Abstract
Bilge wastewater is a high strength, typically saline wastewater, originating from operation of ships. In this study, the treatment of real bilge wastewater was tested using pure isolated aerobic strains and mixed cultures (aerobic and anaerobic). The Chemical Oxygen Demand (COD) and ecotoxicity decrease were monitored over time, while the microbial dynamics alterations in mixed cultures were also recorded. The isolated strains Pseudodonghicola xiamenensis, Halomonas alkaliphila and Vibrio antiquaries were shown to significantly biodegrade bilge wastewater. Reasonable COD removal rates were achieved by aerobic mixed cultures (59%, 9 days), while anaerobic mixed cultures showed lower performance (34%, 51 days). The genus Pseudodonghicola was identified as dominant under aerobic conditions both in the mixed cultures and in the control sample (raw wastewater), after exposure to bilge wastewater, demonstrating natural proliferation of the genus and potential contribution to COD reduction. Biodegradation rates were higher when initial organic load was high, while the toxicity of raw wastewater partially decreased after treatment.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Environmental deterioration and pollution of water bodies are directly caused by waste and wastewater discharges. Waste handling and efficiency of wastewater treatment prior to disposal determines the degree of the environmental impact. In the case of wastewater, industrial effluents can be more important in contributing to water quality decrease as they contain heavier load of pollutants, compared to municipal wastewater, and need intense effort for efficient treatment. Oily industrial wastewater is generated in large volumes by several industrial activities (e.g. petroleum, food and shipping industry), and discharge limits are often hard to achieve even after applying a series of treatment steps (Coca et al. 2011). Bilge wastewater is a type of oily-wastewater, produced from every motorized vessel, and includes all water accumulated in the interior lower part of the ship’s hull (Mclaughlin et al. 2014). Bilge wastewater may contain hydrocarbons, grease, hydraulic fluids, oil additives, cleaning and degreasing solvents, detergents, various metals and other (Andersson et al. 2016). It is described as being highly toxic to marine organisms and can remain highly toxic following treatment (Tiselius and Magnusson 2017). Though discharge of bilge wastewater at sea is prohibited according to MARPOL 73/78 and the European directive 2000/59/EC (Vyrides et al. 2018) and the accidental leakages have been limited in the last decades, marine pollution remains important because of illegal discharges (bilge wastewater, tank cleaning residues, propeller lubricants, cargo chemicals, etc.) (Jägerbrand et al. 2019), contributing to deterioration of the marine environment. According to MARPOL 73/78, vessels above 400 gross tonnes should use an oily-water separator (OWS) to separate oil from the water and discharge the water overboard, provided the effluent contains less than 15 g L−1 of oil. The remaining oily water should be consequently discharged and treated on-shore. Vessels below 400 gross tonnes retain on-board bilge water for subsequent discharge at reception port facilities. Treatment of oily wastewater is challenging and can be effective only if several steps are applied (primary, secondary and tertiary), including an oil separation technique followed by physical or chemical processes that will decrease the organic load, while tertiary treatment (membranes, chemical oxidation) is usually necessary in order to meet discharge limits (Varjani et al. 2020). The main wastewater purification step is secondary treatment and it is the stage where optimization of the selected technique can importantly decrease the cost of tertiary treatment, thereby providing motivation for adequate treatment of the wastewater. Biological treatment is usually selected as a low cost yet efficient secondary treatment process, able to achieve high performance, but easily disrupted by the complexity of the wastewater and its alterations in composition over time (Han et al. 2019). Apart from the high Chemical Oxygen Demand (COD) value (from 3 to 15 g L−1) that bilge wastewater presents (Han et al. 2019) bilge water is usually highly saline, which may inhibit biological activity, leading to poor biological treatment performance (Lefebvre and Moletta 2006).
Several researchers have generated data that could contribute to optimization of biological processes at an industrial level application. An appealing approach is to select appropriate microorganisms highly adapted to the wastewater that would maximize treatment efficiency. Αerobic microorganisms able to grow in bilge wastewater and able to biodegrade organic compounds have been described in the past (Nievas et al. 2006; Cappello et al. 2012; Feknous et al. 2017; Uma and Gandhimathi 2019), while performance in a consortia has also been investigated (Cerqueira et al. 2011). However, when it comes to application, it is usually impossible to operate large scale industrial reactors and fully control the pure culture or consortia. Therefore, interest is growing for open mixed cultures and autochthonous species performance (Castro et al. 2018; Corti-Monzón et al. 2020), where operational conditions can be more convenient and suitable for large scale applications.
There is little information regarding the presence or tracking of isolated or highly efficient microorganisms in autochthonous cultures for oily wastewater degradation, while this approach led to promising results in a recent study examining pharmaceutical compound bioremediation (Fernandes et al. 2020). It is also unknown whether the wastewater characteristics will lead to the same microbial diversity after operation with different starting inocula. Temudo et al. (2008) have indicated no risk of strain degeneration in open mixed cultures, based on natural inocula with a high microbial diversity, operated at continuous process under non-sterile conditions. On the other hand, few studies have examined the biological treatment of bilge wastewater with parallel production of useful by-products. Few studies have focused on energy production, via biogas production, during the anaerobic digestion of bilge wastewater (Li et al. 2017; Vyrides et al. 2018; Morgan-Sagastume et al. 2019) but there is poor description regarding the biodegradation potential and microbial diversity alteration of anaerobic granular sludge when acclimatized to bilge wastewater. In most studies, a specific culturing approach is followed and evaluated while there is lack of studies conducting parallel experiments, comparing different culture types, examined for the same wastewater treatment potential.
In this study, different types of biological cultures and culturing techniques were tested and compared at a small laboratory scale for the treatment of real bilge wastewater. Small scale comparative experiments (conducted on-shore) were used to provide valuable knowledge prior to scale magnification. The treatment efficiency was examined with parallel monitoring of alteration in microbial groups responsible for the biodegradation of the organic load of the wastewater. Furthermore, the potential of biogas production increase was examined through adaptation of the microbial anaerobic culture. The following culturing approaches were examined in small scale laboratory bioreactors: (i) pure aerobic cultures with isolated strains, (ii) consortium of isolated strains under sterile conditions and (iii) mixed cultures obtained with microbial adaptation to the wastewater (aerobic and anaerobic). Real wastewater obtained from the same source for all experiments was used and treatment efficiency was monitored regarding COD and ecotoxicity decrease. Regarding pure strains (used independently and as a consortium), isolation occurred through gradual exposure of initial environmental inocula to bilge wastewater and cultivation on agar plates. To obtain cultures with adapted microbial groups, two conditions were applied: (i) aerobic and (ii) anaerobic. In the first case, nine (9) open mixed aerobic cultures (OMAC) were created by long-term enrichment of liquid cultures with bilge wastewater (150 days of exposure). In the second case, one (1) mixed anaerobic culture (MAnC) with anaerobic granular sludge was created by exposing the biomass to bilge wastewater for 90 days. To the author’s knowledge, this is the first study that systematically compares the biodeterioration of bilge wastewater by different microbial culture types and culturing conditions and provides information regarding the microbial profile of cultures over time. The results of this study enrich the existing information regarding microbial comportment during the treatment of oily wastewater and provide significant information regarding autochthonous-based culture evolution.
Materials and methods
Analytical standards and reagents
All chemicals and reagents used in this study were of high purity, obtained from Sigma-Aldrich (USA), Fluka (Switzerland) and Merck (Germany).
