts. Only the reactions with efficiencies above 90 % and correlation coefficients (r 2) above 0.99 were accepted.
2.5 PCR amplification, clone libraries, and phylogenetic analysis
Twenty microliters of PCR reaction mixture consisted of about 10 to 20 ng of DNA, 0.4 μM of each primer, 0.25 μL of 0.1 % BSA, 0.32 mM of dNTP, 2.5 μL of 10× PCR buffer (Mg2+ Plus), and 1 U rTaq (Takara). PCR conditions of AOA were 1 cycle of 95 °C for 3 min, then 35 cycles of 94 °C for 1 min, 53 °C for 45 s and 72 °C for 1 min, and 72 °C for 10 min. For β-AOB, the condition was 1 cycle of 95 °C for 3 min, then 35 cycles of 94 °C for 1 min, 55 °C for 45 s and 72 °C for 1 min, and 72 °C for 10 min. PCR products of replicates were pooled for cloning. Briefly, three AOA and three AOB amoA clone libraries representing three different vegetation types were constructed. For sequencing, 60 clones for each library were selected. Operational taxonomic unit (OTU) was picked at 97 % similarity level using Mothur 1.19 (Schloss et al. 2009). The DNA sequences representative to each OTU were blasted against reference sequences form the GenBank database for identification. Then, these DNA sequences were deduced to amino acid to construct the neighbor-joining phylogenetic trees using MEGA 6.06 software with 1,000 times bootstrap.
The GenBank accession numbers for the AOB and AOA amoA sequences were KJ778204-KJ778376 and KJ778377-KJ778553, respectively.
2.6 Terminal restriction fragment length polymorphism (T-RFLP) analysis
T-RFLP was used to determine the composition of the nitrifying communities. The forward primers were fluorescently labeled at the 5′ end with 6-carboxyfluorescein (FAM). PCR products were verified through an electrophoresis on a 1 % agarose gel. The expected fragments (631 and 491 bp for AOA and AOB amoA, respectively) were excised from the gel and purified with Wizard SV Gel and PCR Clean-Up System (Promega, Madison, USA). Purified products were digested with RsaI and MspI (New England BioLabs, Beverly, MA) for AOA (Boyle-Yarwood et al. 2008) and RsaI and HhaI (New England BioLabs) for AOB (Jin et al. 2010). All the digestions were performed at 37 °C for 3 h in 20 μL of reaction mixtures according to the instructions. The digestions were terminated in an 80 °C water bath for 20 min.
Digests were then precipitated by ethanol with sodium acetate and dissolved in 10 μL sterilized MilliQ water. Purified digests labeled with fluorescence were detected using a 3730×l DNA Analyzer (Applied Biosystems, USA) by Invitrogen Biotechnology Corporation (Guangzhou, China). GS-ROX500 and GS-LIZ1200 were used as internal standards for AOA and AOB digests, respectively. The size and relative abundance of terminal restriction fragments (T-RFs) were analyzed using GeneMapper software (version 4.1, ABI, USA). T-RFs with a size ≥50 bp were chosen for cluster analysis. T-RFs with relative abundance below 1 % of sum of all peak areas were excluded, and T-RFs within 2 bp were combined together and considered as one T-RF.
2.7 Statistical analysis
Nonmetric multidimensional scaling (nMDS) was performed for T-RFLP data to determine the effect of plant on the community composition. The similarity matrix used for nMDS analysis was calculated from the T-RF relative abundances using the Bray-Curtis coefficient. The analysis of similarity (ANOSIM) based on the Bray-Curtis similarity matrix was used to assess the variations between treatments. The similarity percentages (SIMPER) routine was performed to identify the T-RFs responsible for variations between different treatments or for the major contribution in a single treatment. nMDS, AOSIM, and SIMPER were performed in Primer v5.2.9 software (Primer-e, Plymouth, UK). Diversity indices were analyzed by R software v3.0.2 (R Foundation for Statistical Computing, Vienna, Austria) with the vegan and MASS packages.
Analysis of variance (ANOVA) was performed on the sediment physiochemical properties, abundances, and transcripts of AOA and AOB amoA genes, as well as the diversity indices. Pearson correlations were performed between the sediment physiochemical properties and potential nitrification activity, amoA abundances, the diversity indices and the values of nMDS axes. One-way ANOVA and Pearson correlation tests were performed using SPSS v19.0 software (IBM, USA), and significance was accepted at the 0.05 level (P < 0.05). Data failing to meet the assumptions of ANOVA were log transformed or analyzed by Dunnett’s post hoc test.
