Abstract
This experiment was conducted to provide a better insight into the plant responses to nitric oxide (NO) and selenium nanoparticle (nSe). Chicory seedlings were sprayed with nSe (0, 4, and 40 mg l−1), and/or NO (0 and 25 μM). NO and/or nSe4 improved shoot and root biomass by an average of 32%. The nSe40 adversely influenced shoot and root biomass (mean = 26%), exhibiting moderate toxicity partly relieved by NO. The nSe and NO treatments transcriptionally stimulated the dehydration response element B1A (DREB1A) gene (mean = 29.6-fold). At the transcriptional level, nSe4 or NO moderately upregulated phenylalanine ammonia-lyase (PAL) and hydroxycinnamoyl-CoA quinate transferase (HCT1) genes (mean = sevenfold). The nSe4 + NO, nSe40, and nSe40 + NO groups drastically induced the expression of PAL and HCT1 genes (mean = 30-fold). With a similar trend, hydroxycinnamoyl-CoA Quinate/shikimate hydroxycinnamoyl transferase (HQT1) gene was also upregulated in response to nSe and/or NO (mean = 25-fold). The activities of nitrate reductase and catalase enzymes were also induced in the nSe- and/or NO-treated seedlings. Likewise, the application of these supplements associated with an increase in ascorbate concentration (mean = 31.5%) reduced glutathione (mean = 35%). NO and/or nSe enhanced the PAL activity (mean = 36.4%) and soluble phenols (mean = 40%). The flowering was also influenced by the supplements in dose and compound dependent manner exhibiting the long-time responses. It appears that the nSe-triggered signaling can associate with a plethora of developmental, physiological, and molecular responses at least in part via the fundamental regulatory roles of transcription factors, like DREB1A as one the most significant genes for conferring tolerance in crops.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Severe environmental conditions and various pollutants reduce plant growth rate and yield. Hence, diverse strategies have been employed to find a novel procedure to improve nutritional status, stress tolerance, primary metabolism, secondary metabolism, yield, and biofortification in crops, and industrial/medicinal plants. In this regard, the exogenous utilization of selenium (Se) or nitric oxide (NO) has gained a lot of concern to modify plant growth, metabolism, productivity, and stress tolerance (Djanaguiraman et al. 2018; Safari et al. 2018; Rajaee et al. 2020; Sotoodehnia-Korani et al. 2020). Taking scientific knowledge gaps into account, more convincing studies are required to gain a better insight into the molecular and physiological principles of Se and NO functions/risks in agriculture.
NO, a ubiquitous gasotransmitter signaling molecule, governs plant responses to internal and external cues. Moreover, it has a close interplay with other signaling agents among which hormones, reactive oxygen species, and H2S are of critical significance (Santisree et al. 2018). NO presents a cytoprotective role against oxidative stress at the low concentrations through modifications in transcriptions of stress-responsive genes (Huang et al. 2018). Besides, the fundamental regulatory roles of NO towards transcriptome and proteome are supported by several recent reports in different plant species, including Arabidopsis (Hussain et al. 2016; Imran et al. 2018), and cotton (Huang et al. 2018).
Taking the health of humans into account, it has been emphasized that humans should daily consume Se, especially in its organic forms (selenocysteine and selenomethionine) at the concentration of 55 μg day−1 (Babajani et al. 2019b). Moreover, insufficient Se intake via daily diet (especially seed/plant-originated foods) globally make Se deficiency as a health concern. Hence, the productions of Se-biofortified food and medicine derived from cultivating plants are of great significance. Depending on the plant species, applied concentrations, Se type, plant developmental stage, and treatment method, Se intake can be associated with both positive and negative responses in the plants (Tamaoki and Maruyama-Nakashita 2017; Djanaguiraman et al. 2018; Nazerieh et al. 2018; Babajani et al. 2019a, b; Hussein et al. 2019; Quiterio-Gutiérrez et al. 2019; Zahedi et al. 2019). Nanoscience and technologies are currently offering significant opportunities for the productions of numerous innovative promising fertilizers, pesticides, and so on, thereby improving plant growth, yield, metabolism, and protection (Seddighinia et al. 2020). Moreover, recent scientific reports imply that nano-based products can cause exclusive reactions in a biological system different from bulk counterparts (Moghanloo et al. 2019a, b). It has become evident that Se nanoparticle (nSe) provides a great opportunity for raising Se bioavailability (Hu et al. 2018; Babajani et al. 2019a).
Numerous researchers underlined the potential benefits of individual utilization of Se to promote plant growth and resistance against various stresses, including salinity (Zahedi et al. 2019; Soleymanzadeh et al. 2020), drought (Andrade et al. 2018), and high temperature (Djanaguiraman et al. 2018). Activating the antioxidant system (Andrade et al. 2018; Quiterio-Gutiérrez et al. 2019), enhancing the nutritional status (Babajani et al. 2019a; Zahedi et al. 2019), inducing hormonal changes (Tamaoki and Maruyama-Nakashita 2017; Cao et al. 2018; Babajani et al. 2019a; Zahedi et al. 2019), stimulating primary metabolism (Sotoodehnia-Korani et al. 2020), and secondary metabolism (Babajani et al. 2019a; Sotoodehnia-Korani et al. 2020) are known as the most important physiological mechanisms through which Se utilization can promote plant growth and resistance. It is important to note that nSe treatment may induce epigenetic responses (Rajaee et al. 2020; Sotoodehnia-Korani et al. 2020). However, convincing evidence on Se-mediated molecular changes is rare, and involved mechanisms remain controversial. Likewise, the potential advantages of exogenously applied NO towards improving plant tolerance to stress conditions have supported by several lines of evidence (Majeed et al. 2018; Nabi et al. 2019; Nazerieh et al. 2018; Dai et al. 2020). Besides, beneficial roles of NO in improving stress tolerance have been attributed to several major mechanisms, including modification in primary metabolism (Majeed et al. 2018), transcriptional upregulation in stress-responsive genes (Imran et al. 2018), post-translational modifications of proteins (Huang et al. 2018), activation of the antioxidant system (Akram et al. 2018; Nazerieh et al. 2018), regulation of developmental programs (Nabi et al. 2019), stimulation in primary metabolism (Nazerieh et al. 2018), and induction in secondary metabolism (Akram et al. 2018; Nabi et al. 2019). However, NO (Nazerieh et al. 2018) and Se (Babajani et al. 2019a; Rajaee et al. 2020) at high concentrations can cause cytotoxicity through oxidative/nitrosative stress and metabolic disturbance.