Aerobic pure cultures
Isolation of microorganisms
In order to identify highly adapted microorganisms, several potential sources were chosen and a small amount of liquid or solid material was used for the inoculation of a starting culture. The starting inocula were withdrawn from nine (9) environmental and industrial samples that were (i) previously exposed to oil or bilge wastewater or (ii) had a high microbial diversity (activated sludge, dewatered sludge) (Table S1). The liquid media chosen as a substrate for microorganism growth was a mixture of a minimal microbial growth medium (M9 solution), containing per litre of water: 6 g Na2HPO4, 3 g KH2PO4, 35 g NaCl, 1 g NH4Cl, 1 mL of 100 mM MgSO47H2O, 1 mL of 10 mM CaCl2 (Cappello et al. 2012) and raw bilge wastewater (COD: 1.3–6.2 g L−1; BOD5: 0.2–2.6 g L−1; pH: 6.2–6.7; Salinity: 25–32 ppt; see also Table S5), obtained from a local company (Ecofuel LTD) collecting and treating hazardous oily wastewater from the marine industry. The bilge wastewater was placed in sealed containers under ambient conditions prior to its use in the experiments. For the creation of starting cultures, 250 mL conical glass flasks were filled with 100 mL of liquid media (containing M9 solution; 75% and raw bilge wastewater; 25%) and 5 mL or 5 g of each sample. All flasks were covered with aluminium foil and placed in a shaking incubator at 100 rpm and a constant temperature of 33 °C. Every seven (7) days, the liquid cultures were enriched by retaining 10 mL of the existing liquid in the flask and adding 90 mL of fresh media. The composition of the fresh media was gradually altered over enrichment steps in order to increase bilge wastewater concentration (Table S2). After nine (9) enrichment steps, liquid cultures were used to inoculate agar plates, while enrichment steps were continued to preserve the liquid cultures for use in experiments described in Section 2.3.1. Sterile conditions were applied during this experimental part and all tasks were conducted in a laminar hood, in order to identify and multiply single colonies. Solid media was created by sterilizing (121 °C for 30 min in an autoclave): (i) M9 solution and (ii) bilge wastewater (at the desired ratio each time, see Table S3), with the addition of 20 g L−1 agar for bacteriology. In each petri plate, approximately 30 mL of media was poured and allowed to cool prior to inoculation with 20 μL of each liquid culture, respectively. An appropriate number of plates were inoculated in order to select the most efficient growing colonies. All plates were placed in the dark, at room temperature, and were visually checked until colonies were observed. Every 4–7 days, the colonies were transferred with a loop onto new plates, gradually increasing the concentration of bilge wastewater in the solid substrate, until high concentration of bilge wastewater was achieved. When pure cultures were observed and could be repeatedly multiplied, colonies were collected with sterilized M9 and stored in cryovials containing glycerol until analysed with 16S rRNA gene sequencing for bacteria identification (analysis was performed by Macrogen Europe B.V., Netherlands). The 16s rRNA gene amplification and sequencing were conducted via following universal sequencing primers 785F (5′-GGA TTA GAT ACC CTG GTA-3′) and 907R (5′- CCG TCA ATT CMT TTR AGT TT-3′).
Pure strain biodegradation tests
Following isolation, the strains were grown separately in liquid cultures and then tested for their capacity to biodegrade the organic load encountered in bilge wastewater. All equipment and solvents used were sterilized to exclude contaminations (autoclaved at 121 °C for 20 min). The liquid media used was prepared by adding 0.3 g L−1 phenol, 0.1 g L−1 yeast extract and 0.1 g L−1 glucose to M9 solution (described in Section 2.2.1.). One-hundred fifty (150) millilitre glass serum bottles with a sealing cap were used with a working volume of 50 mL (sterilized liquid media). The appropriate stain was transferred from the agar plate into the serum bottle with a loop (see Section 2.2.1), and sealed. All bottles were incubated at 30 °C and 120 rpm. Pure strain liquid cultures were prepared as described above to be used for biodegradation test with real bilge wastewater. Batch tests were conducted to determine the capacity of pure strains to degrade bilge wastewater individually as well as in a consortium (Exp. 1A and 1B, Table 1 and Table S5). For biodegradation capacity monitoring, Chemical Oxygen Demand (COD) was selected as a reliable and representative parameter quantifying larger part of organic compounds found in this wastewater type (Tran et al. 2015). Experiments were conducted in a low (below 2000 mg L−1) and medium range (2500–3500 mg L−1) of initial COD in wastewater (see Table S5). The growth of strains in pure cultures was monitored by measuring Optical Density at 600nm (OD600). Abiotic degradation was investigated by creating samples without strain addition (control samples). Ιn order to ensure colony abundance, strains were added in the bottles: (i) from liquid cultures (with 1 mL addition), and (ii) from petri plates with the use of a loop. All bottles were incubated at 33 °C and 100 rpm. Sampling was accomplished by piercing the silicone septa of the serum bottle with a sterilized needle connected to a syringe for liquid extraction.
Mixed cultures
Open mixed aerobic culture biodegradation tests
Open cultures were examined for the biodegradation of raw bilge wastewater under aerobic conditions. Nonsterile conditions were applied; however the wastewater was heated (121 °C for 10 min) in order to thermally eliminate volatile compounds. Batch biodegradation experiments were conducted in conical flasks of 250 mL capacity and 100 mL working volume. Samples were created by adding in each flask 90 mL of raw bilge wastewater and 10 mL of the appropriate starting liquid culture (see Section 2.2.2). A flask with 100 mL of bilge wastewater was used (control sample) in order to monitor biodegradation due to microbial growth of autochthonous species present in the wastewater. All flasks were covered with cotton and aluminium foil on top and placed in a shaking incubator at 100 rpm and a constant temperature of 33 °C. COD was monitored over time and experiments were conducted at three initial COD range levels: low (below 2000 mg L−1), medium (2500–3500 mg L−1) and high (higher than 5000 mg L−1). All nine (9) liquid cultures (see Section 2.2.1) were tested in a first experimental cycle with medium COD concentration and in a second one with low COD concentration (Exp. 2A, Table 1). During these experiments and as an open headspace was used, a test was performed in order to ensure that COD removal was not enchased due to volatilization of organic compounds. For this reason, the same initial conditions were applied under two different headspace conditions: open headspace and closed headspace.
Subsequently, two open cultures were chosen in order to examine biodegradation at a high initial COD level and real industrial conditions, whereas sterilization is usually not feasible (Exp. 2B, Table 1). Triplicates of each open culture and control sample were created and monitored over time. During this experiment, the microbial community and diversity were examined and compared with the indigenous species initially present in the wastewater.
Mixed anaerobic culture biodegradation tests
For the determination of anaerobic microorganism’s ability to biodegrade real bilge wastewater, batch tests were performed under anaerobic conditions (Exp. 3A, Table 1). Anaerobic granular sludge withdrawn from a full-scale Internal Circulation (IC) bioreactor treating dairy wastewater operated at pH 7.0–7.5 (Charalambides Christis Ltd, Limassol, Cyprus) was used as inoculum in the present study. Glass serum bottles were used (total volume of 125 mL) filled with 70 mL of bilge wastewater and 55 mL of headspace. The headspace in each serum bottle was flushed with CO2 gas (99.99% purity) for 5 min to remove oxygen before sealing the bottle with a butyl rubber septa and aluminium crimp cap to avoid gas leaks. Three type of samples were created: (i) granular sludge that has already been exposed to bilge wastewater for 90 days (mixed anaerobic culture 1; MAnC 1), (ii) granular sludge that was exposed for the first time to bilge wastewater (mixed anaerobic culture 2; MAnC 2) and (iii) raw bilge wastewater for the monitoring of biomass development (Control). The methane production was monitored in the headspace over 51 days and COD decrease of the wastewater was compared after 51 days of treatment. By the end of the experiment, the microbial profile in each sample was determined and compared with the microbial profile of the initial granular sludge used.
Analytical methods and apparatus
The oxidation of chloride ions present in bilge wastewater was taken into consideration for the analysis of COD, as the salinity of the examined wastewater was high (greater than 25 ppt, see Table S5) (Vyrides and Stuckey 2009). Therefore, a modified method was used (Freire and Sant’Anna 1998). The appropriate dilution of samples took place prior to digestion (in order to obtain the desired salinity in samples) and the appropriate reagent range was used for digestion and quantification. Different calibration curves were created and used for quantification, depending on the salinity of each sample measured. For samples with less than 2000 mg L−1 concentration of chloride, the COD was analysed according to Standard Methods (APHA 2012) . The absorbance of digested samples was measured at 600 nm or 420 nm using a portable spectrophotometer (Hach DR1900). Biochemical Oxygen Demand (BOD) was determined by a five-day BOD test (BOD5) using a respirometric system (Lovibond BD 600). Temperature, salinity and pH were measured using portable instruments (Consort C6030). For the determination of microbial growth, OD600 was measured with a benchtop spectrophotometer at 600 nm (Jenway 7315 UV/Visible spectrophotometer). The gas composition was measured using a gas chromatograph, coupled with a thermal conductivity detector (GC-TCD, Agilent technologies 7820A GC system, Wilmington, DE) according to the method described by Vardanyan et al. (2018).
