Abstract
Alzheimer’s disease(AD) is an age-associated neurodegenerative disease that results in deterioration of memory and cognitive function. As a currently untreatable disorder, AD has emerged as one of the defining biomedical challenges of our time. Thus, new approaches that can examine the cellular and molecular mechanisms underlying age-related AD pathology are sorely needed. One of the hallmarks of Alzheimer’s disease is the hyperphosphorylation of the tau protein. Caenorhabditis elegans have been previously used to study the genetic pathways impacted by tau proteotoxic stress; however, currently, available C. elegans tau models express the human protein solely in neurons, which are unresponsive to global RNA interference (RNAi). This limits powerful RNAi screening methods from being utilized effectively in these disease models. Our goal was to develop a C. elegans tau model that has pronounced tau-induced disease phenotypes in cells that can be modified by feeding RNAi methods. Towards this end, we generated a novel C. elegans transgenic line with codon-optimized human 0N4R V337M tau expressed in the body wall muscle under the myo-3 promoter. Immunoblotting experiments revealed that the expressed tau is phosphorylated on epitopes canonically associated with human AD pathology. The tau line has significantly reduced health metrics, including egg laying, growth rate, paralysis, thrashing frequency, crawling speed, and lifespan. These defects are suppressed by RNAi directed against the tau mRNA. Taken together, our results suggest that this alternative tau genetic model could be a useful tool for uncovering the mechanisms that influence the hyperphosphorylation and toxicity of human tau via RNAi screening and other approaches.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Tau is widely expressed in the mammalian nervous system, functioning in neurons, astrocytes, and oligodendrocytes where it contributes to the assembly and stabilization of microtubules [1]. Tauopathies are clinically, morphologically, and biochemically heterogeneous age-associated neurodegenerative diseases characterized by the deposition of abnormal tau protein in the brain. These include numerous diseases such as Pick’s disease, progressive supranuclear palsy, and primary age-related tauopathy, as well as Alzheimer’s disease (AD) [2]. A major barrier in tauopathy research is a lack of animal models that both age rapidly and recapitulate the key molecular features of tau toxicity.
AD is a currently untreatable disorder, estimated to afflict more than 24 million people in 2014 with that number projected to quadruple by 2050 [3]. Chronological age is the strongest known risk factor for AD [4, 5], and there is growing consensus that biological aging [6] either contributes causally to AD or, at a minimum, creates permissive physiology for AD onset and progression [7]. AD pathology is characterized by the aggregation of protein deposits in various parts of the cerebrum, including the frontal, temporal, and entorhinal cortex as well as the hippocampus. These aggregates notably consist of amyloid-beta 42 (Aβ42) senile plaques and hyperphosphorylated tau (P-tau) neurofibrillary tangles, both of which are accompanied by neuronal death, loss of synaptic connections in the brain, and cerebral atrophy [8,9,10]. Both Aβ42 and P-tau have been proposed as causal mediators of neuronal death and show toxicity in cell and animal-based models. Hyperphosphorylation is the most important posttranslational modification of tau because it directly contributes to the pathogenic depositing of tau, resulting in serious functional impacts [11]. Indeed, tau protein is known to be differentially phosphorylated in normal and AD brains, and the tau tangles characteristic of AD are largely made up of P-tau [12, 13], supporting a potential causal role. For this reason, antibodies specific for AD-associated phosphorylation sites have been developed as a means of diagnosing and characterizing the pathology with phosphorylation epitopes at S202 and S414 being two of the most utilized antibodies for this purpose [14, 15].
While tau has been primarily associated with its role in diseases of the central nervous system, the work of the Human Protein Atlas project has shown that skeletal muscle tissue expresses tau at similar levels to those found in brain tissue [16]. Pathological phosphorylated tau deposits have been associated with muscle disorders, most notably in muscle from patients suffering from sporadic inclusion body myositis (sIBM) [17, 18]. sIBM shares many pathological features with AD including age dependence, aberrant protein folding, excessive oxidative stress, mitochondrial abnormalities, hyperphosphorylation, and gene expression changes [19,20,21,22]. Thus, while brain and muscle are quite different tissues, some of the underlying cellular mechanisms of tau-related pathology may be shared between these tissue types.
The well-developed genetics, short life cycle, and abundance of simple health metrics of the nematode Caenorhabditis elegans make it an attractive platform for genetic screening. Research over the past 20 years has established C. elegans as a useful in vivo system for examining the toxic outcomes caused by overexpression tau and amyloid beta [23,24,25,26,27]. The relative simplicity of the C. elegans makes it an attractive platform for discovering genetic pathways and small molecule compounds that protect against these pathogenic AD-associated proteins. The simple body plan and behaviors of C. elegans facilitate the identification of pathological phenotypes, enabling screening strategies to identify genetic relationships. In addition, the short lifespan of C. elegans makes it a useful model for studying age-related tauopathies.
A major strength of C. elegans as an experimental animal model is the ability to reduce expression of specific genes through feeding bacteria expressing double-stranded RNA [28]. As such, commercial bacterial RNAi libraries have been developed that cover most C. elegans genes [29]. However, effective genetic screening via RNAi has not been feasible with any of the current C. elegans Tau AD models [30,31,32,33,34,35,36] as they express human tau in neurons, which are insensitive to the effects of RNAi feeding [37, 38].