3 Results
3.1 Physiochemical properties of sediments and potential nitrification rate
The physiochemical properties of sediments in different treatments are presented in Table 1. Salinity was significantly higher in the vegetated treatments compared to the bulk treatment (P < 0.05). In contrast, ammonium concentration and pH were higher in the bulk sediment than in the vegetated sediments (P < 0.05). However, other properties showed no significant variation between vegetated and bulk sediments. In addition, no obvious difference was observed between the two plant species for all properties.
The nitrification rates were 0.21 ± 0.07, 0.90 ± 0.01, and 1.26 ± 0.16 mg NO3 − kg−1 dry weight sediment h−1 in bulk, KO, and SA sediment, respectively (Table 1). The nitrification rates in vegetated treatments were four to six times of the bulk treatment. And the nitrification rate in the SA treatment was significantly higher than that in the KO treatment (P < 0.05).
3.2 Abundances of amoA gene and transcript
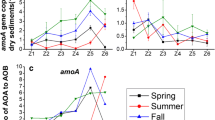
The abundances of amoA genes for AOA and AOB ranged from 1.79 × 106 to 1.26 × 107 and 7.11 × 106 to 3.84 × 107 copies g−1 dry sediment, respectively (Fig. 1a). Transcript abundances of AOA and AOB ranged from 3.76 × 104 to 8.89 × 104 and 1.80 × 107 to 3.73 × 107 copies ng−1 RNA, respectively (Fig. 1b). By calculating the ratios of bacterial to archaeal amoA gene copy numbers and transcripts, it was obvious that the abundance of AOB was significantly higher than AOA abundance, specifically at the mRNA levels where the AOB transcript abundance was about 480 times higher than that of AOA (Fig. 1). In addition, amoA gene abundance for AOB was significantly higher in the vegetated sediments compared to the bulk sediment (Fig. 1a). However, for AOA, there is no significant difference between KO and bulk treatments, while abundance was significantly higher in the SA treatment compared to the bulk treatment (Fig. 1a). Transcript abundance for AOA in both vegetated treatments was higher when compared to the bulk treatment, while no obvious difference was observed between plant species (Fig. 1b). For AOB, only the SA treatment showed a higher abundance when compared to the bulk treatment (Fig. 1b). Abundances of both AOA and AOB amoA genes and transcripts in the SA treatment were significantly higher than those in the bulk sediment, which thereby presented a stronger promoting effect of S. alterniflora when compared with K. obovata.
Abundances of archaea and bacteria amoA genes (a) and transcripts (b) in different vegetation treatments. Error bars represent standard deviation of the mean (n = 3), and different letters above the bars indicate significant differences between three different treatments for AOA and AOB, respectively. The ratios of bacterial to archaeal amoA gene copy numbers and transcripts are shown in boxes above the bars. Bulk, bulk sediment; KO, Kandelia obovata; SA, Spartina alterniflora; dw, dry weight
3.3 Phylogenetic analysis of AOA and AOB
Totally, 177 and 173 sequences were obtained for AOA and AOB phylogenetic analysis, respectively. For AOA clones, 25 OTUs were obtained which formed two major clusters (Fig. 2). Most of the obtained sequences were related to the clones obtained from mangrove and river sediment, while the others were related to the clones from marine and estuarine sediment. Moreover, several OTUs were also related to the cultured archaea genera of Nitrososphaera, Cenarchaeum, and Nitrosopumilus (Fig. 2). OTU1 and OTU2 were the dominant OTUs which occupied about 22 and 41 % of the total clone sequences, respectively.
Neighbor-joining phylogenetic tree based on the archaeal amoA-deduced amino acid sequences (black up-pointing triangle, bulk sediment; black circle, Kandelia obovata; black square, Spartina alterniflora). Numbers after the symbols represent the numbers of sequences in each treatment. Bootstrap values above 50 % are shown
For AOB clones, 31 OTUs were obtained and could be divided into three distinct clusters, affiliating to genera of Nitrosomonas and Nitrosospira (Fig. 3). Most of the AOB clones were related to the Nitrosomonas genus, indicating a predominance of Nitrosomonas over Nitrosospira in sediments.