Next-generation sequencing
Appropriate amount of liquid from open cultures was centrifuged in order to collect 180–220 mg of biomass. Then, the total genomic DNA was extracted through the NucleoSpin DNA stool (Macherey-Nagel, Germany) and was then sent to DNASense Company (Denmark) for sequencing. Bacteria 16S V1–3 rRNA gene sequencing libraries were prepared by a custom protocol. Up to 10 ng of extracted DNA was used as template for PCR amplification of the bacteria 16S V1–3 rRNA gene amplicons. Each PCR reaction (25 μL) contained dNTPs (100 μM of each), MgSO4 (1.5 mM), Platinum Taq DNA polymerase HF (0.5 U/reaction), Platinum High Fidelity buffer (1X) (Thermo Fisher Scientific, USA) and barcoded library adaptors (400 nM of each forward and reverse). PCR was conducted with the following programme: initial denaturation at 95 °C for 2 min, 30 cycles of amplification (95 °C for 20 s, 56 °C for 30 s, 72 °C for 60 s) and a final elongation at 72 °C for 5 min. Duplicate PCR reactions were performed for each sample and the duplicates were pooled after PCR. The adaptors contain 16S V1–3 specific primers: [27F] AGAGTTTGATCCTGGCTCAG and [534R] ATTACCGCGGCTGCTGG (Ward et al. 2012). The resulting amplicon libraries were purified using the standard protocol for Agencourt Ampure XP Beads (Beckman Coulter, USA) with a bead to sample ratio of 4:5. DNA was eluted in 25 μL of nuclease free water (Qiagen, Germany). DNA concentration was measured using Qubit dsDNA HS Assay kit (Thermo Fisher Scientific, USA). Gel electrophoresis using Tapestation 2200 and D1000/High sensitivity D1000 screentapes (Agilent, USA) was used to validate product size and purity of a subset of sequencing libraries. The purified sequencing libraries were pooled in equimolar concentrations and diluted to 6 nM. The samples were paired-end sequenced (2×300 bp) on a MiSeq (Illumina, USA) using a MiSeq Reagent kit v3 (Illumina, USA) and bioinformatics processing was performed by DNASense Company (Denmark).
Acute toxicity screening with Aliivibrio fischeri
The widely applied acute toxicity assay using the marine bacteria A. fischeri was used as a screening test to evaluate the treatment conditions resulting to lower toxicity. The bioluminescence inhibition of the marine bacterium Aliivibrio fischeri was used to infer acute toxic effects, as described elsewhere (Vasquez and Fatta-Kassinos 2013). Lyophilized bacteria were hydrated using a reconstitution solution provided by the kit manufacturer (Microtox, Modern Water, UK). The raw initial wastewater and mixed cultures (aerobic and anaerobic) were tested. Prior to testing, all samples were filtered using a 0.22 μm syringe filter and pH and salinity were adjusted. Serial 1:1 dilutions of the samples were done using a 2% NaCl solution to test four concentrations in the range 9.9–99% and a control in which only bacteria and the 2% NaCl solution were used. An initial inoculum of 10 μL of the bacteria was exposed to 990 μL of samples or NaCl solution for 5 and 15 min. The bioluminescence at time 0, 5 and 15 min was calculated using a self-calibrating photometer (M500, Modern Water UK) to estimate growth inhibition towards the bacteria. Each sample was tested in triplicates and a positive control with phenol was also used. The inhibition of bioluminescence was calculated comparing the bioluminescence in the control experiment with the bioluminescence of each sample. The level of toxicity was then categorized as extremely toxic (Inhibition >90%), highly toxic (70% < Inhibition < 90%), moderately toxic (50% < Inhibition < 70%), medium-low toxic (30% < Inhibition < 50%), low toxic (10% < Inhibition < 30%) and non-toxic (00% < Inhibition < 10%).
Equations
COD removal in batch reactors was calculated according to Eq. (1):
where CODin is the COD value in influent wastewater (mg L−1) and CODout is the value in treated wastewater (mg L−1).
The mass (mg) of COD removed per 10 mL of liquid culture in batch reactors was calculated according to Eq. (2):
where CODin is the COD value in influent wastewater (mg L−1) and CODout is the value in treated wastewater (mg L−1), Vin is the volume of the liquid culture at the beginning of the experiment and Vout is the volume at the end of the experiment.
Bacterial growth rate (μ as h−1) in pure strain cultures was calculated according to Eq. (3):
where logOD1 and logOD2 are the OD600 values at the respective time t1 and t2 (h) (Widdel 2010).
Toxicity was evaluated by determining in the first place the percentage of bioluminescence inhibition (I) according to Eq. (4):
where Ic is the bioluminescence value when Aliivibrio fischeri was exposed to saline control solution and Is is the bioluminescence value when Aliivibrio fischeri was exposed to each test sample (Parmaki et al. 2018).
As described by Parmaki et al. (2018), a linear regression of the inhibition (I), as Γ value (Eq. (5), was logarithmically plotted against the concentration of the compound. Then, the EC50 value, as the concentration that inhibited 50% of the population, was identified and the Toxicity Units (TU50) were calculated as shown in Eq. (6).
Data analysis
For the 16s rRNA result analysis, the software BioEdit 7.2.5.0 was used and for the creation of phylogenetic trees, the software MEGA-X 10.0.5 was used. Phylogenetic trees of microbial isolates were obtained by evolutionary analysis using Maximum Likelihood method and the Jukes–Cantor model. Graphs were created with the software GraphPad Prism. In order to compare the COD values during the second biodegradation test with open cultures, one-way ANOVA was used with Tukey post-test for significant differences between groups.
Results and discussion
Isolation of bilge wastewater degrading aerobic strains
From the nine (9) sources initially selected, three purified aerobic strains could be identified following gradual exposure to bilge wastewater. The following three strains were identified and were able to grow and multiply in bilge wastewater: (i) Halomonas alkaliphila S5a (ACCN: MT374099), (ii) Vibrio antiquarius S8ib (ACCN: MT374098) and (iii) Pseudodonghicola xiamenensis S8iia. (ACCN: ΜT374100), GenBank SUB7324019. For each strain, a partial 16S rRNA sequence was obtained and through sequence alignment and after gene alignment was implemented by the BLAST programme of NCBI (www.ncbi.nlm.nih.gov), the similarities with known strains were determined. Strain S5a presented 100% similarity (identities 1466/1466 base pairs) with Halomonas alkaliphila (CP024811.1), strain S8ib presented 99.93% (identities 1425/1426 base pairs) with Vibrio antiquarius (CP001805.1) and strain S8iia presented 100% similarity (identities 1394/1394) with Pseudodonghicola xiamenensis (NR_043565.1). The isolated strains were therefore named accordingly and their phylogenetic tree is shown in Fig. 1. As described in Fig. 1, H. alkaliphila S5a also presents high similarity with H. venusta while V. antiquarius S8ib is closely related to V. diabolicus, V. alginolyticus and V. parahaemolyticus. Finally, Pseudodonghicola xiamenensis S8iia is closely related to Donghicola eburneus strain.