The goal of this study was to establish and characterize a novel nematode model of tauopathy that is responsive to RNAi treatment, facilitating large-scale genetic screening. We did this through generating a transgenic line that expresses codon-optimized human tau 0N4R in body wall muscle cells instead of neurons. We anticipated that such a model would show significant age-associated paralysis and movement deficits due to muscle cell dysfunction, which are substantially easier phenotypes to assess than the subtle uncoordinated movement caused by neuronal expression of tau. Here, we describe the creation of such transgenic animals and provide an initial characterization, confirming these predictions.
Results
We engineered a transgenic C. elegans line (KAE112) in which codon-optimized human V337M 0N4R tau is expressed in body wall muscles by a myo-3 promoter [39] (Fig. 1A, B). We obtained the protein sequence of human tau 0N4R tau from the NCBI protein resource. To recapitulate the familial pathogenic tau allele V337M, we changed the analogous amino acid from valine to methionine. The protein sequence was then reverse translated, and codon-optimized for robust expression in C. elegans (Supplemental Fig. 1).
Generation of a transgenic C. elegans model of muscle tauopathy. A Schematic of human tau transgene. The tau transgene is the 0N4R V337M tau allele expressed under the myo-3p promoter. The human tau gene was codon-optimized for C. elegans and integrated into the genome. B Tissue expression diagram of tau transgene and mCherry intestinal fluorescent marker
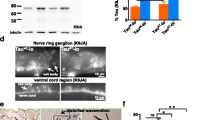
Expression of the transgenic human tau was confirmed via Western blotting against a pan-tau antibody. The blot displayed a strong band at ~ 45 kDa, the expected size [40, 41] of 0N4R tau (Fig. 2A). To determine whether the tau protein is phosphorylated at amino acids associated with human tauopathies, we then conducted immunoblots against phosphorylated S202 and S416. both cases, we found strong immunoreactive protein bands of the expected size (~ 50 kDa) (Fig. 2A). We observed significantly higher tau levels in day 6 adult animals compared to day 1 adults (Fig. 2B) when normalized to total protein (Supplemental Fig. 2). We compared the change in relative intensity between young and old animals and found that the older animals had about 1.5-fold higher expression of total tau and phospho-tau S416 proteins while phospho-tau S202 protein levels in the older cohorts were about 2.3-fold higher (Fig. 2C).
Immunoblot analysis to assess relative expression and phosphorylation of human tau with age. A Fluorescence bands are tau immunohistochemical and shown as a heat map for clarity. Each lane is a separate biological replicate. Whole worm lysate from the myo-3 driven tau line KAE112 displayed clear ~ 50-kDa immunoreactive bands to total tau as well as P-tau S202 and P-tau 416. Immunoreactive bands shown as a heat plot to emphasize differences in abundance. B Quantification of relative tau expression between KAE112 animals at days 1 and 6 of adulthood from blots shown in A. The animals had significantly higher total tau, P-Tau S416, and P-Tau S202 expression levels at day 6 of adulthood compared to day 1 adult animals. The immunohistochemical signals were normalized to amount of total protein loaded in each lane (Supplemental Fig. 2). p-Values were obtained by unpaired two-tailed t-tests with Welch’s correction; * = p-value < 0.05, ** = p-value < 0.01. C Fold increase in tau protein expression. The intensities of the tau signals from day 6 vs day 1 adult animals were calculated as a ratio. p-Values were obtained by unpaired two-tailed t-tests with Welch’s correction; * = p-value < 0.05, ** = p-value < 0.01, n.s. = not significant
We then wanted to determine if KAE112 exhibited phenotypes that could be leveraged for large-scale genetic or pharmacological screening. First, we quantified the number of eggs laid per animal per hour. Consistent with previous reports [42], wild-type animals at the first day of adulthood laid about four eggs per hour (3.68 eggs/worm/h); however, the tau-expressing animals on average laid only one egg per 2 h (0.54 eggs/worm/h) (Fig. 3A). To determine the effect of tau expression on total fecundity, we monitored egg laying across the entire reproductive period. While most eggs were deposited during the second day of adulthood (Fig. 3B), tau animals only produced about 18 eggs total, compared to the expected more than 300 eggs typically produced by a single wild-type animal [42]. Adult animals expressing tau in body wall muscle were also significantly smaller than wild-type animals throughout reproductive adulthood (Fig. 3D).