The coordinated network of different signaling pathways manages the plant responses to environmental factors. Signal/stimulus reception and transduction are usually associated with changes in transcriptions of genes, antioxidant machinery, cell cycle, and chromatin epigenetic modification (Iranbakhsh et al. 2020). Adaptation or acclimation to diverse biotic and abiotic stress factors is mediated through the transcriptional regulation of stress/stimulus-responsive genes. Several families of transcription factors are involved in transcriptional regulation of defense-related genes. As is well supported by recent scientific reports, dehydration-responsive element binding 1A (DREB1A) transcription factor is contributed to regulating plant growth (Kohan-Baghkheirati et al. 2018), increasing yield (Donde et al. 2019), and conferring plant tolerance to abiotic stress conditions, like salinity (Kohan-Baghkheirati et al. 2018), drought (Kudo et al. 2017), and cold (Donde et al. 2019). DREB1A transcription factors are proteins involved in the modulation of a plethora of downstream genes through binding to the DRE (dehydration-responsive element) cis-regulatory DNA sequence (Kohan-Baghkheirati et al. 2018). Taking agricultural aims into account, the DREB1A gene has been characterized among the most significant genes for conferring tolerance in crops counteracting with stresses (Kudo et al. 2017; Donde et al. 2019). Hereby, we selected DREB1A as a target gene to address transactional responses to the nSe and/or NO.
Along with photosynthesis, nitrogen metabolism is a determining factor for plant growth, productivity, metabolism, and stress tolerance. In plants, a mineral nitrogen assimilation is a fundamental event that is mediated through the catalytic actions of key enzymes, such as nitrate reductase, nitrite reductase, and glutamate synthase. Moreover, nitrate reductase is also involved in NO production (Tejada-Jimenez et al. 2019). Few studies focused on the effect of Se or nSe (Babajani et al. 2019a; Bian et al. 2020) and NO (Balotf et al. 2018) on nitrogen metabolism, which further highlights the necessity of conducting more studies. Hereby, we aimed to address the possible changes in nitrate reductase activity as a target point in primary metabolism, following individual or combined applications of nSe and NO.
The pools of reactive oxygen species (ROS) and reactive nitrogen species (RNS) are influenced by both internal and external cues. As is well known, these active agents trigger complex signaling cascades, thereby regulating the activation/inactivation of responsive genes and defense routes in response to environmental factors. However, an imbalance between production and scavenging these active agents can be hazardous to cells in terms of destructions of biomolecules, cellular structures, and organelles. Plant cell employs both enzymatic (like catalase, peroxidase, and superoxide dismutase) and non-enzymatic (like carotenoids, ascorbate, glutathione, tocopherol, and flavonoids) antioxidants to protect the cell against oxidative stress. Ascorbate and glutathione are non-enzymatic antioxidants which serve in maintaining redox balance in plant cells under diverse stress conditions. Moreover, the ascorbate-glutathione cycle (one of the most significant components of the antioxidant machinery) in the cytoplasm, mitochondria, and chloroplasts not only counteracts with the oxidative burst but also contributes to the modulation of plant developmental and metabolism programs (Pandey et al. 2015). Furthermore, the transcriptional upregulation in the ascorbate and glutathione pathway has become evident in response to diverse environmental factors (Pandey et al. 2015). Many researchers confirmed that Se fertilization in forms of selenate or selenite in a dose-dependent manner makes changes in the antioxidant system (Andrade et al. 2018; Quiterio-Gutiérrez et al. 2019). However, evidence on the nSe-associated potential changes in the antioxidant system, especially the non-enzymatic component, is rare and needs to be further explored. NO is also involved in the modulation of enzymatic antioxidants and ascorbate/glutathione cycle, mainly through post-translational modifications of proteins (Huang et al. 2018). Therefore, monitoring the nSe- and/or NO-mediated changes in the enzymatic and non-enzymatic antioxidants is one of the objectives of this study.
Cichorium intybus L. (chicory) as one of the most important medicinal herbs possesses pharmaceutical valuable secondary metabolites, especially chlorogenic acid (monocaffeoyl quinic acid), isochlorogenic acid (dicaffeoylquinic acid), caftaric acid (monocaffeoyl tartaric acid), cichoric acid (dicaffeoyl tartaric acid), caffeic acid, and flavonoids (Legrand et al. 2016). The considerable wound healing, antidiabetic, hypolipidemic, anti-inflammatory, anti-allergic, anti-tumor, antimicrobial, antioxidative, anthelmintic, gastroprotective, and hepato-protective functions of chicory-derived extract/medicines have been well illustrated. As it is highlighted in Fig. 1, phenylalanine ammonia-lyase (PAL), hydroxycinnamoyl-CoA: quinate hydroxycinnamoyl transferases (HQTs), and hydroxycinnamoyl-CoA: shikimate/quinate hydroxycinnamoyl transferases (HCTs) are the major checkpoints during the production of these valuable secondary metabolites. Moreover, PAL is contributed to the generation of salicylic acid (SA) which is a significant bioactive signaling agent (Sheteiwy et al. 2019). Among plant secondary metabolites, phenolic substances are of critical importance contributing to a plethora of biological functions, including antioxidant activity, signaling events, and defense responses (Sheteiwy et al. 2019). Considering antioxidant, anti-senescence, anti-inflammatory, and anti-cancer activities of diverse phenolic metabolites, the quantity and quality of these secondary metabolites in plant-derived foods and medicine are among the most important qualitative factors which are considered in the food and pharmaceutical industries. In this regard, salicylic acid, a ubiquitous phenylpropanoid derivative involves major signaling cascades through which cellular transcriptional program is remodeled in response to internal and external environmental stimuli or stress conditions. We, therefore, aimed to monitor the nSe- and NO-associated changes in the expression of HCT1, HQT1, and PAL genes as target points in secondary metabolism and defense system.