Phylogenetic trees of microbial isolates obtained by evolutionary analysis using Maximum Likelihood method and the Jukes–Cantor model. The percentage of trees in which the associated taxa clustered together is shown next to the branches. Bootstrap method was used as test of phylogeny with 100 replications. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. Evolutionary analyses were conducted in MEGA X. (a) Halomonas alkaliphila S5a, (b) Vibrio antiquarius S8ib and (c) Pseudodonghicola xiamenensis S8iia
The strain Halomonas alkaliphila S5a, was isolated from raw bilge wastewater (Table S1) and is a Gram-negative halophilic proteobacteria. Halophilic bacteria have the well-known advantages of surviving in high salinity environments and at the same time degrade contaminants, rendering these bacteria suitable and desirable for treatment processes dealing with saline wastewater (Zhuang et al. 2010). They have also been studied for their capacity to degrade hydrocarbons (Castillo-Carvajal et al. 2014). In previous studies, the biodegradation of saline petrochemical wastewater by Halomonas alkaliphila grown in a consortium was investigated (Ahmadi et al. 2017), as well as its nitrification and denitrification capacity under high salinity concentration (Wang et al. 2017).
The second strain, Vibrio antiquarius S8ib, was isolated from an oily mud sample. Vibrio is a genus of Gram-negative bacteria, typically found in seawater. Vibrio antiquarius has been recently isolated from a mesophilic bacterial community associated with hydrothermal vents (East Pacific Rise; Hasan et al. 2015) and from clam and oysters in Korea; Ruditapes philippinarum (Dahanayake et al. 2019) and Crassostrea gigas (Dahanayake et al. 2018). As far as, we know no records exist regarding its capacity to biodegrade organic compounds present in wastewater.
The third strain, Pseudodonghicola xiamenensis S8iia was also isolated from an oily mud sample. As described by Hameed et al. (2014), Pseudodonghicola xiamenensis is a Gram-negative, non-pigmented, strictly aerobic bacterium. It has been previously isolated in China from surface seawater (Hameed et al. 2014) as well as from oil-contaminated surface water (Tan et al. 2009). Therefore, its potential to survive in oil-contaminated water is known but no biodegradation tests have been reported previously.
Bilge wastewater biodegradation under sterile conditions
Single strain biodegradation tests
Each strain was tested individually for its capacity to biodegrade organic compounds present in real saline bilge wastewater (Exp. 1A, Table 1). The highest growth rate for all strains was observed within the first 28 h of contact time and the highest OD600 value was observed at 28 h of growth for Halomonas alkaliphila S5a and Vibrio antiquarius S8ib and at 76 h for Pseudodonghicola xiamenensis S8iia (Fig. 2a and b). The larger part of COD removal (greater than 88.5%) occurred within the 76 first hours of contact time, while the total COD reduction after 10 days of treatment was 72.5%, 61.0% and 77.6% for Halomonas alkaliphila S5a, Vibrio antiquarius S8ib and Pseudodonghicola xiamenensis S8iia, respectively (Fig. 2a and b). No significant decrease of COD was identified in the control sample (less than 4% of COD reduction after 10 days), in which abiotic conditions were implemented and sterilized wastewater was used. In addition, by the end of the experiment, the gas composition in the headspace of each sample was examined. It was found that the O2/N2 (%) in the samples with pure strains was lower, compared to the control sample, more likely due to O2 utilization by microorganisms for bilge biodegradation. Carbon dioxide (end product) in the samples with pure strains was 6 times higher compared to the control sample (Table S4), indicating degradation due to biological activity.
Taking into consideration that the BOD5/COD ratio of the wastewater used was 0.32 (Table S5) and the wastewater examined is classified as medium biodegradable (Bouknana et al. 2014), the performance of the three strains is found to be satisfactory and promising. In a similar study (Ahmadi et al. 2017), a consortium containing the strain Halomonas alkaliphila was tested in an activated sludge lab-scale bioreactor treating saline petrochemical wastewater. The highest COD removal rate achieved was 78.7% for 4 days of contact time (HRT, hydraulic retention time) at a relatively low initial COD (1356 mg L−1) and a similar salinity level with this study. As far as we know, no relative study has been conducted for the strains Vibrio antiquarius and Pseudodonghicola xiamenensis.
Consortium biodegradation test
Τhe performance of a consortium including all three strains was examined and compared with the performance of each strain individually (Exp. 1B, Table 1). In this experiment, bilge wastewater from another sampling period was used with a lower initial COD level and lower BOD5/COD ratio equal to 0.13 (Table S5). During this experiment, lower biodegradation potential of individual strains was observed, in comparison with the Exp. 1A (Table 1). Though COD in this case was relatively lower, the wastewater is still characterised as refractory wastewater due to the high content of non-biodegradable compounds (Tran et al. 2015). It is highly probable that the metabolic activity of the stains was lower due to different wastewater compositions, with fewer easily biodegradable compounds (e.g. low-molecular-weight PAHs) (Nikolopoulou et al. 2013). It is also possible that due to stress factors, the microorganism released soluble microbial products, resulting in a final increase of soluble COD (Lefebvre and Moletta 2006), decreasing thus the recorded biodegradation.
There were no significant differences exhibited between the performance of single strain and consortium treatment, in terms of COD decrease (Fig. 2c). It is likely that the organic compounds able to be degraded are fully utilized in a similar rate and the remaining refractory organic compounds can no longer be degraded biologically. There could also exist antagonistic trends within the consortium due to proteolytic substances produced by the metabolic pathway of each strain (Feichtmayer et al. 2017), inhibiting optimal growth in the consortium, thus biodegradation. A similar trend was observed by Cerqueira et al. (2011) who compared pure and mixed bacteria cultures for the degradation of oily sludge and observed closely related performance of the consortium compared with the single strains in the saturated fraction of the sludge.
Open mixed aerobic cultures: performance and microbial diversity
Biodegradation tests
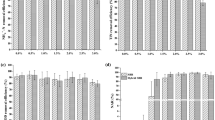
The biodegradation of raw bilge wastewater, from different sampling periods, having different initial COD (low, medium and high) was tested by open mixed aerobic cultures (OMAC) under non-sterile conditions (Exp. 2A, Table 1) in order to reproduce real industrial conditions. All nine starting liquid cultures were examined and compared for their capacity to biodegrade bilge wastewater in order to identify the most efficient open cultures. Two degradation tests were performed at a medium and low initial COD level and the COD decrease was monitored over time (Fig. S6). During low organic loading test, lower biodegradation rates were recorded, as previously observed using sterile conditions and pure cultures (see Section 3.2.1). Unexpectedly, the COD removal observed in the control sample (containing only raw bilge wastewater) was at a similar level with removal observed in the cultures, indicating that autochthonous species present in the wastewater may grow and contribute to biodegradation. Since there was no exceptional performance observed in a specific OMAC, the two cultures from which microorganisms were isolated were chosen for the performance of a high organic load biodegradation test (OMAC 1 and OMAC 8, originating from raw bilge wastewater and an oily mud sample). Total COD removal rates reached 59.9 ± 3.0%, 58.5 ± 1.2% and 58.0 ± 3.3% for the two open cultures (OMAC 1, OMAC 8) and the control sample, respectively (Fig. 3a, b). The removal recorded under high organic load after 9 days of treatment was lower, compared to previous experiments whereas after 3 days of treatment of medium range organic load wastewater, COD removal varied between 61 and 73%. (Exp. 2Ai, Table 1). Furthermore, during this experiment, a significant part of biodegradation occurred between the 6th and 9th day, whereas in previous experiments with low and medium initial COD, the larger part of biodegradation took place prior to the 5th day of treatment. This could be explained due to the longer adaptation time needed for the biodegradation to begin and to the greater availability of biodegradable organic compounds in the wastewater prolonging the biodegradation period. Comparing the two OMAC and control sample (Fig. 3a) and after 9 days of treatment, only one open culture demonstrated a statistically lower average COD value (OMAC 1).