Comparison of health metrics between wild type and tau line KAE112. A Influence of tau-RNAi on the number of eggs laid by wild type and KAE112 day 2 adults. The number eggs laid over a 3-h period was converted into number of eggs laid per animal per hour. B KAE112 fecundity time course. The total number of eggs laid each day between day 2 and day 6 of adulthood was counted. C, D Influence of tau-RNAi on growth rate differences between wild type and KAE112 after 48 h from egg lay. tau-RNAi treatment suppressed the slower grown of the KAE112 line but did not impact the growth of wild-type animals. D Growth of wild type and KAE112 across 6 days of adulthood. KAE112 animals had slower growth than wild type, but the size differences decreased with age. E, F tau-RNAi treatment restored crawling speed in tau worms to wild-type speeds but did not affect wild-type animals. Worms were quantified on their second day of adulthood. G Comparison of the onset of paralysis between wild type and KAE112 animals. KAE112 worms became paralyzed significantly earlier than wild type. F Crawling speed of wild type and KAE112. Crawling speed was quantified through measuring the trajectories of worms for 30 s immediately after being placed on a fresh bacterial lawn. KAE112 animals displayed significantly lower crawling speeds than wild type. G Comparison of paralysis rate between KAE112 and wild-type worms. KAE112 animals became paralyzed in about half the time required of wild type. H Comparison of thrashing between wild type and KAE112 animals. Both at larval stage 4 and day 4 adult KAE112 animals displayed significantly lower rate of thrashing rates than wild type. I Manual lifespan analysis of tau RNAi on KAE and N2 animals. The KAE112 line fed empty vector RNAi was significantly shorter lived than wild-type animals or KAE112 animals fed tau RNAi while there was not a significant difference between wild-type animals fed empty vector or tau RNAi containing bacteria. J cco-1 and daf-2 RNAi treatment of the KAE112 line. Both cco-1 and daf-2 RNAi extended the lifespan of KAE112 animals by about 5 days over the empty vector RNAi treatment. For all panels, each data point or line represents three biological replicates each containing at least 50 animals. p-Values were obtained by using unpaired two-tailed t-tests with Welch’s correction; **** = p-value < 0.0001, * = p-value < 0.05
As tauopathies have been shown to result from the loss of microtubule stability due to hyperphosphorylation of tau, we hypothesized that tau pathogenicity in body wall muscles should result in pronounced age-associated movement defects. Muscle dysfunction in C. elegans is often assessed through the quantification of crawling speed, paralysis onset, and the frequency with which animals enact body bends (thrashes) while navigating a liquid environment. Animals expressing tau showed a significant defect in crawling speed on days 2–8 of adulthood (Fig. 3F). We also found that the tau worms displayed age-associated onset of paralysis roughly twice as quickly as wild type (Fig. 3G) and that their frequency of body bends was significantly decreased (Fig. 3H). However, it should be noted that a significant fraction of the tau animals died during the time course of these experiments, potentially biasing the analysis toward survivors. To address this, we conducted lifespan analysis and found that the median age of death of the tau animals (10 days of adulthood) was about half that of wild-type animals (19.5 days of adulthood) (Fig. 3I).
Intrigued by this strong difference in lifespan, we sought to determine whether two known longevity pathways, inhibition of mitochondrial electron transport chain function and reduced insulin-like signaling, increase survival of the tau-expressing animals. We treated KAE112 animals with RNAi clones directed toward the cytochrome c oxidase subunit cco-1 or the insulin-like receptor daf-2 from the L1 larval stage and saw a marked increase in median lifespan from 10 to 15 days (Fig. 3J). Feeding the tau-expressing animals RNAi targeted to the tau transgene suppressed the egg laying, growth, and crawling speed deficits while not influencing these metrics in wild-type worms (Fig. 3A, C, E, I).
Discussion
The goal of this study was to develop a C. elegans tauopathy model with significant phenotypic deficits that is amenable for high-throughput RNAi or pharmacological screening. As C. elegans neurons are resistant to feeding RNAi methods and animals expressing tau in neurons show mostly subtle lifespan and uncoordinated movement defects, we chose to express tau in body wall muscle cells using the myo-3 promoter. To ensure robust expression of the transgene, we codon-optimized the 0N4R (v337M) allele of human tau sequence for C. elegans and generated an integrated overexpression line. The rescue of the health phenotypes by RNAi knock-down of the tau transgene demonstrates that the observed deficits directly result from tau expression. We further demonstrate that two known longevity pathways, inhibition of mitochondrial electron transport chain function and reduced insulin-like signaling, can increase survival in the tau-expressing animals.
While expressing a dementia-associated protein non-neuronally may seem counterintuitive, a great deal has been learned about the genetic pathways that influence amyloid-beta toxicity from studying a C. elegans AD model that expresses amyloid-beta[1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42] in the body wall musculature [43]. The amenability for interrogating the genetic pathways impacted by amyloid-beta via RNAi has led to this line being utilized in over 40 studies. These have uncovered mitochondrial function as an important regulator of amyloid-beta toxicity [44], validated systemic approaches to human brain protein networks [45], determined the impacts of aging and longevity-promoting interventions on amyloid-beta aggregation [46,47,48], and identified numerous potential therapeutic compounds [49, 50]. The tau line we described in this study could be used in analogous studies to define general mechanisms of proteotoxicity as well as tau-specific modifiers. As human tau has been shown to be highly expressed in striated muscle and play a role in muscle diseases such as sIBM, the KAE112 line described in this study could potentially also be utilized as an invertebrate model of this disease. We note that, while this study investigated the consequences of only one particular isoform of human tau (0N4R V337M), in the future, other forms of tau could also be studied using this muscle-expression paradigm.