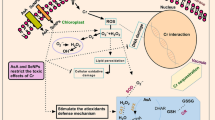
A schematic design on metabolic routes contributes to the production of hydroxycinnamic derivatives in chicory. The involvements of PAL (phenylalanine ammonia-lyase), HQTs (hydroxycinnamoyl-CoA: quinate hydroxycinnamoyl transferases), and HCTs (hydroxycinnamoyl CoA: shikimate/quinate hydroxycinnamoyl transferases) in production of the significant secondary metabolites are highlighted
Within this framework, we aimed to explore whether foliar applications of nSe and NO can associate with modifications in (I) seedling growth, (II) physiology, (III) expression of DREB1A transcription factor, (IV) transcription patterns of HCT1, HQT1, and PAL genes, and (V) flowering in C. intybus (chicory).
Material and methods
Chicory (C. intybus) seeds (PAKANBAZR, ISFAHAN, Iran) were grown in a pot containing peat and perlite (1:1). Thirty-five-day old seedlings were grouped into eight treatment groups with six independent replications. The nanoscale Se was supplied in form of stock red solution (1000 mg l−1 containing 0.1% polyvinylpyrrolidone (PVP) as a stabilizer) with traits, including zeta potential of − 16.32 mV (the negative surface charge), APS of 10–45 nm, density of 3.89 g cm−3, and spherical morphology from the company (NanoSany Corporation, Iranian Nanomaterials Pioneers Company, Mashhad City, Khorasan Province, Iran). The treatments were selected based on the pretest experiment. Chicory seedlings were sprayed with different concentrations of nSe (0, 4, and 40 mg l−1), corresponding doses of Se (IV) oxide (SeO2) as a bulk Se, and/or NO (sodium nitroprusside (SNP; Merck) as a NO donor; 0 and 25 μM) for six times with 1-week intervals. Seven days after the last sprays, seedlings of three replications (pots) were harvested and subjected to further growth, physiological, and molecular analysis, while the other three pots for each treatment group were harvested 2 months after the last sprays to evaluate flowering at plant reproductive stage (long-term effects of the treatments).
Transcriptions of target genes
Expression patterns of the target genes were estimated in leaves. Liquid nitrogen-grounded samples were subjected to RNA purification in Trizol-based protocol (GeneAll Biotechnology Co, South Korea). Next, the RNA purity was validated according to the recorded absorbance ratio (260/280 nm) with nanodrop (Thermo Scientific™NanoDrop Model 2000c). After that, complementary DNA (cDNA) was synthesized using a thermocycler (PEQLAB, 96Grad). The forward and reverse primer sequences for the target genes, including phenylalanine ammonia lyase (PAL; KP752086), hydroxycinnamoyl-CoA quinate transferase (HCT1 gene; KT222891), hydroxycinnamoyl-CoA quinate/shikimate hydroxycinnamoyl transferase (HQT1; KT222893), and DREB1A transcription factor as well as elongation factor as a housekeeping gene, are presented in Table 1. The transcription profiles of the target genes were assessed based on SYBR green (GeneAll, South Korea) and the real-time quantitative PCR approach (Applied Biosystems StepOne™ Real-Time PCR). Next, according to the CT method, fold differences were calculated using 2−ΔΔCT to determine the variations in expressions of the genes (Ghasempour et al. 2019).
Activities of target enzymes
Well-powdered leaves in liquid nitrogen were subjected to enzyme extraction using the phosphate buffer (100 mM; pH of 7.3) and centrifugation at 4 °C. According to the procedure described by Babajani et al. (2019a), the nitrate reductase activity was measured and expressed in micromole nitrite per hour per gram fresh mass (μmol NO2 –h −1g−1 fw). Besides, catalase activity was quantified based on the method represented by Sheteiwy et al. (2019) and expressed in unit enzyme per gram fresh weight (unit Eg−1 fw). Also, PAL activity was determined using the protocol of Beaudoin-Eagan and Thorpe (1985) and expressed in microgram cinnamate per min per gram fresh weight (μg Cin min−1 g−1 fw).
Quantifications of ascorbate, reduced glutathione, and soluble phenols
Quantification of ascorbate and reduced glutathione concentrations in leaves were spectrophotometrically performed according to the reduction reaction of Mo (VI) to Mo (V) at an acidic pH condition in the way the reaction mixture consisted of leaf extract, 4 mM (NH4)6Mo7O24.4H2O, 28 mM NaH2PO4, and 0.6 M H2SO4. The samples were incubated at 95 °C for 90 min and then cooled. Finally, the absorbance for each sample was monitored at 695 nm. The concentrations of ascorbate and reduced glutathione were measured using a molar absorption coefficient of 3400 and 2700 M−1 cm−1, respectively (Babajani et al. 2019a). Total soluble phenols in the ethanolic leaf extracts were also estimated by the application of Folin-Ciocalteu reagent and the standard curve of a standard compound, tannic acid (Nazerieh et al. 2018).
Statistical analysis
The experimental design was completely randomized. All data were subjected to analysis of variance (two-way ANOVA) using SPSS software. The mean values of three independent replications were submitted to variance analysis by the Duncan test at a level of 5% of probability. The correlation coefficient (r) was also evaluated to address the potential correlation between the expression of genes.
Results
The NO, BSe4, nSe4, and nSe4 + NO treatments significantly improved shoot biomass by 31.3%, 21.5%, 38.4%, and 74.6%, respectively (Fig. 2a), while the BSe40, nSe40, and nSe40 + NO adversely influenced shoot biomass by 42.6%, 38%, and 24.9%, respectively (Fig. 2a). With a similar trend, the NO (17.5%), BSe4 (12.7%), nSe4 (21%), and nSe4 + NO (38%) treatments led to the significant increase in root fresh mass when compared with the control (Fig. 2b). BSe40, nSe40, and nSe40 + NO decreased root fresh mass by 23.5%, 18%, and 11% (Fig. 2b).