Neighbor-joining phylogenetic tree based on the bacterial amoA-deduced amino acid sequences (black up-pointing triangle, bulk sediment; black circle, Kandelia obovata; black square, Spartina alterniflora). Numbers after the symbols represent the numbers of sequences in each treatment. Bootstrap values above 50 % are shown
3.4 T-RFLP analysis of AOA and AOB
The relative abundance of T-RFs in a T-RFLP profile was used to analyze the differences between samples (Fig. 4). There were 9 and 17 T-RFs with a relative abundance above 1 % for AOA and AOB amoA genes, respectively. The lowest AOA and AOB richness both occurred in the SA treatment, which was consistent with the values of Shannon and Simpson indices (Table 2). The AOB evenness values showed a similar trend to richness. However, the AOA evenness was significantly higher in the bulk treatment than in the vegetated treatments.
SIMPER analysis showed that the dominant AOA T-RFs in the bulk, KO, and SA treatments were 298, 56, and 56 bp, contributing about 29.1, 49.6, and 74.1 %, respectively. For AOB, T-RF of 97 bp dominated in all treatments, accounting for 48.7, 46.6, and 79.2 % in the bulk, KO, and SA treatments, respectively.
nMDS analysis showed that both AOA and AOB community compositions formed three distinct groups with three replicates comprising of the three vegetation types: bulk, KO, and SA (Fig. 5). ANOSIM revealed that treatments significantly differed in community composition for both AOA and AOB (AOA, global R = 0.992, significance level = 0.004; AOB, global R = 0.827, significance level = 0.004). SIMPER results pointed to the fact that the T-RF of 56 bp was the major contributor to the variations for AOA communities, while it was 64 and 97 bp T-RFs that contributed most to the AOB communities.
3.5 Correlations of environmental factors with PNR, gene copies and transcripts, diversity, and structure of ammonia oxidizer communities
Pearson’s correlation analysis was performed to find out whether there were some important environmental factors influencing the PNR, abundances, and community structures of AOA and AOB (Table 3). Results showed that salinity, pH, and ammonium were the main factors affecting nitrifying process in the sediment, since they were significantly correlated with PNR (P < 0.01), amoA transcripts of both AOA and AOB (P < 0.01), Simpson index and evenness of AOA (P < 0.05), axis 1 of AOA (P < 0.01), and axis 2 of AOB (P < 0.05) (Table 3). However, it seemed that other properties had no strong correlation with these determined parameters. Furthermore, PNR was also significantly and positively correlated with amoA transcripts of both AOA (P < 0.01) and AOB (P < 0.001) (Table 3). However, PNR was only correlated with nMDS axis 1 of AOA (P < 0.001) (Table 3).
4 Discussion
4.1 Sediment properties as potential drivers for ammonia-oxidizing process
In this study, plants significantly altered the sediment salinity, pH, and ammonium concentration (Table 1), which are important factors in shaping the ecological niches of AOA (Erguder et al. 2009) and as well influencing the diversity and abundance of AOB (Li et al. 2011; Zhang et al. 2012; Li and Gu 2013). Our results are in agreement with these studies given the salinity, pH, and ammonium concentration were all correlated to the PNR, amoA transcript numbers, and community diversities and structures of ammonia-oxidizing microorganisms (Table 3). This indicated the potential roles of these properties in driving the ammonia oxidation. Also, mechanisms on how plants affected these properties have been commendably explained by previous studies (Berg and Smalla 2009; Philippot et al. 2009, 2013a). It is demonstrated that root uptake may lead to the fluctuation of ammonium and nitrate concentrations in the rhizosphere. Salinity in the rhizosphere is possibly changed by the plant root activities. The release and assimilation of ions by roots and the fact that root exudates usually include compounds such as organic acids and amino acids could result in the change of pH. Furthermore, if sufficient oxygen is introduced by the plants, reduced sulfur compounds might be oxidized to sulfuric acid, thus leading to a lower pH in the vegetated sediments. Moreover, these changing environmental factors, in turn, will influence the abundances and compositions of bacteria and archaea, the final driving force for the nitrification activity. In this study, variations in the nitrification activity were most probably due to changes in the transcript abundance of ammonia oxidizers as revealed by correlation analysis (Table 3). However, community structure showed a minor effect on the activity even though AOA axis 1 was significantly correlated with PNR (Table 3). Hence, we suggest that the shifts on the abundances and community structures of ammonia oxidizers could be associated with changes of these important properties caused by plants, and variations in the abundances further resulted in the shifts of nitrification activity.