Although the promoter region of myo-3 is routinely used as a body wall muscle-specific genetic tool [51,52,53,54], this promoter also induces weak expression in the vulva muscles, as well as the gonad. Thus, the results of the egg-laying assay suggest that even relatively low levels tau 0N4R V337M protein can severely disrupt cellular function. Rather than being a drawback, the expression in these tissues gives this genetic model further utility because the ~ 30-fold decrease in egg laying (Fig. 3A, B) can be used as a robust phenotype for screening genetic or pharmacological suppressors of tau toxicity.
Previous work with C. elegans neuronal tau models has shown that tau protein abundance increases with age and is phosphorylated at several different epitopes associated with AD pathology in humans [30, 32, 55]. Here, we show that non-neuronal tissues also accumulate tau with age and can phosphorylate the protein at epitopes associated with AD in humans. A wealth of literature has identified kinases and phosphatases that influence tau in cell culture and mammalian systems [56, 57]. However, it is not known whether the C. elegans orthologs of these genes can modify human tau in similar ways. Understanding the kinases and phosphatases that modify tau in C. elegans would give a richer biochemical context to the disease model and open up a new avenue for conducting hypothesis-driven biochemical studies of tau pathogenicity. The RNAi tractability of the non-neuronal model we generated could be leveraged to systematically screen candidate kinases and phosphatases that influence the phosphorylation status of the different clinically relevant tau epitopes. The abundance of tau in our model (Fig. 2A) facilitates the straightforward assessment of tau phosphorylation status. It will be of interest to expand on these findings by testing other antibodies specific for phosphorylated epitopes canonically associated with AD, as well as identify phosphorylation changes with age. It would also be useful to determine the degree of tau aggregation in this model and determining age-related changes.
We are particularly interested in identifying modifiers of the age-associated movement and survival defects observed in the KAE112 line. While manual lifespan analysis can be laborious, it is possible to conduct analogous experiments in a high-throughput manner by utilizing automatic time-lapse imaging platforms such as the WormBot [58]. We are also interested in understanding the interactions between tau and other disease-associated proteins. For instance, previous work combining amyloid-beta and tau in C. elegans neurons has identified a synergistic effect resulting in higher toxic protein expression (59) as well as other gene expression and behavioral changes [60]. Therefore, a logical next step would be to conduct an analogous experiment by crossing body wall muscle expressed tau and amyloid-beta lines. Likewise, our tau model could be crossed with body wall muscle models of alpha-synuclein, huntingtin 513, or poly-Q [61,62,63]. Body wall proteotoxicity models have an advantage over the neuronal models in that any synergistic effects could be interrogated via RNAi screening. In addition, since body wall muscle tissue is more abundant than neuronal tissue, obtaining sufficient material for high-quality omics studies is significantly easier.
Conclusion
In this study, we have described a novel non-neuronal C. elegans tau model that is responsive to RNAi by bacterial feeding. Our model displays high tau protein expression which is phosphorylated at epitopes canonically associated with human AD and increases in abundance with age. The health phenotypes we identified are straightforward to assess and easy to distinguish from wild-type worms; thus, they can be leveraged for high-throughput screening. Taken together, our results suggest that this novel non-neuronal tauopathy model has the potential to serve as a powerful genetic tool for identifying genetic pathways and pharmacological agents that mitigate the pathological phosphorylation and cellular toxicity of tau.
Methods
Strains
For this study, we used the N2 wild-type line as well as KAE112 tau model we generated. Strains were cultured and maintained using standard methods [64]. All worm lines were maintained on NGM plates seeded with live OP50 bacteria and maintained at 20 °C. All experimental procedures were conducted at 20 °C. The wild-type C. elegans line as well as the OP50 and HT115 bacterial strains were provided by the Caenorhabditis Genetics Center (CGC), which is funded by the NIH Office of Research Infrastructure Programs (P40 OD010440). KAE112 will be made available to the scientific community through the CGC.
Cloning
We utilized the Gateway cloning system to engineer KAE112 with human 0N4R tau V337M expression in muscle and gonad. The protein sequence of human isoform C which encodes the 0N4R allele was obtained from the NCBI protein resource (NP_058518.2). The 279th amino acid on isoform 3 (corresponding to 337 amino acids on 3N4R isoform 1) was then changed from valine to methionine. This protein sequence was then reverse translated using Expasy [65]. The translated DNA sequence was codon-optimized for C. elegans using the webtool Codon Adaptor [66] with no introns and a codon adaptation index of 1.00. This sequence was synthesized with corresponding donor plasmid pDONR221 Gateway insertion sequences added on the 5′ and 3′ ends. Gel purification was used to purify the synthesized construct, and it was inserted into pDONR 221 using BP Clonase II (Thermo Fisher, Cat #11,789,020). The gene was then combined with 5′ promoter and 3′ untranslated sequences via Gateway Cloning using the LR Clonase II plus enzyme (Thermo Fisher, Cat #12,538,120). The pDONR 221 vector containing the tau sequence was combined with p_K12F2.1_93, the pDONR 4–1 containing the promoter region of myo-3 obtained from the C. elegans promoterome (Source BioScience, Cat # DFCIp742D074D), the pDONR 2–3 vector pCM5.37 containing the unc-54 UTR region (Addgene, Cat # 17,253), and the destination vector pCFJ150 (Addgene, Cat #19,329). The destination plasmid pHL17 was then verified via Sanger sequencing.