BSe, nSe, and/or NO-mediated changes in biomass accumulation in the shoot (a) and root (b) of chicory seedlings. C, control; NO, NO of 25 μM, BSe4 BSe at 4 mg l−1; nSe4, nSe at 4 mg l−1; nSe4 + NO, nSe at 4 mg l−1 + NO; BSe40, BSe at 40 mg l−1; nSe40, nSe at 40 mg l−1; nSe40 + NO, nSe at 40 mgl−1 + NO
The NO, nSe4, and nSe4 + NO treatments moderately upregulated expression of DREB1A gene by 22.4-, 15-, and 31.2-fold, respectively (Fig. 3a). However, nSe4 and nSe4 + NO treatments led to the drastic upregulation in DREB1A by 43.7- and 36-fold. The individual treatments of nSe4 and NO led to slight upregulation in the expression of the PAL gene by five- and 9.1-fold, respectively (Fig. 3b), while the nSe4 + NO, nSe40, and nSe40 + NO groups displayed the significantly higher expression of the PAL gene by 24.8-, 19.3-, and 31.9-fold, respectively (Fig. 3b). With a similar trend, the slight increase in expression of the HCT1 gene resulted from the individual nSe4 or NO treatments by 4.6- and 9.4-fold (Fig. 3c). However, the nSe4 + NO, nSe40, and nSe40 + NO treatment groups drastically induced expression of HCT1 gene by 28-, 39.9-, and 37.3-fold (Fig. 3c). NO, nSe4, nSe4 + NO, nSe40, and nSe40 + NO treatments also upregulated HQT1 gene by 19-, 17.1-, 23.2-, 31.3-, and 38.5-fold, respectively (Fig. 3d). As it can be noticed in Fig. 3e, there was a significant correlation among expressions of the explored target genes.
The activities of nitrate reductase enzyme were enhanced by NO (62.2%), nSe4 (60.5%), nSe4 + NO (twofold), and nSe40 + NO (56.4%) treatments (Fig. 4a). However, the observed slight increase (21.8%) in nitrate reductase activity in response to the nSe40 treatment was not statistically significant relative to the control. Moreover, the individual applications of NO (62.2%) or nSe4 (60.5%) moderately induced the activity of catalase enzyme by 62.2% and 60.5%, respectively, in comparison with the control (Fig. 4b). However, drastic induction in catalase activity was observed in the nSe4 + NO (twofold), nSe40 (2.1-fold), and nSe40 + NO (twofold) treatment groups (Fig. 4b). The NO, nSe4, nSe4 + NO, nSe40, and nSe40 + NO treatments were also associated with significant increases in ascorbate concentration by 23.5%, 42%, 43.6%, 36.2%, and 48.9%, respectively, over than the control (Fig. 4c). Similarly, the NO (25%), nSe4 (34%), nSe4 + NO (41%), nSe40 (32%), and nSe40 + NO (42.9%) groups exhibited the significant higher contents of reduced glutathione when compared with the control (Fig. 4d). The NO, nSe4, nSe4 + NO, nSe40, and nSe40 + NO treatments significantly stimulated the activity of PAL enzyme by 28.3%, 35.4%, 46.5%, 41.5%, and 37.3%, respectively (Fig. 4e). With a similar trend, the soluble phenols were enhanced in response to the NO (20.7%), nSe4 (21.9%), nSe4 + NO (38.6%), nSe40 (29.3%), and nSe40 + NO (41.4%) treatments (Fig. 4f). The analysis of variance revealed that the effects of NO, nSe, and their interaction (NO*nSe) on almost all dependent variables were statistically significant (Table 2).
At the reproductive stage, the long-time effects of the applied treatments of nSe or NO were evaluated based on the numbers of produced flowers (Fig. 5). The NO, BSe4, nSe4, and nSe4 + NO treatments were associated with increases in the production of flowers by 45%, 23.8%, 50%, and 90%, respectively, over the control (Fig. 5), while the BSe40, nSe40, and nSe40 + NO treatments produced fewer numbers of flowers (Fig. 5), exhibiting the long-time phytotoxicity.
Discussion
The foliar application of NO and/or nSe mediated significant changes in plant growth, physiology, gene expression, and flowering dependent on the supplement doses. Individual NO or nSe4 supplementation enhanced growth and flowering, while the simultaneous application (nSe4 + NO) exhibited to be even more effective. However, the application of nSe at a high concentration was associated with phytotoxicity risk. Based on the best of our knowledge, this is the first report addressing nSe and NO-associated functions or risks in chicory. Furthermore, the observations manifest that plant behavior to nSe is partly distinctive from that of the bulk which can be attributed to differences in influx, metabolism, interactions with biomolecules, and physicochemical properties of nanoparticles relative to the bulk counterpart (Hu et al. 2018; Babajani et al. 2019a; Sotoodehnia-Korani et al. 2020). In line with our results, nSe at an optimized dose modified growth and physiology in Melissa officinalis (Babajani et al. 2019a), strawberry (Zahedi et al. 2019), Arachis hypogaea (Hussein et al. 2019), bitter melon (Rajaee et al. 2020), and pepper (Sotoodehnia-Korani et al. 2020). However, its phytotoxicity at high concentrations has also become evident (Babajani et al. 2019a, b; Rajaee et al. 2020; Sotoodehnia-Korani et al. 2020). Likewise, exogenous NO increased biomass in plant species, like Brassica oleracea (Munawar et al. 2019), peppermint (Nazerieh et al. 2018), and citrus (Khoshbakht et al. 2018). In this regard, the Se-mediated improvement in photosynthesis performance (Hajiboland et al. 2019; Soleymanzadeh et al. 2020), nutrition (Babajani et al. 2019a; Zahedi et al. 2019), nitrogen metabolism (Rajaee et al. 2020; Sotoodehnia-Korani et al. 2020), and transcriptional regulation (Rajaee et al. 2020; Sotoodehnia-Korani et al. 2020) are considered as main mechanisms. Similar mechanisms, including modifications in plant nutrition, photosynthesis, phytohormones, and transcriptions of genes, have been reported for exogenously applied NO (Khoshbakht et al. 2018; Huang et al. 2018; Nazerieh et al. 2018; Munawar et al. 2019; Nabi et al. 2019).