4.2 Activities and abundances of ammonia oxidizers promoted by plants
Results of PNR measurements and quantification of amoA gene and transcript indicated that the existence of plants could stimulate nitrifying activity and ammonia oxidizer abundances in mangrove sediments (Table 1 and Fig. 1). In line with our results, several studies have revealed the promoting effect on nitrification by vegetation as shown by the enhancement of nitrifying activities or AOA and AOB abundances in wetland and rice paddy soils which are both under flooded conditions (Briones et al. 2002; Ruiz-Rueda et al. 2009; Li et al. 2011). Being subject to daily tidal changes, mangrove is under prolonged flooding conditions which may restrain the decomposition of soil organic carbon, leading to high carbon storage. This carbon sequestration may benefit the microbial activities in sediments, and this benefit may be further amplified by plants due to their strategies to transport oxygen to the roots and surrounding environments (Philippot et al. 2009). However, this benefit might be offset in other upland soils, as revealed by some other studies reporting the negative effects on nitrification in the rhizosphere (Lensi et al. 1992; Norton and Firestone 1996; Priha et al. 1999; Herman et al. 2006), given that relative lack of organic sources could result in the competition between plants and microbes. Higher nitrification rates, in addition to root uptake, may lead to the decrease of ammonium concentration, which would in turn limit the ammonia oxidizers. Ammonia oxidation was promoted by plants even though a distinct decrease of ammonium concentration was observed in the vegetated sediments compared to the bulk sediment (Table 1), which suggests that wetland plants could stimulate the nitrification in conditions where nitrogen is not limited.
In the present study, S. alterniflora showed a greater promoting effect on nitrification rates than K. obovata. This might be due to the difference in root physiology that S. alterniflora belongs to the grass family with developed root system which can extend to as deep as 100 cm in the sediment, while K. obovata is a kind of tree whose brace root or buttress root is less developed owing to their adaption to the living environment by rooting superficially (Wang et al. 2006; Pi et al. 2009). S. alterniflora is believed to be inclined to flooding as its shoot growth was stimulated when waterlogging was imposed in addition to salinity (Colmer and Flowers 2008). Species with higher tolerance of waterlogging tend to form deeper roots and possess higher root porosity (Colmer 2003), which can produce sufficient oxygen to the rhizosphere. Another study revealed that S. alterniflora invasion significantly enhanced the organic carbon and total nitrogen levels in the soil compared with bare flat and native plant communities in a costal wetland (Yang et al. 2013). Oxygen and nutrient sources are augmenters for ammonia oxidizers; hence, higher nitrification rate was observed in SA treatment in this study. Owing to the various physiology of different plants, previous studies also observed the difference in nitrification rates with different plant species (Reddy et al. 1989; Wheatley et al. 1990; Steltzer and Bowman 1998; Tsiknia et al. 2013). Moreover, abundance of ammonia oxidizers was found to be different with eight biological wastewater treatment plants (Gao et al. 2013). It is, however, yet poorly understood on the principles of the plant species in differentiating the niche of ammonia oxidizers in mangrove wetlands. Nevertheless, the present study still provides strong evidence on the microbe–nutrient–plant relationship in mangrove systems as revealed by Holguin et al. (2001). Additionally, S. alterniflora is considered to be highly invasive around the world (An et al. 2007) and to strongly impact the diversities and abundances of AOA and AOB in salt marsh sediment (Moin et al. 2009). Given that S. alterniflora might significantly disturb the nitrogen balance in the field taking plant coverage into account, we therefore suggest that how this invasive plant drives the microbial processes should come into notice in future studies.