To generate the custom feeding RNAi vector for the codon-optimized 0N4R tau gene, 400 bp of the gene corresponding to nucleotides 220–620 was inserted into the L4440 RNAi feeding vector (Addgene, Cat # 1654) using restriction digest cloning using the HindIII and NheI sites which flank the RNAi producing sequence. The resulting plasmid pHL18 was verified by Sanger sequencing. We then transformed HT115 bacteria, used as the genetic background for the Arhinger and Vidal C. elegans RNAi libraries, with the pHL18 plasmid [67].
Generation of transgenic C. elegans line
The N2 wild-type strain was obtained from the a construct for the intestinal fluorescent reporter ges-1p::mCherry::unc-54 UTR and injected into the gonads of N2 wild-type worms. The extrachromosomal array was integrated by UV irradiation and then backcrossed with N2 6 times yielding strain KAE112, genotype N2; seaIs201[myo-3p::htau(0N4R;V337M)::unc-54;vha-6p::mCherry:unc-54].
Immunoblotting
Synchronized populations of wild-type and muscle tau lines were grown to the second day of adulthood. Animals were washed off plates, concentrated by centrifugation at 17 kG for 30 s, and fractured by three freeze–thaw cycles in liquid nitrogen. The fractured worm biomass was then centrifuged at 17 kG for 15 min at 4 °C to pellet. Fifty microliters of worm pellet was mixed with 150 µL of Laemmli buffer containing 2-mercaptoethanol and heated to 95 °C for 10 min. Samples were centrifuged at 17 kG for 10 min at 4 °C, and the supernatant was transferred to a new tube. Protein was separated on SDS-PAGE BOLT gels 10% and then transferred to 0.4-µm PVDF filter membrane. Transfer of protein was quantified by No-Stain (Thermo Fisher, Waltham MA, USA; Cat #A44717). Membranes were then blocked with 5% dry milk in TBST with 0.1% Tween for 1 h and incubated primary antibodies. These included phospho-tau (Cell Signaling Technology, Danvers MA USA; P-tau S396 clone PHF13, cat#9632 T; P-tau S202 clone D4H7E, Cat # 39357 T; P-tau S416 clone D7U2P, Cat # 15013 T; P-tau S404 clone D2Z46, Cat # 35834 T) and pan-tau (Millipore Sigma, Burlington MA, USA; clone PC1C8, Cat# MAB3420) antibodies (all at 1:2000 dilution) for 16 h on a rocker at 4 °C followed by secondary anti-mouse or anti-rabbit antibodies (1:5000 dilution) (Cell Signaling Technology, Danvers MA USA, Cat #s 7076P2 and 7074P2) on a rocker for 60 min at 20 °C. The membranes were then developed with Pierce ECL substrate (Thermo Fisher, Waltham MA, USA; Cat #32,109) and imaged on an iBright VL750 gel imager (Thermo Fisher, Waltham MA, USA; Cat #A44116). The intensity of the total protein and immuno-blots was integrated with ImageJ [68].
Growth assays
To compare growth rates of N2 muscle tau worms, we bleached synchronized lines and then propagated them from egg for 48 h at 20 °C and then imaged dozens of animals for each condition. then quantified it using ImageJ to quantify the length by trancing the midline of each animal [69]. The pixel lengths were converted to microns through comparison to a ruler imaged at the same magnification. Statistical significance was calculated using unpaired, parametric t-tests with Welch’s correction using GraphPad Prism software.
Egg-laying assays
N2 and Tau OE lines were bleach synchronized and plated at the L1 stage onto 10-cm NGM plates seeded with OP50. Animals were allowed to mature until they were day 1 adults that had begun laying eggs. The assay plates were placed into a 20 ℃ incubator for at least 30 min prior to testing to control the temperature of the assay. Five animals were transferred to a plate with 10 plates assayed per biological replicate. The worms were then allowed to lay eggs at 20 °C for 3 h after which the eggs were then counted. The assay was repeated on three different days. Statistical significance was calculated using unpaired, parametric t-tests with Welch’s correction using GraphPad Prism software.
Crawling speed assays
To quantify crawling speed, at least 10 living animals were placed in the middle of a 6-cm plate seeded with OP50 bacteria immediately prior to imaging. Their movements were then recorded with an AmScope camera MU503B (AmScope, Irvine CA, USA; Cat # MU503) paired with an AmScope dissecting microscope. The animal positions were imaged every second for 30 s. The images were then imported into ImageJ as a stack and converted into a minimum intensity projection. The length of each minimum intensity trail was then traced and quantified. Animals that stopped along the border of the bacterial lawn were excluded. The track lengths were converted to mm using the 1-cm scale image and the crawling speed (mm/s) was calculated. For each replicate, at least 50 animals were analyzed with at least three biological replicates recorded for each genotype/time point combination. Statistical significance was calculated using unpaired, parametric t-tests with Welch’s correction using GraphPad Prism software.