The nSe and NO treatments upregulated DREB1A transcription factor, PAL, HCT1, and HQT1 genes. According to the best of our knowledge, this is the first evidence on the nSe- and NO-triggered upregulation in DREB1A, PAL, HCT1, and HQT1 genes. Taking the signal recognition and transduction into account, plant hormones, concentrations of ROS/RNS (especially H2O2 and NO), transcription factors, and their interplay are among major elements through which chromatin ultrastructure, gene accessibility, and/or transcriptions of genes can be governed. The Se-associated changes in endogenous concentrations of ROS, especially H2O2, (Feigl et al. 2019) and NO (Feigl et al. 2019; Hajiboland et al. 2019) have become evident. H2O2 and NO are major signaling molecules by which signaling cascades can be triggered (Iranbakhsh et al. 2020). Considering knowledge gaps, it is, therefore, necessary to monitor potential fluctuations in endogenous NO, H2O2, and H2S following nSe and NO application in future studies. One of the most vital regulation layers of genes is mediated through the superfamilies of transcription factors, regulatory proteins interacting with specific promoter sequences of their genes, and a multitude of downstream genes. Transcriptome analysis revealed that a plethora of transcription factors, like WRKYs, MYBs, HSFs (Hussain et al. 2016; Huang et al. 2018; Imran et al. 2018), are NO-responsive. Moreover, the NO-associated modifications in transcriptome had been observed in Arabidopsis (Hussain et al. 2016; Imran et al. 2018) and chickpea (Santisree et al. 2018). However, scientific reports on the nSe-associated changes in the expression profile of transcription factors and their responsive downstream genes are rare, and the necessity of further researches is crystal clear. The expression of the DREB1A gene in the beneficial treatments (nSe4, NO, and nSe + NO groups) were significantly lower (70% to twofold) than the nSe40-treated plants. However, this upregulation could be a sign of a slight transient Se stress and redox-based activation of plant immunity response. Moreover, this mechanism (upregulation in DREB1A) can explain how Se application at low concentrations may improve plant growth and stress tolerance; more molecular studies are needed. However, it should be warned that the nSe40 treatment at high concentration was associated with long-time moderate toxicity in terms of a decrease in biomass and flowering. The growth-promoting effects of the nSe4 and NO treatments can be explained by the increased nitrate reductase activity (a key enzyme in nitrogen primary metabolism) and enhanced antioxidant system, thereby improving photosynthesis efficiency and protection against photoinhibition event; the trade-offs are necessary for coordinating plant growth rate and adaptation to stress. In line with our results, numerous studies have reported that the application of Se or NO can stimulate the defense system as well as promote plant growth and yield (Santisree et al. 2018; Quiterio-Gutiérrez et al. 2019; Rajaee et al. 2020; Sotoodehnia-Korani et al. 2020). Transcriptome analyses in Arabidopsis revealed that DREB transcription factors are Se-responsive genes (Van Hoewyk et al. 2008). In rice, Se led to overexpression of DREB2A and NAC5 transcription factors (Khattab et al. 2014). As another consistent evidence, Se in the form of selenite upregulated ERF96 (ethylene-responsive element-binding factor transcription factors), thereby inducing antioxidant system in Arabidopsis (Jiang et al. 2020). In bitter melon, the nSe application in culture medium led to upregulation in the WRKY1 transcription factor which is involved in secondary metabolism and growth regulation (Rajaee et al. 2020). Moreover, the exposure to nSe resulted in upregulation in bZIP and WRKY1 transcription factors in pepper (Sotoodehnia-Korani et al. 2020) which is consistent with our result. Furthermore, the nSe treatments in wheat transcriptionally stimulated heat shock factor A4A (HSFA4A; an anti-apoptosis agent) which is involved in H2O2 signal reception and signaling (Safari et al. 2018), implying redox-based regulation. Moreover, one important regulation level is epigenetic responses (histone/DNA methylation or acetylation). Recently, convincing reports showed that nSe supplementation was associated with an epigenetic response through cytosine DNA methylation in Capsicum annuum (Sotoodehnia-Korani et al. 2020) and Momordica charantia (Rajaee et al. 2020). Moreover, DREB1A is involved in the regulation of the metabolism of diverse phytohormones, like auxin (Xu et al. 2017). Therefore, the nSe- and/or NO-mediated induction in the expression of DREB1A transcription factor can be regarded as a mechanism by which nSe or NO application can modify growth and confer plant tolerance to stress conditions. Growth and stimulus/stress responses are coordinated through the interplay of signaling cascades and the involvement of a multitude of endogenous factors such as hormones, nutrition, transcriptome, and proteome profiles. At suitable low doses, nSe and NO applications can lead to the promotion in growth and primary metabolism as well as induction in the immune system and secondary metabolism, while these supplements at the high doses can be hazardous to cells through oxidative/nitrosative stress, nutritional perturbation, metabolic disturbance, and abnormalities in the meristem (Sotoodehnia-Korani et al. 2020). Besides, convincing reports highlighted the Se-mediated changes in phytohormones, especially ethylene, jasmonic acid, and salicylic acid (Tamaoki and Maruyama-Nakashita 2017; Cao et al. 2018). Likewise, nSe was associated with modification in salicylic acid (Soleymanzadeh et al. 2020), auxin (Babajani et al. 2019a), and abscisic acid (Zahedi et al. 2019) which are implicated in modulating organogenesis, growth, defense system, metabolism, and gene expression. It appears that the nSe-mediated alterations in endogenous NO along with changes in the generation, distribution, and/or signaling of hormones, in turn, trigger a plethora of developmental, physiological, and molecular responses at least in part via responsive transcription factors, like DREB1A. Taking agricultural aims into account, the DREB1A gene has been characterized among the most significant genes for conferring tolerance in crops (Kudo et al. 2017). According to the results, nSe-triggered signaling remodels transcription program through which the immune system, primary/secondary metabolism, growth, flowering, and stress tolerance can be modified. However, it is necessary to warn that this compound can be associated with cytotoxicity at high doses.