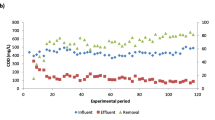
(a) COD values during bilge treatment with open mixed aerobic cultures (OMAC) and nonsterile conditions (Exp. 2B), (b) COD removal during aerobic bilge treatment with open mixed aerobic cultures (OMAC) and nonsterile conditions (Exp. 2B) and (c) CH4 production and COD removal during treatment of bilge with mixed anaerobic cultures (MAnC) and nonsterile conditions (Exp 3A)
Abiotic degradation of bilge wastewater was previously tested, indicating that no significant degradation occurs under sterile conditions and closed headspace (see Section 3.2.1.). However, this experiment took place with an open headspace and a nonsterile environment and a parallel experiment was conducted with the same initial and working conditions with a close headspace in order to test possible decrease of COD in open headspace bottles due to volatilization. The deterioration recorded in both cases was the same, regardless of the headspace condition (Fig. S7), therefore it was concluded that the headspace condition did not influence the COD removal observed. Volatilization was not expected to occur as partial sterilization of bilge was accomplished in order to thermally eliminate volatile compounds. In a similar study where volatile compounds were not eliminated prior to treatment, low deterioration due to volatilization was observed (11.4% decrease of the total hydrocarbon amount, after 14 days of treatment) (Nievas et al. 2006).
Ecotoxicological assessment
As presented in Table 2, raw bilge wastewater was extremely toxic (>90%) in all tested concentrations after 5 and 15 min of exposure. By the end of the treatment period (9 days), the control sample was less toxic than the raw bilge wastewater but still considered as highly toxic (>70%) in most of the concentrations evaluated. After 3 days of treatment, effluents from both OMAC and control sample were slightly less toxic compared to initial wastewater. Between the three testing conditions, samples from the OMAC 1 presented higher toxicity while the same toxicity levels were recorded for OMAC 8 and control sample. Interestingly, after nine (9) days of treatment, samples from OMAC 1 had moderate toxicity at concentrations of 9.9%, 24.8% and 49.5%; whereas samples from OMAC 8 remained highly toxic at concentration of 49.5% and above and the control sample remained highly toxic at concentration of 24.8% and above. This finding indicates that the consortium in OMAC 1 was able to break down the toxic transformation products that were created. All samples tested were at least moderately toxic even after 9 days of treatment, indicating that more treatment steps or longer treatment periods should be considered in future studies.
Aerobic microbial profile
The relative abundance of microorganisms in raw wastewater and cultures that were tested under high organic load is presented in Fig. 4 (Table S9). At the end of the experiment (Exp. 2B, Table 1), both open cultures had similarities, with Alphaproteobacteria being the dominant class (94%) and genus Pseudodonghicola being the dominant genus with 53.6% and 47.2% relative abundance in OMAC 1 and OMAC 8, respectively. It is worth mentioning that in the control sample (which was exposed to bilge for 9 days with an open headspace) Alphaproteobacteria was the dominant class (79%) and Ochrobactrum and Pseudodonghicola were the most abundant genera (56.0% and 19.7%, respectively). The high abundance of Pseudodonghicola in open mixed aerobic cultures (Table S9) is in line with the isolated strain (Section 3.1) and this points out that aerobic treatment of bilge wastewater favours the growth of Alphaproteobacteria and Pseudodonghicola. The higher abundance of Pseudodonghicola in OMAC 1 may be related to better performance of this culture regarding COD removal as well as higher toxicity decrease compared to OMAC 8. It is worth mentioning that Pseudodonghicola xiamenensis S8iia was the strain that exhibited better performance when cultured in single strain cultures (Section 3.2.1). Regarding the other strains isolated, Halomonas and Vibrio genera, they did not have a high relative abundance in the examined open mixed aerobic cultures and this could have been due to isolation procedure (e.g. favourable growth in agar plates but not in real wastewater).
Microbial profile of: (i) initial bilge wastewater (bilge t=0), (ii) open mixed aerobic culture 1(OMAC 1 t=9d), (iii) open mixed aerobic culture 8 (OMAC 8 t=9d) and (iv) control sample (control t=9d) (0.2 cutoff). Class and genus classification is presented in bars and class is also presented in pie charts as a percentage
Celeribacter (from Alphaproteobacteria class), the second genus recorded in high abundance in open mixed aerobic cultures (19.8% in OMAC 1 and 31.7% in OMAC 8) is a well-known genus containing Gram-negative, moderately halophilic, rod-shaped bacteria, encountered and isolated from marine environments (Oh et al. 2015). Celeribacter species are known to biodegrade PAHs and are frequently encountered in bioremediation activities (Cao et al. 2015; Jami et al. 2016). Marispirillum genus (also from the Alphaproteobacteria class) was found in OMAC 1 at 12.6% and OMAC 8 at 3.4% and have been reported for their ability to degrade hydrocarbons (Kim et al. 2010) as well as for their capacity to reduce surface tension and emulsify oils (Gomes et al. 2018).
Regarding the initial wastewater used at the beginning of the biodegradation experiment (for the biodegradation test, 90% of wastewater was used and 10% of liquid culture) Epsilonproteobacteria was the dominant class with Sulfurimonas and Sulfurospirillum, the most abundant genus. Epsilonproteobacteria are found in anoxic and acidic marine environments (Ng and Chiu 2020), and though they were the dominant class in the wastewater used for the biodegradation test, they were not identified in the final culture after 9 days of treatment. The reason for the initial presence of Epsilonproteobacteria could be that prior to use, the wastewater was stored in anoxic sealed bottles, under conditions enhancing their survival and growth. Contrariwise, Alphaproteobacteria were not present in the initial wastewater but had a strong presence in the final cultures and control sample afterwards bilge wastewater was exposed to aerobic batch conditions. Alphaproteobacteria are able to degrade hydrocarbons under aerobic conditions (Cappello et al. 2016; Chen et al. 2019) and they are commonly found in seawater (Rajeev et al. 2019). Similar dynamics are described by Procópio (2020), whereas Alphaproteobacteria is the dominant class in biofilms exposed to oil and chemical surfactants in a marine environment, with Gammaproteobacteria being the second more abundant class. On the other hand, Nisenbaum et al. (2020) enriched a microbial consortium for bilge biodegradation and the Gammaproteobacteria was the dominant class (58%) whereas the Alphaproteobacteria were the second dominant class with 28% relative abundance. The difference between our study and the study of Nisenbaum et al. (2020) could be due to the different compositions of bilge wastewater and the incubation conditions of the consortium.
Mixed anaerobic cultures: performance and microbial diversity
Anaerobic digestion
Bilge wastewater biodegradation under anaerobic conditions (Exp. 3A, Table 1), was tested in lab scale bioreactors under batch mode. After 51 days of treatment, the maximum COD removal rate observed was 38%, achieved by the mixed anaerobic culture containing granular sludge that has been previously exposed to bilge wastewater for 90 days (MAnC 1). Anaerobic treatment of bilge wastewater exhibited lower performance compared to aerobic treatment (38% COD removal after 51 days versus 58–60% after 9 days, see Fig. 3b, c), while wastewater of the same COD range was used (high range). Regarding methane production, the culture, previously exposed to bilge (MAnC 1) was able to produce a higher amount of methane (42.9 mL CH4 in 51 days), indicating that microorganisms could perform better after longer exposure to the wastewater. In all samples, more than 70% of methane production occurred within the first 14 days of treatment, while the percentage of methane in the produced biogas remained low (higher value recorded: 47%, on day 31 for MAnC 1). It is worth mentioning that in the control sample containing only bilge wastewater (nonsterile conditions) a small COD removal was observed and 18% CH4 production. Similar studies reported 28% COD removal in 13 days (Vyrides et al. 2018) and 67±6% COD removal with biogas production with a CH4 content up to 79±6 % in 30 days (Morgan-Sagastume et al. 2019). Regarding ecotoxicity tests, as already described in Section 3.3.2, raw bilge wastewater was extremely toxic. Bilge wastewater treated under anaerobic conditions for 51 days was significantly less toxic than the initial wastewater, with a TU50 ranging between 4 and 10. This finding demonstrates that after an extended treatment, toxic compounds are significantly decomposed.
Anaerobic microbial profile
The relative abundance of bacteria and archaea in the granular sludge before and after exposure to bilge wastewater, as well as the groups developed in the wastewater under anaerobic conditions are presented in Fig. 5 (Table S10). The more efficient culture for the treatment of bilge wastewater (MAnC1) presented differentiations in its microbial profile, further discussed below.