RNAi experiments
Single bacterial colonies were transferred to 10 mL LB + carbenicillin 50 µg/mL and 10 µg/mL tetracycline and grown overnight at 37 °C. Three milliliters of the overnight culture was then transferred into 9 mL of fresh LB + 1 mM IPTG, and 50 µg/mL carbenicillin and grown for 4°h at 37 °C. Bacteria were centrifuged at 3700 rpm for 20 min, and the supernatant was decanted. The bacterial pellets were then resuspended in 650 µL of fresh LB + 1 mM IPTG, and 50 µg/mL carbenicillin; 200 µL of the resuspended pellet was added to NGM plates containing 1 mM IPTG and 50 µM of carbenicillin. Animals were placed on RNAi at L1 larval stage. For the egg-laying assays, RNAi treatment was initiated on the parental generation.
Paralysis assays
Paralysis was assessed by standard methods [43]. Briefly, the worms were bleach synchronized and cultivated on NGM plates with OP50 bacterial lawns. These worms were maintained at 20 °C for until they reached late-stage larval L4 stage and then moved to a fresh NGM OP50 plate each day thereafter. The newly relocated animals were then assessed. The worms that failed to crawl at least one body length from deposition were gently prodded with a platinum wire to induce movement. Prodded worms that still failed to relocate at least one body length away were then considered paralyzed, counted, and removed from plate. By only recorded confirmed paralysis events, any worms that fled the agar plate during the course of the experiment were not considered.
Swimming body bend assays
Worm cohorts were bleach synchronized and then maintained on NGM plates seeded with OP50 bacteria. Subsequently, larval stage 4 animals were visually assessed and transferred to a fresh plate. Some of these transferred animals were then immediately analyzed for body bend rate, and the others were transferred daily to fresh plates until they reached day 4 adults. The thrashing rate was determined by standard methods [70]. For each recording, a single animal was transferred to a 10 µL drop of M9 buffer placed on a siliconized microscope slide. Each worm was allowed to acclimate for 30 s, and then, each thrashing cycle was manually recorded using a cell counter. A single thrash was defined strictly as a C-shaped, head-to-tail movement.
Lifespan analysis
Manual lifespan analysis was carried out according to standard methods [71]. Briefly, animals were bleach synchronized and then cultivated on their respective RNAi bacteria seeded on NGM plates with carbenicillin (50 µg/ml) and tetracycline (10 µg/mL). Animals were then visually assessed to be late L4 stage and manually transferred to analogous plates also containing 1 µM FuDR. Worms were then maintained on these plates throughout the experiment and manually examined each day for death events. Only confirmed death events were quantified; therefore, they were not influenced; the metrics are not influenced by worms lost off the plates. All death events were counted with no censoring. Each biological replicate contained at least three plates each with 30 + worms each. The data from three biological replicates was compiled and statistical significance between each experimental condition via pairwise p-value analysis with Bonferroni correction.
References
Lee VM, Goedert M, Trojanowski JQ. Neurodegenerative tauopathies. Annu Rev Neurosci. 2001;24:1121–59.
Kovacs GG. Invited review: neuropathology of tauopathies: principles and practice. Neuropathol Appl Neurobiol. 2015;41:3–23.
Reitz C, Mayeux R. Alzheimer disease: epidemiology, diagnostic criteria, risk factors and biomarkers. Biochem Pharmacol. 2014;88:640–51.
Ikeda T, Yamada M. Risk factors for Alzheimer’s disease. Brain Nerve. 2010;62:679–90.
Kaeberlein M. Time for a new strategy in the war on Alzheimer’s disease. Public Policy & Aging Report. 2019;29:119–22.
Lopez-Otin C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell. 2013;153:1194–217.
M. Kaeberlein, V. Galvan, Rapamycin and Alzheimer's disease: time for a clinical trial? Sci Transl Med 11 (2019)
Crary JF. Primary age-related tauopathy and the amyloid cascade hypothesis: the exception that proves the rule? J Neurol Neuromedicine. 2016;1:53–7.
Montine TJ, et al. National Institute on Aging-Alzheimer’s Association guidelines for the neuropathologic assessment of Alzheimer’s disease: a practical approach. Acta Neuropathol. 2012;123:1–11.
Montine TJ, et al. Multisite assessment of NIA-AA guidelines for the neuropathologic evaluation of Alzheimer’s disease. Alzheimers Dement. 2016;12:164–9.
Iqbal K, Liu F, Gong CX, Alonso AdC, Grundke-Iqbal I. Mechanisms of tau-induced neurodegeneration. Acta neuropathologica. 2009;118:53–69.
Braak H, Braak E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 1991;82:239–59.
Thal DR, Rüb U, Orantes M, Braak H. Phases of Aβ-deposition in the human brain and its relevance for the development of AD. Neurology. 2002;58:1791–800.
Buée L, Bussière T, Buée-Scherrer V, Delacourte A, Hof PR. Tau protein isoforms, phosphorylation and role in neurodegenerative disorders. Brain Res Rev. 2000;33:95–130.
Šimić G, et al. Tau protein hyperphosphorylation and aggregation in Alzheimer’s disease and other tauopathies, and possible neuroprotective strategies. Biomolecules. 2016;6:6.
Pontén F, Jirström K, Uhlen M. The Human Protein Atlas—a tool for pathology. The Journal of Pathology: A Journal of the Pathological Society of Great Britain and Ireland. 2008;216:387–93.