The observed significant positive correlations among the DREB1A transcription factor and the studied target genes in secondary metabolism and defense system (PAL, HCT1, and HQT1) implying the potential crosstalk between this transcription factor and regulation of secondary metabolism. With a similar trend, nSe enhanced PAL activity and concentrations of soluble phenols, indicating the induced secondary metabolism. In this regard, Se/nSe treatments upregulated diverse genes, including PAL and chalcone synthase in Brassica juncea (Handa et al. 2019), rosmarinic acid synthase (RAS), and hydroxyphenylpyruvate reductase (HPPR) genes in Melissa officinalis (Babajani et al. 2019a), as well as PAL and 4-coumarate: CoA-ligase (4CL) in bitter melon (Rajaee et al. 2020). Apart from nSe, the exogenously applied NO in chickpea transcriptionally upregulated chalcone synthase (Santisree et al. 2018), implying the NO contributions into the secondary metabolism and the ROS scavenging. PAL enzyme is a major marker in the production of phenylpropanoid-derived secondary metabolites, among which salicylic acid (a signaling hormone) contributes to the modulation of defense machinery and confers stress tolerance. The foliar application of nSe in strawberry led to an increase in salicylic acid, induction in antioxidants, a rise in secondary metabolism, and promotion in salinity tolerance (Soleymanzadeh et al. 2020). Consistent with our reports, exogenous application of NO (Akram et al. 2018; Santisree et al. 2018; Nabi et al. 2019) or nSe (Babajani et al. 2019a; Handa et al. 2019; Sotoodehnia-Korani et al. 2020) mediated induction in secondary metabolism and production of secondary metabolites. Taking phytochemistry of chicory into account, chlorogenic acid, isochlorogenic acid, caftaric acid, cichoric acid, caffeic acid, and flavonoids are phenylpropanoid derivatives which confer a multitude of pharmaceutical functions in chicory. Hence, the concentration and quality of these substances are among the most important qualitative factors considered in the food and pharmaceutical industries. Hereby, this study provides molecular evidence on the potential functions of nSe and NO to stimulate the production of secondary metabolites in medicinal plants, like chicory. A schematic design on the potential mechanisms involved in plant responses to nSe and NO is presented in Fig. 6.
The results showed the potential advantageous of NO and nSe at a suitable concentration towards nitrogen metabolism through modification in nitrate reductase activity which is a key index in primary metabolism. Moreover, nitrate reductase is also involved in NO production (Tejada-Jimenez et al. 2019). In line with our results, nSe promoted nitrate reductase in Momordica charantia (Rajaee et al. 2020), lettuce (Bian et al. 2020), peppermint (Nazerieh et al. 2018), pepper (Sotoodehnia-Korani et al. 2020), and Melisa officinalis (Babajani et al. 2019a). The observed similarity in plant responses to individual treatments of NO or nSe indicates that nSe signaling is mediated at least to some extent through endogenous NO; it needs to be further explored. Our observation indicated that nSe at a low dose and/or NO improved flowering. However, nSe adversely influenced flowering at the high dose, which can be attributed to the nSe40-mediated restriction in growth, perturbations in nutrition, potential changes in hormonal balances, and overexpression of DREB1A. In plants, over-transcription of DREB1A, however, may associate with dwarfism morphology and delayed flowering (Kudo et al. 2017). As is well known, there is close coordination between vegetative developmental stage, primary metabolism, and flowering (Seddighinia et al. 2020).
The increase in the generation of non-enzymatic antioxidant compounds, like carotenoids, glutathione, ascorbic acid, phenolics, flavonoids, and tocopherols, is a significant mechanism by which plant cell can protect themselves against oxidative burst. The observed stimulating influence of nSe and NO on both enzymatic and non-enzymatic antioxidant machinery can be also attributed to the regulatory roles of the DREB1A transcription factor. In this concern, several lines of evidence confirm the close interplay among DREB1A expression, ROS homeostasis, and antioxidant machinery in plants (Kohan-Baghkheirati et al. 2018; Bhalani et al. 2019). In Chrysanthemum morifolium, the close correlation among DREB1A expression, concentrations of non-enzymatic, activities of enzymatic antioxidants, and stress tolerance have been confirmed (Elansary et al. 2020). The nSe application enhanced non-enzymatic antioxidants, including ascorbate, glutathione, and phenols in tomato plants (Quiterio-Gutiérrez et al. 2019) which is in agreement with our findings. It was observed that NO partly alleviated the moderate toxicity of nSe at the high dose. In line with our results, Nazerieh et al. (2018) investigated the interaction of NO and nSe in peppermint and found that NO has a considerable efficiency to protect plants against the risk-associated with nanoparticles. Likewise, the exogenously applied NO alleviated the Se toxicity in rice through promoting antioxidant machinery and regulating metabolism-related genes (Dai et al. 2020).
Conclusion
Taken together, the individual and/or combined treatments of nSe and NO influenced growth, enzymatic and non-enzymatic (ascorbate and reduced glutathione) antioxidant system, nitrogen metabolism, secondary metabolism, transcription of genes (DREB1A, HQT1, HCT1, and PAL), and flowering in chicory seedlings. According to our results, it appears that the nSe-triggered signaling can associate with a plethora of developmental, physiological, and molecular responses at least in part via the fundamental regulatory roles of transcription factors, like DREB1A. This experiment highlights both potential functions and risks associated with the nSe application. This study underlined the nSe- and NO-mediated transcriptional modification in secondary metabolism with a close positive correlation with the DREB1A transcription factor. Moreover, this study points out the necessity of monitoring molecular behaviors of genes (especially transcription factors) in response to the nanoparticles and their potential crosslink with metabolic pathways.