4.3 Community structures of ammonia oxidizers shaped by plants
This study revealed the influence of plants on AOA and AOB community structures. Our result compares favorably with earlier observations. It has been demonstrated that vegetation influenced the community structure of nitrifying and denitrifying bacteria in the root zone sediments in constructed wetlands, but the plant species effect was limited to ammonia-oxidizing bacteria community (Ruiz-Rueda et al. 2009). In the Mai Po mangrove wetland, Li et al. (2011) suggested that it was AOB not AOA communities that were grouped by distances between sampling sites and mangrove trees, which indicated the effect of mangrove trees on AOA and AOB community structures. In a black mangrove in the USA, Laanbroek et al. (2012) showed that AOB community structures were shaped by vegetation coverage with which different mangrove habitats were formed. Furthermore, the plant species effect on ammonia-oxidizing community structures was observed in grassland soils as well (Patra et al. 2006). These plant-dependent effects might result from specific linkages between plants and microbes with respect to their preferences to each other. By this way, specific microenvironment was created and specific microorganisms were selected, as shown in this study that AOA T-RF 56 and AOB T-RF 97 flourished in vegetated and S. alterniflora sediment, respectively (Fig. 4). In addition, denitrifier communities were also convinced to be easily shifted by these two wetland plants in mangrove sediment in our previous study (Wang et al. 2014). Consequently, plant exhibited a great capacity to shape the niches of microbes involved in nitrogen cycling as well the potential in affecting the nitrogen balance. The plant effect also occurred with the change of sediment properties. However, disentangling mechanism and linkage between different plant species and these microbes is a challenging task that warrants further investigations.
Phylogenetic analysis showed that most of the AOA clone sequences were related to the uncultured archaea clones, and hence, information is limited on how these microbes respond to the environmental factors. This suggests that isolation and characterization of cultivable archaeal nitrifiers are still required in the near future. The predominance of Nitrosomonas over Nitrosospira genus for AOB clones might be due to the better adaption of Nitrosomonas to the flooded habitats. This was demonstrated by Laanbroek and Speksnijder (2008). They showed that the Nitrosomonas oligotropha lineage dominated in the habitats which were more frequently flooded and active in relation to ammonium oxidation in tidal freshwater wetlands, while the Nitrosospira lineage was more abundant in less frequently flooded and less active nitrifying habitats.
In this study, S. alterniflora decreased the community diversities of ammonia oxidizers but still showed a promoting effect on nitrification activity and nitrifier abundances. In contrast to our study, Philippot et al. found that diversity loss lowered potential denitrification rate involved in nitrogen cycling, whereas the abundances of corresponding denitrifying genes showed no variations (Philippot et al. 2013b). The different results might be due to the fact that microbes with strong nitrification activity had been selected by plants in this study, while dilution method as applied by Philippot et al. could only select microbes by chance.
4.4 Relative importance of AOA and AOB in mangrove sediment
The relative contribution of AOA and AOB to soil nitrification remains a matter of debate. It is believed that soil pH has a considerable effect on the ecology of ammonia oxidizers (Yao et al. 2011). The community and phylogeny of ammonia oxidizers have been related to a soil pH gradient (4.9–7.5), with a decrease of archaeal amoA gene and transcript abundance and an increase of bacterial amoA gene transcripts with increasing soil pH (Nicol et al. 2008). Studies suggest that archaea is able to survive under extreme pH and salinity and even low ammonia availability (Erguder et al. 2009; Martens-Habbena et al. 2009). Hence, their strong adaption to the environment may lead to their flourishment in highly acidic soils and other major ecosystems. It is, however, possible that AOA might lose these competitive advantages in some certain environments, such as grassland soil, nitrogen-rich agricultural soil, and mangrove sediment. Surveys in the Mai Po mangrove wetland have revealed the dominance of AOB over AOA in sediments (Li et al. 2011; Li and Gu 2013); here, we exhibit stronger evidence on this dominance as the transcript abundance of AOB was exponentially greater than that of AOA. The neutral or slightly acidic condition and the available ammonia in the sediment might be more inclined to the growth of AOB. Nevertheless, few studies have detected the ammonia oxidizers on mRNA levels which could provide more useful and accurate information. Hence, our study reiterates the importance of the relationship between AOA and AOB and emphasizes the role of AOB in driving biogeochemical nitrogen cycling in the coastal wetlands.
5 Conclusions
The present study reveals the significant impact of plant on the ammonia-oxidizing process in the mangrove sediment. Plants could promote the nitrifying activities and abundances of both amoA gene and transcript. It was further demonstrated that vegetation along with two plant species significantly shaped the communities of AOA and AOB. These shifts on nitrifying process are associated with the variations of important sediment properties caused by the plants. It was therefore suggested that plant could be the key factor in controlling the nitrogen balance in mangrove systems. AOB was found to be dominant in mangrove wetlands, indicating their significant role on the global nitrogen cycling. However, information about the relationship between ammonia oxidizers and other microbes in mangrove sediment is still limited. Most of the researches, including this one, studying the ecology of ammonia oxidizers in mangrove have been limited in Southeast Asia. Hence, further studies should focus on the nitrogen transformation in mangroves from various regions worldwide, thus better understanding the spatial distribution and variation of microbes involved in biogeochemical cycling at global scale.