Microbial profile (Bacteria and Archaea) of mixed anaerobic cultures in: (i) mixed anaerobic culture with granular sludge, after 141 days exposure to bilge (MAnC 1), (ii) mixed anaerobic culture with granular sludge, after 51-day exposure to bilge (MAnC 2), (iii) suspended biomass formed after 51-day exposure to bilge (Control), and (iv) initial granular sludge used for mixed anaerobic cultures, not being exposed to bilge (GS) (2% cutoff). Class and genus classification is presented in bars and class is also presented in pie charts
Regarding bacteria identified, granular sludge forming the mixed anaerobic cultures demonstrated a large variety of species in terms of genus classification (with more than 60 genus identified with over than 0.5% relative abundance) while in the suspended biomass created in the initial wastewater (Control), a lower abundance was observed. In all mixed anaerobic cultures (MAnC 1, MAnC 2 and initial granular sludge used as inocula), some of the most abundant bacteria groups (class and genus) were Gammaproteobacteria (Nitrosococcus), Thermotogae (Mesotoga), Actinobacteria (Brooklawnia), Deltaproteobacteria (Syntrophorhabdus) and Nitrospira and Anaerolineae uncultured genus. Nitrosococcus genus are Ammonia Oxidizing Bacteria (AOB), adapted to saline and acidic environments, regularly found in marine environments but less commonly detected in waste water treatment reactors (Fumasoli et al. 2017). Mesotoga are frequently found in oil-polluted marine mesophilic anaerobic environments (Nesbø et al. 2019). Brooklawnia species are nonmotile, mesophilic, neutrophilic facultative anaerobic bacteria that have been previously identified in chlorosolvent-contaminated environments (da Costa et al. 2015) and Syntrophorhabdus are as well nonmotile, mesophilic but strictly anaerobic bacteria, metabolizing simple aromatic compounds (Kuever 2014). On the other hand, in the control sample (bilge wastewater exposed to anaerobic conditions for 51 days where biomass was developed without any initial inocula), Alphaproteobacteria, Bacteroidia and Thermotogae were the dominant bacteria classes found and Petrotoga, a thermophilic anaerobic bacteria proven to degrade oil (Daryasafar et al. 2014), was a dominant genus. It is worth mentioning that the MAnC1 was the culture with greater performance (higher COD reduction and CH4 production) and for this culture, the class Deltaproteobacteria was present at a higher percentage compared to other cultures and samples (MAnC2, Control, GS).
Regarding the identification of archaea, in the initial anaerobic granular sludge used for the creation of mixed anaerobic cultures Methanomicrobia class was the dominant class, with the genus Methanosaeta and Methanolinea dominating and the genus Methanosacrina being observed at less than 0.5%. In anaerobic MAnC 1 and MAnC 2, Methanomicrobia was as well the dominant class but in this case, the genus Methanosacrina (43 and 39% relative abundance, respectively) was recorded as being the most abundant, followed by Methanosaeta and Methanolinea (Fig. 5). For the most efficient culture (MAnC1), we can observe that Thermoplasmata class was recorded at a higher percentage in comparison to other cultures and samples (MAnC2, Control, GS). Methanosarcina are of great ecological significance as they are the only organisms known to utilize acetate, methylamines and methanol to methane, carbon dioxide and ammonia. According to a review by De Vrieze et al. (2012), Methanosarcina is a very robust methanogen and can withstand various stressful conditions such as high salinity, organic loading rate, pH, acetate and others and therefore is not surprisingly has become the dominant genus in anaerobic granular sludge exposed to bilge wastewater.
The exposure of bilge under anaerobic conditions (without any initial inoculum) pointed out lower CH4 generation (Fig. 3). Microbial profile analysis in the end of the experiment showed the presence of Methanohalophilus at 90.7% relative abundance. This is the first study that showed the presence of indigenous methanogens in bilge wastewater and future studies can focus on isolation of this strain.
Conclusions
Biological treatment of real bilge wastewater was tested using several microbial cultures under various conditions. Aerobic conditions were found more suitable regarding COD reduction. Pure aerobic cultures with isolated strains Halomonas alkaliphila S5a, Vibrio antiquarius S8ib and Pseudodonghicola xiamenensis S8iia were found to have the highest COD removal rate after ten (10) days of treatment, under medium organic loading (72.5%, 61.0% and 77.6%, respectively). Open mixed aerobic cultures had satisfactory COD removal rates after nine (9) days of treatment and high organic loading (from 58.0 to 59.9% COD removal). The treatment efficiency decreased importantly when low organic loading conditions were tested. Under sterile conditions, no significant decrease of the COD occurred over time in the control sample (raw bilge) while under nonsterile conditions, the COD decreased importantly due to biodegradation. The microbial profile analysis in the end of the open mixed aerobic culture experiment revealed that Alphaproteobacteria, Pseudodonghicola were the dominant class and genus, respectively, both in the cultures and the control sample. The fact that Pseudodonghicola was isolated from bilge wastewater but also was the dominant genus identified both in the control sample and in the open mixed cultures shows that it is naturally thriving during aerobic treatment of the examined bilge wastewater. Anaerobic digestion, after 51 days of treatment, achieved poor COD removal rates (from 18 to 38%), while biogas production and methane percentage were low. Contrariwise, the toxicity decrease was important. By the end of bilge wastewater anaerobic treatment (51 days), Methanosacrina genus exhibited the higher relative abundance in granular sludge and Methanohalophilus was identified in the control sample (raw bilge).
Data availability
The datasets used and/or analysed during the current study are available from the corresponding author upon reasonable request.
References
Ahmadi M, Jorfi S, Kujlu R, Ghafari S, Soltani RDC, Haghighifard NJ (2017) A novel salt-tolerant bacterial consortium for biodegradation of saline and recalcitrant petrochemical wastewater. J Environ Manag 191:198–208. https://doi.org/10.1016/j.jenvman.2017.01.010
Andersson, Karin, Selma Brynolf, J. Fredrik Lindgren, and Magda Wilewska-Bien. 2016. Shipping and Environment. Springer.