References
Akram N, Iqbal M, Muhammad A, Ashraf M, Al-Qurainy F, Shafiq S (2018) Aminolevulinic acid and nitric oxide regulate oxidative defense and secondary metabolisms in canola (Brassica napus L.) under drought stress. Protoplasma 255:163–174
Andrade F, Da Silva G, Guimarães K, Barreto H, De Souza K, Guilherme L, Faquin V (2018) Selenium protects rice plants from water deficit stress. Ecotoxicol Environ Saf 164:562–570
Babajani A, Iranbakhsh A, Ardebili ZO, Eslami B (2019a) Differential growth, nutrition, physiology, and gene expression in Melissa officinalis mediated by zinc oxide and elemental selenium nanoparticles. Environ Sci Pollut Res 26:24430–24444
Babajani A, Iranbakhsh A, Ardebili ZO, Eslami B (2019b) Seed priming with non-thermal plasma modified plant reactions to selenium or zinc oxide nanoparticles: cold plasma as a novel emerging tool for plant science. Plasma Chem Plasma Process 39:21–34
Balotf S, Islam S, Kavoosi G, Kholdebarin B, Juhasz A, Ma W (2018) How exogenous nitric oxide regulates nitrogen assimilation in wheat seedlings under different nitrogen sources and levels. PLoS One 13:e0190269. https://doi.org/10.1371/journal.pone.0190269
Beaudoin-Eagan LD, Thorpe TA (1985) Tyrosine and phenylalanine ammonia-lyase activities during shoot initiation in tobacco callus cultures. Plant Physiol 78:438–441
Bhalani H, Thankappan R, Mishra GP, Sarkar T, Bosamia TC, Dobaria J (2019) Regulation of antioxidant mechanisms by AtDREB1A improves soil-moisture deficit stress tolerance in transgenic peanut (Arachis hypogaea L.). PLoS One 14:e0216706. https://doi.org/10.1371/journal.pone.0216706
Bian Z, Bo L, Cheng R, Yu W, Tao L, Yang Q (2020) Selenium distribution and nitrate metabolism in hydroponic lettuce (Lactuca sativa L.): effects of selenium forms and light spectra. J Integr Agric 19:133–144. https://doi.org/10.1371/journal.pone.0216706
Cao D, Liu Y, Ma L, Jin X, Guo G, Tan R, Liu Z, Zheng L, Ye F, Liu W (2018) Transcriptome analysis of differentially expressed genes involved in selenium accumulation in tea plant (Camellia sinensis). PLoS One 13:6
Dai Z, Rizwan M, Gao F, Yuan Y, Huang H, Hossain M, Xiong S, Cao M, Liu Y, Tu S (2020) Nitric oxide alleviates selenium toxicity in rice by regulating antioxidation, selenium uptake, speciation and gene expression. Environ Pollut 257:113540
Djanaguiraman M, Belliraj N, Bossmann SH, Prasad PV (2018) High-temperature stress alleviation by selenium nanoparticle treatment in grain sorghum. ACS Omega 3:2479–2491
Donde R, Gupta M, Gouda G, Kumar J, Vadde R, Sahoo K, Dash S, Behera L (2019) Computational characterization of structural and functional roles of DREB1A, DREB1B and DREB1C in enhancing cold tolerance in rice plant. Amino Acids 51:839–853. https://doi.org/10.1007/s00726-019-02727-0
Elansary HO, Abdel-Hamid AM, Yessoufou K, Al-Mana FA, El-Ansary DO, Mahmoud EA, Al-Yafrasi MA (2020) Physiological and molecular characterization of water-stressed Chrysanthemum under robinin and chitosan treatment. Acta Physiol Plant 42:31
Feigl G, Horváth E, Molnár Á, Oláh D, Poór P, Kolbert Z (2019) Ethylene-nitric oxide interplay during selenium-induced lateral root emergence in Arabidopsis. J Plant Growth Regul 38:1481–1488
Ghasempour M, Iranbakhsh A, Ebadi M, Ardebili ZO (2019) Multi-walled carbon nanotubes improved growth, anatomy, physiology, secondary metabolism, and callus performance in Catharanthus roseus: an in vitro study. 3 Biotech 9(11):404
Hajiboland R, Rahmat S, Zeinalzadeh N, Farsad-Akhtar N, Hosseinpour-Feizi MA (2019) Senescence is delayed by selenium in oilseed rape plants. J Trace Elem Med Biol 55:96–106
Handa N, Kohli SK, Sharma A, Thukral AK, Bhardwaj R, AbdoAllah EF, Alqarawi A, Ahmad P (2019) Selenium modulates dynamics of antioxidative defence expression, photosynthetic attributes and secondary metabolites to mitigate chromium toxicity in Brassica juncea L. plants. Environ Exp Bot 161:180–192
Hu T, Li H, Li J, Zhao G, Wu W, Liu L, Wang Q, Guo Y (2018) Absorption and bio-transformation of selenium nanoparticles by wheat seedlings (Triticum aestivum L.). Front Plant Sci 9:597
Huang J, Wei H, Li L, Yu S (2018) Transcriptome analysis of nitric oxide-responsive genes in upland cotton (Gossypium hirsutum). PLoS ONE 13(3):e0192367
Hussain A, Mun BG, Imran QM, Lee SU, Adamu TA, Shahid M, Kim KM, Yun BW (2016) Nitric oxide mediated transcriptome profiling reveals activation of multiple regulatory pathways in Arabidopsis thaliana. Front Plant Sci 7:975
Hussein H, Darwesh OM, Mekki B (2019) Environmentally friendly nano-selenium to improve antioxidant system and growth of groundnut cultivars under sandy soil conditions. Biocatal Agric Biotechnol 18:101080
Imran QM, Hussain A, Lee SU, Mun BG, Falak N, Loake GJ, Yun BW (2018) Transcriptome profile of NO-induced Arabidopsis transcription factor genes suggests their putative regulatory role in multiple biological processes. Sci Rep 8:771. https://doi.org/10.1038/s41598-017-18850-5
Iranbakhsh A, Ardebili ZO, Molaei H, Ardebili NO, Amini M (2020) Cold plasma up-regulated expressions of WRKY1 transcription factor and genes involved in biosynthesis of cannabinoids in Hemp (Cannabis sativa L.). Plasma Chem Plasma Process 40:527–537. https://doi.org/10.1007/s11090-020-10058-2
Jiang L, Yang J, Liu C, Chen Z, Yao Z, Cao S (2020) Overexpression of ethylene response factor ERF96 gene enhances selenium tolerance in Arabidopsis. Plant Physiol Biochem 149:294–300
Khattab HI, Emam MA, Emam MM, Helal NM, Mohamed MR (2014) Effect of selenium and silicon on transcription factors NAC5 and DREB2A involved in drought-responsive gene expression in rice. Biol Plant 58:265–273
Khoshbakht D, Asghari MR, Haghighi M (2018) Effects of foliar applications of nitric oxide and spermidine on chlorophyll fluorescence, photosynthesis and antioxidant enzyme activities of citrus seedlings under salinity stress. Photosynthetica 56:1313–1325