Maurage CA, et al. Tau aggregates are abnormally phosphorylated in inclusion body myositis and have an immunoelectrophoretic profile distinct from other tauopathies. Neuropathol Appl Neurobiol. 2004;30:624–34.
Askanas V, Engel WK, Bilak M, Alvarez RB, Selkoe DJ. Twisted tubulofilaments of inclusion body myositis muscle resemble paired helical filaments of Alzheimer brain and contain hyperphosphorylated tau. Am J Pathol. 1994;144:177–87.
Askanas V, Engel WK. Inclusion-body myositis: muscle-fiber molecular pathology and possible pathogenic significance of its similarity to Alzheimer’s and Parkinson’s disease brains. Acta Neuropathol. 2008;116:583–95.
D. Levacic, L. R. Peddareddygari, D. Nochlin, L. R. Sharer, R. P. Grewal, Inclusion-body myositis associated with Alzheimer’s disease. Case reports in medicine 2013 (2013)
Liu QY, et al. Molecular events linking cholesterol to Alzheimer’s disease and inclusion body myositis in a rabbit model. Am J Neurodegener Dis. 2016;5:74.
Kannanayakal TJ, Mendell JR, Kuret J. Casein kinase 1 alpha associates with the tau-bearing lesions of inclusion body myositis. Neurosci Lett. 2008;431:141–5.
D. Paul, S. Chipurupalli, A. Justin, K. Raja, S. K. Mohankumar, Caenorhabditis elegans as a possible model to screen anti-Alzheimer's therapeutics. Journal of Pharmacological and Toxicological Methods, 106932 (2020)
Godini R, Pocock R, Fallahi H. Caenorhabditis elegans hub genes that respond to amyloid beta are homologs of genes involved in human Alzheimer’s disease. Plos one. 2019;14:e0219486.
Griffin EF, Caldwell KA, Caldwell GA. Genetic and pharmacological discovery for Alzheimer’s disease using Caenorhabditis elegans. ACS Chem Neurosci. 2017;8:2596–606.
Alexander AG, Marfil V, Li C. Use of Caenorhabditis elegans as a model to study Alzheimer’s disease and other neurodegenerative diseases. Front Genet. 2014;5:279.
Ewald CY, Li C. Understanding the molecular basis of Alzheimer’s disease using a Caenorhabditis elegans model system. Brain Struct Funct. 2010;214:263–83.
Timmons L, Court DL, Fire A. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene. 2001;263:103–12.
Kamath RS, Ahringer J. Genome-wide RNAi screening in Caenorhabditis elegans. Methods. 2003;30:313–21.
Kraemer BC, et al. Neurodegeneration and defective neurotransmission in a Caenorhabditis elegans model of tauopathy. Proc Natl Acad Sci. 2003;100:9980–5.
Miyasaka T, et al. Progressive neurodegeneration in C. elegans model of tauopathy. Neurobiology of disease. 2005;20:372–83.
Brandt R, Gergou A, Wacker I, Fath T, Hutter H. A Caenorhabditis elegans model of tau hyperphosphorylation: induction of developmental defects by transgenic overexpression of Alzheimer’s disease-like modified tau. Neurobiol Aging. 2009;30:22–33.
Pir GJ, Choudhary B, Mandelkow E, Mandelkow E-M. Tau mutant A152T, a risk factor for FTD/PSP, induces neuronal dysfunction and reduced lifespan independently of aggregation in a C. elegans Tauopathy model. Molecular neurodegeneration. 2016;11:1–21.
Butler VJ, et al. Tau/MAPT disease-associated variant A152T alters tau function and toxicity via impaired retrograde axonal transport. Hum Mol Genet. 2019;28:1498–514.
Miyasaka T, et al. Curcumin improves tau-induced neuronal dysfunction of nematodes. Neurobiol Aging. 2016;39:69–81.
Morelli F, et al. V363I and V363A mutated tau affect aggregation and neuronal dysfunction differently in C. elegans. Neurobiology of disease. 2018;117:226–34.
Kamath RS, et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–7.
Feinberg EH, Hunter CP. Transport of dsRNA into cells by the transmembrane protein SID-1. Science. 2003;301:1545–7.
Okkema PG, Harrison SW, Plunger V, Aryana A, Fire A. Sequence requirements for myosin gene expression and regulation in Caenorhabditis elegans. Genetics. 1993;135:385–404.
L. Wu et al., Human tau isoform aggregation and selective detection of misfolded tau from post-mortem Alzheimer’s disease brains. bioRxiv, 2020. 2019.2012. 2031.876946
He Z, et al. Transmission of tauopathy strains is independent of their isoform composition. Nat Commun. 2020;11:1–18.
Byerly L, Cassada R, Russell R. The life cycle of the nematode Caenorhabditis elegans: I Wild-type growth and reproduction. Developmental biology. 1976;51:23–33.
McColl G, et al. Utility of an improved model of amyloid-beta (Aβ 1–42) toxicity in Caenorhabditis elegans for drug screening for Alzheimer’s disease. Mol Neurodegener. 2012;7:1–9.
Sorrentino V, et al. Enhancing mitochondrial proteostasis reduces amyloid-β proteotoxicity. Nature. 2017;552:187–93.