References
Alves RJE, Wanek W, Zappe A, Richter A, Svenning MM, Schleper C, Urich T (2013) Nitrification rates in Arctic soils are associated with functionally distinct populations of ammonia-oxidizing archaea. ISME J 7:1620–1631
An S, Gu B, Zhou C, Wang Z, Deng Z, Zhi Y, Li H, Chen L, Yu D, Liu Y (2007) Spartina invasion in China: implications for invasive species management and future research. Weed Res 47:183–191
Angeloni NL, Jankowski KJ, Tuchman NC, Kelly JJ (2006) Effects of an invasive cattail species (Typha × glauca) on sediment nitrogen and microbial community composition in a freshwater wetland. FEMS Microbiol Lett 263:86–92
Berg G, Smalla K (2009) Plant species and soil type cooperatively shape the structure and function of microbial communities in the rhizosphere. FEMS Microbiol Ecol 68:1–13
Boyle-Yarwood SA, Bottomley PJ, Myrold DD (2008) Community composition of ammonia-oxidizing bacteria and archaea in soils under stands of red alder and Douglas fir in Oregon. Environ Microbiol 10:2956–2965
Briones AM, Okabe S, Umemiya Y, Ramsing N-B, Reichardt W, Okuyama H (2002) Influence of different cultivars on populations of ammonia-oxidizing bacteria in the root environment of rice. Appl Environ Microbiol 68:3067–3075
Cao H, Li M, Hong Y, Gu J-D (2011) Diversity and abundance of ammonia-oxidizing archaea and bacteria in polluted mangrove sediment. Syst Appl Microbiol 34:513–523
Colmer T (2003) Long-distance transport of gases in plants: a perspective on internal aeration and radial oxygen loss from roots. Plant Cell Environ 26:17–36
Colmer TD, Flowers TJ (2008) Flooding tolerance in halophytes. New Phytol 179:964–974
Di H, Cameron K, Shen JP, Winefield C, O’Callaghan M, Bowatte S, He J (2009) Nitrification driven by bacteria and not archaea in nitrogen-rich grassland soils. Nat Geosci 2:621–624
Erguder TH, Boon N, Wittebolle L, Marzorati M, Verstraete W (2009) Environmental factors shaping the ecological niches of ammonia-oxidizing archaea. FEMS Microbiol Rev 33:855–869
Francis CA, Roberts KJ, Beman JM, Santoro AE, Oakley BB (2005) Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. Proc Natl Acad Sci U S A 102:14683–14688
Gao J-F, Luo X, Wu G-X, Li T, Peng Y-Z (2013) Quantitative analyses of the composition and abundance of ammonia-oxidizing archaea and ammonia-oxidizing bacteria in eight full-scale biological wastewater treatment plants. Bioresour Technol 138:285–296
Gubry-Rangin C, Nicol GW, Prosser JI (2010) Archaea rather than bacteria control nitrification in two agricultural acidic soils. FEMS Microbiol Ecol 74:566–574
Hart SC, Stark JM, Davidson EA, Firestone MK (1994). Nitrogen mineralization, immobilization, and nitrification. In: Weaver RW, Angle S, Bottomley P, Bezdicek D, Smith S, Tabatabi A, Wollum A (eds) Methods of soil analysis: microbiological and biochemical properties. Soil Sci Soc Am, Madison, pp 985–1018.