Kohan-Baghkheirati E, Bagherieh-Najjar M, Abdolzadeh A, Geisler-Lee J (2018) Altered DREB1A gene expression in Arabidopsis thaliana leads to change in root growth, antioxidant enzymes activity, and response to salinity but not to cold. J Genet Resour 4:90–104
Kudo M, Kidokoro S, Yoshida T, Mizoi J, Todaka D, Fernie AR, Shinozaki K, Yamaguchi-Shinozaki K (2017) Double overexpression of DREB and PIF transcription factors improves drought stress tolerance and cell elongation in transgenic plants. Plant Biotechnol J 15:458–471
Legrand G, Delporte M, Khelifi C, Harant A, Vuylsteker C, Mörchen M, Hance P, Hilbert JL, Gagneul D (2016) Identification and characterization of five BAHD acyltransferases involved in hydroxycinnamoyl ester metabolism in chicory. Front Plant Sci 7:741
Majeed S, Nawaz F, Naeem M, Ashraf MY (2018) Effect of exogenous nitric oxide on sulfur and nitrate assimilation pathway enzymes in maize (Zea mays L.) under drought stress. Acta Physiol Plant 40:206
Moghanloo M, Iranbakhsh A, Ebadi M, Ardebili ZO (2019a) Differential physiology and expression of phenylalanine ammonia lyase (PAL) and universal stress protein (USP) in the endangered species Astragalus fridae following seed priming with cold plasma and manipulation of culture medium with silica nanoparticles. 3 Biotech 9:288
Moghanloo M, Iranbakhsh A, Ebadi M, Satari TN, Ardebili ZO (2019b) Seed priming with cold plasma and supplementation of culture medium with silicon nanoparticle modified growth, physiology, and anatomy in Astragalus fridae as an endangered species. Acta Physiol Plant 41:54
Munawar A, Akram NA, Ahmad A, Ashraf M (2019) Nitric oxide regulates oxidative defense system, key metabolites and growth of broccoli (Brassica oleracea L.) plants under water limited conditions. Sci Hortic 254:7–13
Nabi R, Tayade R, Hussain A, Kulkarni KP, Imran QM, Mun B, Yun BW (2019) Nitric oxide regulates plant responses to drought, salinity, and heavy metal stress. Environ Exp Bot 161:120–133
Nazerieh H, Oraghi Ardebili Z, Iranbakhsh A (2018) Potential benefits and toxicity of nanoselenium and nitric oxide in peppermint. Acta Agric Slov 111:357–368
Pandey P, Singh J, Achary V, Reddy M (2015) Redox homeostasis via gene families of ascorbate-glutathione pathway. Front Environ Sci 3:25
Quiterio-Gutiérrez T, Ortega-Ortiz H, Cadenas-Pliego G, Hernández-Fuentes AD, Sandoval-Rangel A, Benavides-Mendoza A, Cabrera-De La Fuente M, Juárez-Maldonado A (2019) The application of selenium and copper nanoparticles modifies the biochemical responses of tomato plants under stress by Alternaria solani. Int J Mol Sci 20(8):1950
Rajaee S, Iranbakhsh A, Ebadi M, Majd A, Ardebili ZO (2020) Red elemental selenium nanoparticles mediated substantial variations in growth, tissue differentiation, metabolism, gene transcription, epigenetic cytosine DNA methylation, and callogenesis in bitter melon (Momordica charantia); an invitro experiment. PLoS One 15(7):e0235556
Safari M, Ardebili ZO, Iranbakhsh A (2018) Selenium nano-particle induced alterations in expression patterns of heat shock factor A4A(HSFA4A), and high molecular weight glutenin subunit 1Bx (Glu-1Bx) and enhanced nitrate reductase activity in wheat (Triticum aestivum L.). Acta Physiol Plant 40:117
Santisree P, Bhatnagar-Mathur P, Sharma K (2018) Molecular insights into the functional role of nitric oxide (NO) as a signal for plant responses in chickpea. Funct Plant Biol 45:267–283
Seddighinia FS, Iranbakhsh A, Ardebili ZO, Satari TN, Soleimanpour S (2020) Seed priming with cold plasma and multi-walled carbon nanotubes modified growth, tissue differentiation, anatomy, and yield in bitter melon (Momordica charantia). J Plant Growth Regul 39:87–98. https://doi.org/10.1007/s00344-019-09965-2
Sheteiwy MS, An J, Yin M, Jia X, Guan Y, He F, Hu J (2019) Cold plasma treatment and exogenous salicylic acid priming enhances salinity tolerance of Oryza sativa seedlings. Protoplasma 256:79–99
Soleymanzadeh R, Iranbakhsh A, Habibi G, Ardebili ZO (2020) Selenium nanoparticle protected strawberry against salt stress through modifications in salicylic acid, ion homeostasis, antioxidant machinery, and photosynthesis performance. Acta Biol Cracov Ser Bot. https://doi.org/10.24425/abcsb.2019.127751
Sotoodehnia-Korani S, Iranbakhsh A, Ebadi M, Majd A, Ardebili ZO (2020) Selenium nanoparticles induced variations in growth, morphology, anatomy, biochemistry, gene expression, and epigenetic DNA methylation in Capsicum annuum; an in vitro study. Environ Pollut 5:114727. https://doi.org/10.1016/j.envpol.2020.114727
Tamaoki M, Maruyama-Nakashita A (2017) Molecular mechanisms of selenium responses and resistance in plants. In: Selenium in plants. Springer, Cham, pp 35-51
Tejada-Jimenez M, Llamas A, Galván A, Fernández E (2019) Role of nitrate reductase in NO production in photosynthetic eukaryotes. Plants 8:56
Van Hoewyk D, Takahashi H, Inoue E, Hess A, Tamaoki M, Pilon-Smits EA (2008) Transcriptome analyses give insights into selenium-stress responses and selenium tolerance mechanisms in Arabidopsis. Physiol Plant 132:236–253
Xu L, Li F, Han L, Song G, Zhang X (2017) Overexpression of Arabidopsis DREB1A gene in transgenic Poa pratensis: Impacts on osmotic adjustment and hormone metabolism under drought. Int Turf Soc Res J 13:527–36
Zahedi SM, Abdelrahman M, Hosseini MS, Hoveizeh NF, Tran LSP (2019) Alleviation of the effect of salinity on growth and yield of strawberry by foliar spray of selenium-nanoparticles. Environ Pollut 253:246–258
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible Editor: Gangrong Shi
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Abedi, S., Iranbakhsh, A., Oraghi Ardebili, Z. et al. Nitric oxide and selenium nanoparticles confer changes in growth, metabolism, antioxidant machinery, gene expression, and flowering in chicory (Cichorium intybus L.): potential benefits and risk assessment. Environ Sci Pollut Res 28, 3136–3148 (2021). https://doi.org/10.1007/s11356-020-10706-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11356-020-10706-2