Bouknana D, Hammouti B, Salghi R, Jodeh S, Zarrouk A, Warad I, Aouniti A, Sbaa M (2014) Physicochemical characterization of olive oil mill wastewaters in the eastern region of Morocco. J Mater Environ Sci 5(4):1039–1058
Cao J, Lai Q, Yuan J, Shao Z (2015) Genomic and metabolic analysis of fluoranthene degradation pathway in Celeribacter indicus P73 T. Sci Rep 5:1–12. https://doi.org/10.1038/srep07741
Cappello S, Santisi S, Calogero R, Hassanshahian M, Yakimov MM (2012) Characterisation of oil-degrading bacteria isolated from bilge water. Water Air Soil Pollut 223(6):3219–3226. https://doi.org/10.1007/s11270-012-1103-y
Cappello S, Volta A, Santisi S, Morici C, Mancini G, Quatrini P, Genovese M, Yakimov MM, Torregrossa M (2016) Oil-degrading bacteria from a membrane bioreactor (BF-MBR) system for treatment of saline oily waste: isolation, identification and characterization of the biotechnological potential. Int Biodeterior Biodegrad 110:235–244. https://doi.org/10.1016/j.ibiod.2015.12.028
Castillo-Carvajal LC, Sanz-Martín JL, Barragán-Huerta BE (2014) Biodegradation of organic pollutants in saline wastewater by halophilic microorganisms: a review. Environ Sci Pollut Res 21(16):9578–9588. https://doi.org/10.1007/s11356-014-3036-z
Castro AR, Silva PTS, Castro PJG, Eliana A, Rosário M, Domingues M, Pereira MA (2018) Tuning culturing conditions towards the production of neutral lipids from lubricant-based wastewater in open mixed bacterial communities. Water Res 144:532–542. https://doi.org/10.1016/j.watres.2018.07.068
Cerqueira VS, Hollenbach EB, Maboni F, Vainstein MH, Camargo FAO, do Carmo Peralba MR, Bento FM (2011) Biodegradation potential of oily sludge by pure and mixed bacterial cultures. Bioresour Technol 102(23):11003–11010. https://doi.org/10.1016/j.biortech.2011.09.074
Chen C, Ming J, Yoza BA, Liang J, Li QX, Guo H, Liu Z, Deng J, Wang Q (2019) Characterization of aerobic granular sludge used for the treatment of petroleum wastewater. Bioresour Technol 271(September 2018):353–359. https://doi.org/10.1016/j.biortech.2018.09.132
Coca J, Gutierrez G, Benito J (2011) Treatment of oily wastewater. In: Coca-Prados J, Gutiérrez-Cervelló G (eds) Water purification and management. Springer Netherlands, Dordrecht, pp 1–55
Corti-Monzón G, Nisenbaum M, Villegas-Plazas M, Junca H, Murialdo S (2020) Enrichment and characterization of a bilge microbial consortium with oil in water-emulsions breaking ability for oily wastewater treatment. Biodegradation 31:57–72. 0123456789. https://doi.org/10.1007/s10532-020-09894-y
da Costa MS, Rainey FA, Moe WM (2015) Brooklawnia. In: Bergey’s manual of systematics of archaea and bacteria. Society, American Cancer, pp 1–5. https://doi.org/10.1002/9781118960608.gbm00161
Dahanayake PS, Hossain S, Wickramanayake MVKS, Wimalasena SHMP, Heo GJ (2019) Manila clam (Ruditapes philippinarum) marketed in Korea as a source of Vibrios harbouring virulence and β-lactam resistance genes. Lett Appl Microbiol 71(1):46–53. 1–8. https://doi.org/10.1111/lam.13229
Dahanayake PS, De Silva BCJ, Hossain S, Shin GW, Heo GJ (2018) Occurrence, virulence factors, and antimicrobial susceptibility patterns of Vibrio spp. isolated from live oyster (Crassostrea gigas) in Korea. J Food Saf 38(5):1–8. https://doi.org/10.1111/jfs.12490
Daryasafar A, Azad EG, Ghahfarokhi AK, Mousavi SF (2014) Simulation studies on growth and death of microorganisms using the oil-degrading bacteria Petrotoga sp. Chem Eng Technol 37(12):2152–2164. https://doi.org/10.1002/ceat.201400129
Feichtmayer J, Deng L, Griebler C (2017) Antagonistic microbial interactions : contributions and potential applications for controlling pathogens in the aquatic systems. Front Microbiol 8(November):2192, 1–14. https://doi.org/10.3389/fmicb.2017.02192
Feknous N, Branes Z, Rouabhia K, Batisson I, Amblard C (2017) Isolation characterization and growth of locally isolated hydrocarbonoclastic marine bacteria (Eastern Algerian Coast). Environ Monit Assess 189(2):49. https://doi.org/10.1007/s10661-016-5758-5
Fernandes JP, Patrícia D, Marisa C, Almeida R, Carvalho MF, Mucha AP (2020) Potential of bacterial consortia obtained from different environments for bioremediation of paroxetine and bezafibrate. J Environ Chem Eng 8(4):103881. https://doi.org/10.1016/j.jece.2020.103881
Freire DDC, Sant’Anna GL (1998) A proposed method modification for the determination of COD in saline waters. Environ Technol (United Kingdom) 19(12):1243–1247. https://doi.org/10.1080/09593331908616784
Fumasoli A, Bürgmann H, Weissbrodt DG, Wells GF, Beck K, Mohn J, Morgenroth E, Udert KM (2017) Growth of nitrosococcus-related ammonia oxidizing bacteria coincides with extremely low pH values in wastewater with high ammonia content. Environ Sci Technol 51(12):6857–6866. https://doi.org/10.1021/acs.est.7b00392
Gomes MB, Gonzales-Limache EE, Sousa STP, Dellagnezze BM, Sartoratto A, Silva LCF, Gieg LM, Valoni E, Souza RS, Torres APR, Sousa MP, de Paula SO, Silva CC, Oliveira VM (2018) Exploring the potential of halophilic bacteria from oil terminal environments for biosurfactant production and hydrocarbon degradation under high-salinity conditions. Int Biodeterior Biodegrad 126:231–242. https://doi.org/10.1016/j.ibiod.2016.08.014
Hameed A, Shahina M, Lin SY, Nakayan P, Liu YC, Lai WA, Hsu YH (2014) Youngimonas vesicularis Gen. Nov., Sp. Nov., of the family Rhodobacteraceae, isolated from surface seawater, reclassification of Donghicola xiamenensis Tan et al. 2009 as Pseudodonghicola xiamenensis Gen. Nov., Comb. Nov. and Emended Description of the Ge. Int J Syst Evol Microbiol 64(PART 8):2729–2737. https://doi.org/10.1099/ijs.0.060962-0
Han M, Zhang J, Chu W, Chen J, Zhou G (2019) Research progress and prospects of marine oily wastewater treatment: a review. Water (Switzerland) 11(12):1–29. https://doi.org/10.3390/w11122517
Hasan NA, Grim CJ, Lipp EK, Rivera ING, Chun J, Haley BJ, Taviani E, Choi SY, Hoq M, Munk AC, Brettin TS, Bruce D, Challacombe JF, Detter JC, Han CS, Eisen JA, Huq A, Colwell RR (2015) Deep-sea hydrothermal vent bacteria related to human pathogenic vibrio species. Proc Natl Acad Sci U S A 112(21):E2813–E2819. https://doi.org/10.1073/pnas.1503928112
Jägerbrand AK, Brutemark A, Svedén JB, Gren IM (2019) A review on the environmental impacts of shipping on aquatic and nearshore ecosystems. Sci Total Environ 695:133637. https://doi.org/10.1016/j.scitotenv.2019.133637
Jami M, Lai Q, Ghanbari M, Moghadam MS, Kneifel W, Domig KJ (2016) Celeribacter persicus sp. Nov., a polycyclicaromatic-hydrocarbon-degrading bacterium isolated from mangrove soil. Int J Syst Evol Microbiol 66(4):1875–1880. https://doi.org/10.1099/ijsem.0.000961
Kim PI, Ryu J, Kim YH, Chi Y-T (2010) Production of biosurfactant lipopeptides Iturin A, fengycin, and surfactin A from Bacillus subtilis CMB32 for control of Colletotrichum gloeosporioides. J Microbiol Biotechnol 20(1):138–145. https://doi.org/10.4014/jmb.0905.05007
Kuever J (2014) The Family Syntrophorhabdaceae. In: Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F (eds) The prokaryotes: Deltaproteobacteria and Epsilonproteobacteria. Springer Berlin Heidelberg, Berlin, Heidelberg, pp 301–303. https://doi.org/10.1007/978-3-642-39044-9_403
Lefebvre O, Moletta R (2006) Treatment of organic pollution in industrial saline wastewater: a literature review. Water Res 40(20):3671–3682. https://doi.org/10.1016/j.watres.2006.08.027
Li J, Sun S, Yan P, Fang L, Yu Y, Xiang Y, Wang D, Gong Y, Gong Y, Zhang Z (2017) Microbial communities in the functional areas of a biofilm reactor with anaerobic–aerobic process for oily wastewater treatment. Bioresour Technol 238:7–15. https://doi.org/10.1016/j.biortech.2017.04.033
Mclaughlin C, Falatko D, Danesi R, Albert R (2014) Characterizing shipboard bilgewater effluent before and after treatment. Environ Sci Pollut Res 21:5637–5652. https://doi.org/10.1007/s11356-013-2443-x