Head IM, Hiorns WD, Embley TM, McCarthy AJ, Saunders JR (1993) The phylogeny of autotrophic ammonia-oxidizing bacteria as determined by analysis of 16S ribosomal RNA gene sequences. J Gen Microbiol 139:1147–1153
Herman D, Johnson K, Jaeger C, Schwartz E, Firestone M (2006) Root influence on nitrogen mineralization and nitrification in rhizosphere soil. Soil Sci Soc Am J 70:1504–1511
Holguin G, Vazquez P, Bashan Y (2001) The role of sediment microorganisms in the productivity, conservation, and rehabilitation of mangrove ecosystems: an overview. Biol Fertil Soils 33:265–278
Jia Z, Conrad R (2009) Bacteria rather than Archaea dominate microbial ammonia oxidation in an agricultural soil. Environ Microbiol 11:1658–1671
Jin T, Zhang T, Yan Q (2010) Characterization and quantification of ammonia-oxidizing archaea (AOA) and bacteria (AOB) in a nitrogen-removing reactor using T-RFLP and qPCR. Appl Microbiol Biotechnol 87:1167–1176
Könneke M, Bernhard AE, José R, Walker CB, Waterbury JB, Stahl DA (2005) Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 437:543–546
Laanbroek HJ, Speksnijder AG (2008) Niche separation of ammonia-oxidizing bacteria across a tidal freshwater marsh. Environ Microbiol 10:3017–3025
Laanbroek HJ, Keijzer RM, Verhoeven JT, Whigham DF (2012) The distribution of ammonia-oxidizing betaproteobacteria in stands of black mangroves (Avicennia germinans). Front Microbiol 3:153
Leininger S, Urich T, Schloter M, Schwark L, Qi J, Nicol G, Prosser J, Schuster S, Schleper C (2006) Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature 442:806–809
Lensi R, Domenach A, Abbadie L (1992) Field study of nitrification and denitrification in a wet savanna of West Africa (Lamto, Côte d'Ivoire). Plant Soil 147:107–113
Li M, Gu J-D (2013) Community structure and transcript responses of anammox bacteria, AOA, and AOB in mangrove sediment microcosms amended with ammonium and nitrite. Appl Microbiol Biotechnol 97:9859–9874
Li M, Cao H, Hong Y, Gu J-D (2011) Spatial distribution and abundances of ammonia-oxidizing archaea (AOA) and ammonia-oxidizing bacteria (AOB) in mangrove sediments. Appl Microbiol Biotechnol 89:1243–1254
Martens-Habbena W, Berube PM, Urakawa H, de La Torre JR, Stahl DA (2009) Ammonia oxidation kinetics determine niche separation of nitrifying Archaea and Bacteria. Nature 461:976–979
Moin NS, Nelson KA, Bush A, Bernhard AE (2009) Distribution and diversity of archaeal and bacterial ammonia oxidizers in salt marsh sediments. Appl Environ Microbiol 75:7461–7468
Nicol GW, Leininger S, Schleper C, Prosser JI (2008) The influence of soil pH on the diversity, abundance and transcriptional activity of ammonia oxidizing archaea and bacteria. Environ Microbiol 10:2966–2978
Norton JM, Firestone MK (1996) N dynamics in the rhizosphere of Pinus ponderosa seedlings. Soil Biol Biochem 28:351–362
Patra AK, Abbadie L, Clays-Josserand A, Degrange V, Grayston SJ, Guillaumaud N, Loiseau P, Louault F, Mahmood S, Nazaret S (2006) Effects of management regime and plant species on the enzyme activity and genetic structure of N-fixing, denitrifying and nitrifying bacterial communities in grassland soils. Environ Microbiol 8:1005–1016
Philippot L, Hallin S, Börjesson G, Baggs E (2009) Biochemical cycling in the rhizosphere having an impact on global change. Plant Soil 321:61–81
Philippot L, Raaijmakers JM, Lemanceau P, van der Putten WH (2013a) Going back to the roots: the microbial ecology of the rhizosphere. Nat Rev Microbiol 11:789–799
Philippot L, Spor A, Henault C, Bru D, Bizouard F, Jones CM, Sarr A, Maron PA (2013b) Loss in microbial diversity affects nitrogen cycling in soil. ISME J 7:1609–1619
Pi N, Tam N, Wu Y, Wong M (2009) Root anatomy and spatial pattern of radial oxygen loss of eight true mangrove species. Aquat Bot 90:222–230
Priha O, Grayston SJ, Pennanen T, Smolander A (1999) Microbial activities related to C and N cycling and microbial community structure in the rhizospheres of Pinus sylvestris, Picea abies and Betula pendula seedlings in an organic and mineral soil. FEMS Microbiol Ecol 30:187–199
Purkhold U, Wagner M, Timmermann G, Pommerening-Röser A, Koops H-P (2003) 16S rRNA and amoA-based phylogeny of 12 novel betaproteobacterial ammonia-oxidizing isolates: extension of the dataset and proposal of a new lineage within the nitrosomonads. Int J Syst Evol Microbiol 53:1485–1494