Mukherjee S, et al. Systems biology approach to late-onset Alzheimer’s disease genome-wide association study identifies novel candidate genes validated using brain expression data and Caenorhabditis elegans experiments. Alzheimers Dement. 2017;13:1133–42.
Groh N, et al. Age-dependent protein aggregation initiates amyloid-β aggregation. Frontiers in aging neuroscience. 2017;9:138.
Steinkraus KA, et al. Dietary restriction suppresses proteotoxicity and enhances longevity by an hsf-1-dependent mechanism in Caenorhabditis elegans. Aging Cell. 2008;7:394–404.
Mehta R, et al. Proteasomal regulation of the hypoxic response modulates aging in C. elegans. Science. 2009;324:1196–8.
Chia S, et al. Systematic development of small molecules to inhibit specific microscopic steps of amyloid-beta42 aggregation in Alzheimer’s disease. Biophys J . 2018;114:225a.
Chen X, Barclay JW, Burgoyne RD, Morgan A. Using C. elegans to discover therapeutic compounds for ageing-associated neurodegenerative diseases. Chemistry Central Journal. 2015;9:1–20.
Genes G. C. G. Silva-García et al., Single-copy knock-in loci for defined gene expression in Caenorhabditis elegans. G3. Genetics. 2019;9:2195–8.
Qadota H, et al. Establishment of a tissue-specific RNAi system in C. elegans. Gene. 2007;400:166–73.
Genes G. J. S. Watts et al., New strains for tissue-specific RNAi studies in Caenorhabditis elegans. G3. Genetics. 2020;10:4167–76.
Davis MW, Morton JJ, Carroll D, Jorgensen EM. Gene activation using FLP recombinase in C. elegans. PLoS Genet. 2008;4:e1000028.
Guha S, Johnson GVW, Nehrke K. The crosstalk between pathological tau phosphorylation and mitochondrial dysfunction as a key to understanding and treating Alzheimer’s disease. Mol Neurobiol. 2020. https://doi.org/10.1007/s12035-020-02084-0.
Kimura T, Sharma G, Ishiguro K, Hisanaga S-I. Phospho-tau bar code: analysis of phosphoisotypes of tau and its application to tauopathy. Front Neurosci. 2018;12:44.
Noble W, Hanger DP, Miller CC, Lovestone S. The importance of tau phosphorylation for neurodegenerative diseases. Front Neurol. 2013;4:83.
Pitt JN, et al. WormBot, an open-source robotics platform for survival and behavior analysis in C. elegans. GeroScience. 2019;41:961–73.
Benbow SJ, Strovas TJ, Darvas M, Saxton A, Kraemer BC. Synergistic toxicity between tau and amyloid drives neuronal dysfunction and neurodegeneration in transgenic C. elegans. Human molecular genetics. 2020;29:495–505.
Wang C, Saar V, Leung KL, Chen L, Wong G. Human amyloid β peptide and tau co-expression impairs behavior and causes specific gene expression changes in Caenorhabditis elegans. Neurobiol Dis. 2018;109:88–101.
Van Ham TJ, et al. C. elegans model identifies genetic modifiers of α-synuclein inclusion formation during aging. PLoS Genet. 2008;4:e1000027.
Lee AL, Ung HM, Sands LP, Kikis EA. A new Caenorhabditis elegans model of human huntingtin 513 aggregation and toxicity in body wall muscles. PloS one. 2017;12:e0173644.
Jia K, Hart AC, Levine B. Autophagy genes protect against disease caused by polyglutamine expansion proteins in Caenorhabditis elegans. Autophagy. 2007;3:21–5.
Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94.
Artimo P, et al. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012;40:W597–603.
Redemann S, et al. Codon adaptation–based control of protein expression in C. elegans. Nature methods. 2011;8:250–2.
Rual J-F, et al. Toward improving Caenorhabditis elegans phenome mapping with an ORFeome-based RNAi library. Genome Res. 2004;14:2162–8.
Schindelin J, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–82.
Rueden CT, Eliceiri KW. ImageJ for the next generation of scientific image data. Microsc Microanal. 2019;25:142–3.
M. Koopman, R. I. Seinstra, E. A. Nollen, C. elegans as a model for synucleinopathies and other neurodegenerative diseases: tools and techniques in Alpha-Synuclein. (Springer, 2019), pp. 93–112
G. L. Sutphin, M. Kaeberlein, Measuring Caenorhabditis elegans life span on solid media. Journal of visualized experiments: JoVE (2009)
Funding
This work was supported by NIH grants P30AG013280 and R01AG061132 to MK and NIH grant F32AG054098 to JCR.
Author information
Authors and Affiliations
Contributions
JCR conceived the study, carried out the experiments unless otherwise indicated, and wrote the manuscript. HL generated the tau expression construct. RKC and TL generated the transgenic nematode line and carried out experiments; SF and SMM characterized the crawling speed and egg laying. AN quantified the nematode lengths. MK supervised the study and wrote the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
About this article
Cite this article
Russell, J.C., Lei, H., Chaliparambil, R.K. et al. Generation and characterization of a tractable C. elegans model of tauopathy. GeroScience 43, 2621–2631 (2021). https://doi.org/10.1007/s11357-021-00436-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11357-021-00436-9