Morgan-Sagastume F, Jacobsson S, Olsson LE, Carlsson M, Gyllenhammar M, Sárvári Horváth I (2019) Anaerobic treatment of oil-contaminated wastewater with methane production using anaerobic moving bed biofilm reactors. Water Res 163:114851. https://doi.org/10.1016/j.watres.2019.07.018
Nesbø CL, Charchuk R, Pollo SMJ, Budwill K, Kublanov IV, Haverkamp THA, Foght J (2019) Genomic analysis of the mesophilic Thermotogae genus Mesotoga reveals phylogeographic structure and genomic determinants of its distinct metabolism. Environ Microbiol 21(1):456–470. https://doi.org/10.1111/1462-2920.14477
Ng JCY, Chiu JMY (2020) Changes in biofilm bacterial communities in response to combined effects of hypoxia, ocean acidification and nutrients from aquaculture activity in three fathoms cove. Mar Pollut Bull 156(October 2019):111256. https://doi.org/10.1016/j.marpolbul.2020.111256
Nievas ML, Commendatore MG, Olivera NL, Esteves JL, Bucalá V (2006) Biodegradation of Bilge waste from Patagonia with an indigenous microbial community. Bioresour Technol 97(18):2280–2290. https://doi.org/10.1016/j.biortech.2005.10.042
Nikolopoulou M, Pasadakis N, Kalogerakis N (2013) Evaluation of autochthonous bioaugmentation and biostimulation during microcosm-simulated oil spills. Mar Pollut Bull 72(1):165–173. https://doi.org/10.1016/j.marpolbul.2013.04.007
Nisenbaum M, Corti-Monzón G, Villegas-Plazas M, Junca H, Mangani A, Patat ML, González JF, Murialdo SE (2020) Enrichment and key features of a robust and consistent indigenous marine-cognate microbial consortium growing on oily bilge wastewaters. Biodegradation 31(1–2):91–108. https://doi.org/10.1007/s10532-020-09896-w
Oh YT, Avedoza C, Lee SS, Jeong SE, Jia B, Jeon CO (2015) Celeribacter naphthalenivorans Sp. Nov., a naphthalene-degrading bacterium from tidal flat sediment. Int J Syst Evol Microbiol 65(9):3073–3078. https://doi.org/10.1099/ijs.0.000381
Parmaki S, Vyrides I, Vasquez MI, Hartman V, Zacharia I, Hadjiadamou I, Catarina B.M. Barbeitos, et al. (2018) Bioconversion of alkaloids to high-value chemicals: comparative analysis of newly isolated lupanine degrading strains. Chemosphere. 193:50–59. https://doi.org/10.1016/j.chemosphere.2017.10.165
Procópio L (2020) Changes in microbial community in the presence of oil and chemical dispersant and their effects on the corrosion of API 5L steel coupons in a marine-simulated microcosm. Appl Microbiol Biotechnol 104(14):6397–6411. https://doi.org/10.1007/s00253-020-10688-8
Rajeev M, Sushmitha TJ, Toleti SR, Pandian SK (2019) Culture dependent and independent analysis and appraisal of early stage biofilm-forming bacterial community composition in the southern coastal seawater of India. Sci Total Environ 666:308–320. https://doi.org/10.1016/j.scitotenv.2019.02.171
APHA (2012) Standard Methods for the Examination of Water and Wastewater / Prepared and Published Jointly by American Public Health Association, American Water Works Association, Water Environment Federation
Tan T, Wang B, Shao Z (2009) Donghicola xiamenensis Sp. Nov., a marine bacterium isolated from seawater of the Taiwan Strait in China. Int J Syst Evol Microbiol 59(5):1143–1147. https://doi.org/10.1099/ijs.0.000901-0
Temudo MF, Muyzer G, Kleerebezem R, Van Loosdrecht MCM (2008) Diversity of microbial communities in open mixed culture fermentations: impact of the pH and carbon source. Appl Microbiol Biotechnol 80(6):1121–1130. https://doi.org/10.1007/s00253-008-1669-x
Tiselius P, Magnusson K (2017) Toxicity of treated bilge water: the need for revised regulatory control. Mar Pollut Bull 114(2):860–866. https://doi.org/10.1016/j.marpolbul.2016.11.010
Tran NH, Ngo HH, Urase T, Gin KYH (2015) A critical review on characterization strategies of organic matter for wastewater and water treatment processes. Bioresour Technol 193:523–533. https://doi.org/10.1016/j.biortech.2015.06.091
Uma V, Gandhimathi R (2019) Organic removal and synthesis of biopolymer from synthetic oily bilge water using the novel mixed bacterial consortium. Bioresour Technol 273(October 2018):169–176. https://doi.org/10.1016/j.biortech.2018.11.003
Vardanyan A, Kafa N, Konstantinidis V, Shin SG, Vyrides I (2018) Phosphorus dissolution from dewatered anaerobic sludge: effect of pHs, microorganisms, and sequential extraction. Bioresour Technol 249(February):464–472. https://doi.org/10.1016/J.BIORTECH.2017.09.188
Varjani S, Joshi R, Srivastava VK, Ngo HH, Guo W (2020) Treatment of wastewater from petroleum industry: current practices and perspectives. Environ Sci Pollut Res 27(22):27172–27180. https://doi.org/10.1007/s11356-019-04725-x
Vasquez MI, Fatta-Kassinos D (2013) Is the evaluation of ‘traditional’ physicochemical parameters sufficient to explain the potential toxicity of the treated wastewater at sewage treatment plants? Environ Sci Pollut Res 20(6):3516–3528. https://doi.org/10.1007/s11356-013-1637-6
De Vrieze J, Hennebel T, Boon N, Verstraete W (2012) Methanosarcina: the rediscovered methanogen for heavy duty biomethanation. Bioresour Technol 112:1–9. https://doi.org/10.1016/j.biortech.2012.02.079
Vyrides I, Stuckey DC (2009) A modified method for the determination of chemical oxygen demand (COD) for samples with high salinity and low organics. Bioresour Technol 100(2):979–982. https://doi.org/10.1016/j.biortech.2008.06.038
Vyrides I, Drakou EM, Ioannou S, Michael F, Gatidou G, Stasinakis AS (2018) Biodegradation of bilge water: batch test under anaerobic and aerobic conditions and performance of three pilot aerobic moving bed biofilm reactors (MBBRs) at different filling fractions. J Environ Manag 217:356–362. https://doi.org/10.1016/j.jenvman.2018.03.086
Wang T, Li J, Zhang LH, Yu Y, Zhu YM (2017) Simultaneous heterotrophic nitrification and aerobic denitrification at high concentrations of NaCl and ammonia nitrogen by Halomonas bacteria. Water Sci Technol 76(2):386–395. https://doi.org/10.2166/wst.2017.214
Ward DV, Gevers D, Giannoukos G, Earl AM, Methé BA, Sodergren E et al (2012) Evaluation of 16s rDNA-based community profiling for human microbiome research. PLoS One 7:e39315
Widdel F (2010) Theory and measurement of bacterial growth. Di Dalam Grundpraktikum Mikrobiologie 4:1–11
Zhuang X, Han Z, Bai Z, Zhuang G, Shim H (2010) Progress in decontamination by halophilic microorganisms in saline wastewater and soil. Environ Pollut 158(5):1119–1126. https://doi.org/10.1016/j.envpol.2010.01.007
Acknowledgements
The author A.A.M. would like to thank Stella Parmaki for her support in 16s rRNA result analysis and phylogenetic tree creation.
Funding
This work was co-funded by the European Regional Development Fund and the Republic of Cyprus through the Research and Innovation Foundation (Project: OPPORTUNITY/0916/MSCA/0006). Research Program Acronym: MicrobEatBilge.
Author information
Authors and Affiliations
Contributions
Aikaterini A. Mazioti: Methodology, investigation, validation, visualization, writing - original draft preparation. Vyrides, Ioannis: Conceptualization, methodology, project administration, supervision, writing - review and editing. Vasquez, Marlen I.: Investigation, writing - review and editing.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Responsible Editor: Gerald Thouand
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
ESM 1
(PDF 206 kb)
Rights and permissions
About this article
Cite this article
Mazioti, A.A., Vasquez, M.I. & Vyrides, I. Comparison of different cultures and culturing conditions for the biological deterioration of organic load from real saline bilge wastewater: microbial diversity insights and ecotoxicity assessment. Environ Sci Pollut Res 28, 36506–36522 (2021). https://doi.org/10.1007/s11356-021-13153-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11356-021-13153-9