Purvaja R, Ramesh R, Ray A, Rixen T (2008) Nitrogen cycling: a review of the processes, transformations and fluxes in coastal ecosystems. Curr Sci India 94:1419–1438
Reddy K, Patrick W, Lindau C (1989) Nitrification-denitrification at the plant root-sediment interface in wetlands. Limnol Oceanogr 34:1004–1013
Rotthauwe JH, Witzel KP, Liesack W (1997) The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl Environ Microbiol 63:4704–4712
Ruiz-Rueda O, Hallin S, Bañeras L (2009) Structure and function of denitrifying and nitrifying bacterial communities in relation to the plant species in a constructed wetland. FEMS Microbiol Ecol 67:308–319
Schleper C, Jurgens G, Jonuscheit M (2005) Genomic studies of uncultivated archaea. Nat Rev Microbiol 3:479–488
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ (2009) Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541
Skiba M, George T, Baggs E, Daniell T (2011) Plant influence on nitrification. Biochem Soc Trans 39:275–278
Steltzer H, Bowman WD (1998) Original articles: differential influence of plant species on soil nitrogen transformations within moist meadow alpine tundra. Ecosystems 1:464–474
Tsiknia M, Tzanakakis VA, Paranychianakis NV (2013) Insights on the role of vegetation on nitrogen cycling in effluent irrigated lands. Appl Soil Ecol 64:104–111
Wang Q, An S, Ma Z, Zhao B, Chen J, Li B (2006) Invasive spartina alterniflora: biology, ecology and management. Acta Phytotaxonomica Sinica 44:559–588
Wang H-T, Su J-Q, Zheng T-L, Yang X-R (2014) Impacts of vegetation, tidal process, and depth on the activities, abundances, and community compositions of denitrifiers in mangrove sediment. Appl Microbiol Biotechnol 98:9375–9387
Ward BB, O'Mullan GD (2002) Worldwide distribution of Nitrosococcus oceani, a marine ammonia-oxidizing γ-proteobacterium, detected by PCR and sequencing of 16S rRNA and amoA genes. Appl Environ Microbiol 68:4153–4157
Wedin DA, Tilman D (1990) Species effects on nitrogen cycling: a test with perennial grasses. Oecologia 84:433–441
Wheatley R, Ritz K, Griffiths B (1990) Microbial biomass and mineral N transformations in soil planted with barley, ryegrass, pea or turnip. Plant Soil 127:157–167
Yang W, Zhao H, Chen X, Yin S, Cheng X, An S (2013) Consequences of short-term C4 plant Spartina alterniflora invasions for soil organic carbon dynamics in a coastal wetland of Eastern China. Ecol Eng 61:50–57
Yao H, Gao Y, Nicol GW, Campbell CD, Prosser JI, Zhang L, Han W, Singh BK (2011) Links between ammonia oxidizer community structure, abundance, and nitrification potential in acidic soils. Appl Environ Microbiol 77:4618–4625
Yu X, Yang J, Liu L, Tian Y, Yu Z (2014) Effects of Spartina alterniflora invasion on biogenic elements in a subtropical coastal mangrove wetland. Environ Sci Pollut Res. doi:10.1007/s11356-014-3568-2
Zhang Q-F, Peng J-J, Chen Q, Li X-F, Xu C-Y, Yin H-B, Yu S (2011) Impacts of Spartina alterniflora invasion on abundance and composition of ammonia oxidizers in estuarine sediment. J Soils Sediments 11:1020–1031
Zhang L-M, Hu H-W, Shen J-P, He J-Z (2012) Ammonia-oxidizing archaea have more important role than ammonia-oxidizing bacteria in ammonia oxidation of strongly acidic soils. ISME J 6:1032–1045
Acknowledgments
This study was financially supported by the Strategic Priority Research Program of Chinese Academy of Sciences (XDB15020302, XDB15020402), the Natural Science Foundation of China (31270153, 41376119), and the International Science & Technology Cooperation Program of China (2011DFB91710). We would like to thank Prof. Yong-guan Zhu from the Institute of Urban Environment, Chinese Academy of Sciences for valuable suggestions on the earlier version of this manuscript.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Responsible editor: Jizheng He
Rights and permissions
About this article
Cite this article
Wang, H., Su, J., Zheng, T. et al. Insights into the role of plant on ammonia-oxidizing bacteria and archaea in the mangrove ecosystem. J Soils Sediments 15, 1212–1223 (2015). https://doi.org/10.1007/s11368-015-1074-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11368-015-1074-x