Abstract
Ammonia oxidation is mainly performed by ammonia-oxidizing archaea (AOA) and ammonia-oxidizing bacteria (AOB). Allylthiourea (ATU) has been found to specifically inhibit ammonia oxidation. However, the effect of ATU on AOA and AOB transcription has been infrequently studied. In the present study, we examined the responses of AOA and AOB activity and DNA/cDNA community structure to ATU exposure. The ammonia oxidation activity in the 100-mg/L ATU group was 4.3% of that in the control group after 7 days. When exposed to ATU, the gene abundance of AOA was favored compared with that of AOB, and there were no statistically significant differences in the abundance of AOB amoA in DNA and cDNA between the two groups. Compared with the control group, the gene abundance of AOA significantly increased by 5.23 times, while the transcription of AOA significantly decreased by 0.70 times. Moreover, the transcriptional ratio of AOA in the ATU group was only 0.05 times as high as that in the control group. ATU selectively affected AOB and completely inhibited Nitrosomonas europaea and Bacterium amoA.22.HaldeII.kultur at the genetic level. Under ATU exposure, all AOA clusters were transcribed, but three AOB clusters were not transcribed. Our results indicated that the ammonia oxidation potential of the soil of water level fluctuation areas, based on ATU inhibition, was associated mainly with AOA amoA gene abundance and AOB community shifts in DNA and cDNA.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
The ammonia oxidation process is the first and rate-limiting step of nitrification, which is an important part of the global nitrogen cycle (Shen et al. 2012). Ammonia oxidation is mainly performed by two kinds of microorganisms, ammonia-oxidizing archaea (AOA) and ammonia-oxidizing bacteria (AOB) (You et al. 2009). These two groups of microorganisms are abundant in the environment and play a vital role in the removal of ammonia. In recent years, it has been found that the abundance of AOA is much higher than that of AOB in oceans (Wuchter et al. 2006; De Corte et al. 2009), soils (Leininger et al. 2006), and estuaries (Caffrey et al. 2007) and that AOA might play an important role in nitrification.
The water level fluctuation zone is a special wetland ecosystem. It is the transitional region between terrestrial and aquatic ecosystems. Because of periodic fluctuations in water level, soil in the water level fluctuation zone experiences alternating oxygenation and hypoxia. Microorganisms are the main drivers of nitrogen transformation in the soil of the water level fluctuation zone. The alternation of oxygenation and hypoxia in the water level fluctuation zone will affect the abundance and community of ammonia-oxidizing microorganisms (Pett-Ridge et al. 2013; Liu et al. 2015). Therefore, it is important to study ammonia-oxidizing microorganisms in the soil of the water level fluctuation zone. Many articles have researched AOA and AOB in environments such as oceans (Newell et al. 2011; Beman et al. 2012), soils (He et al. 2007; Zhang et al. 2012; Jung et al. 2014), sediments (Beman and Francis 2006; Yang et al. 2016), and wetlands (Molina et al. 2018). Nevertheless, there are few studies on AOA and AOB in the water level fluctuation zone.
Soil in the water level fluctuation zone is very suitable for cultivation and farming. The Three Gorges water level fluctuation zone is covered by plants such as maize (Zhang et al. 2016; Liu et al. 2017). From the perspective of agriculture, maintaining ammonium nitrogen is beneficial to plant growth, while nitrification might lead to a large loss of ammonia (Abbasi and Adams 1998). Nitrification inhibitors can selectively inhibit the activity of nitrifying microorganisms and effectively slow down the transformation from ammonia to nitrate. Therefore, nitrification inhibitors are commonly used in agricultural soil (Moir et al. 2007; Clough et al. 2007). Allylthiourea (ATU) is a widely used nitrification inhibitor that selectively inhibits the oxidation of ammonia (Shen et al. 2012). Ammonia monooxygenase (AMO), the key enzyme for ammonia oxidation by AOA and AOB, contains copper. ATU is a copper-chelating agent that can consume copper ions from AMO and thus inhibit ammonia oxidation (Shen et al. 2013). There are many molecular and cellular differences between AOA and AOB, which might lead to different responses of AOA and AOB to ATU. Many studies have reported the effects of ATU on AOA and AOB activities (Taylor et al. 2010; Jung et al. 2011; Yu et al. 2014; Srithep et al. 2018). In recent years, with the development of sequencing technology, researchers have studied the influence of ATU on the community structure of AOA and AOB by high-throughput sequencing. Wang et al. (2017) reported that the AOB-related genera Nitrosomonas and Nitrosococcus were the genera most affected by ATU according to high-throughput pyrosequencing analysis.
The effects of ATU on AOA and AOB have been studied mostly based on the quantification of AOA and AOB amoA genes (Lehtovirta-Morley et al. 2013; Wang and Gu 2014). However, there is no obvious causal relationship between the gene abundance of AOA and AOB and the activity of ammonia oxidation (Prosser and Nicol 2008). Theoretically, the functional gene amoA could represent the potential for microbial ammonia oxidation. However, the presence of functional genes does not indicate whether nitrification occurs because these genes might not be transcribed or their mRNA/enzymes might be inactivated. Furthermore, previous research has suggested that amoA gene transcription has a quicker response (1–2 h) to the recovery of ammonia oxidation activity compared with its copy numbers in DNA (Aoi et al. 2004). The transcription of DNA can reflect the activities of microorganisms more directly than the DNA itself, so RNA-based studies could solve this problem more effectively. At present, researchers mainly study the effects of ATU on AOA and AOB at the genetic level but rarely study the response of AOA and AOB to ATU at the transcriptional level.
Many studies have reported the effects of ATU on enzymes and genes of AOA and AOB, but the effects on transcription have been less studied. It has been reported that the gene abundance of AOA and AOB is altered during ATU exposure (Lehtovirta-Morley et al. 2013; Wang and Gu 2014). ATU inhibits ammonia oxidation activity, while AOA and AOB amoA genes have been detected in DNA. In this case, the transcriptional activity of AOA and AOB has rarely been discussed. There are few studies on the responses and mechanisms of AOA and AOB to ATU at the transcriptional level. This study intended to explore the transcriptional activity of AOA and AOB when ATU inhibited ammonia oxidation activity, that is, when AMO was inactivated, to study the responses and mechanisms of AOA and AOB transcription to ATU exposure. In the present study, we investigated the responses of AOA and AOB activity and community structure to ATU exposure in the soil of the Three Gorges water level fluctuation zone at the genetic and transcriptional levels. We explored the response of ammonia-oxidizing microorganisms to ATU exposure by quantitative polymerase chain reaction (qPCR) and high-throughput sequencing of the AOA and AOB amoA genes in DNA and cDNA.
Methods
Culture experiment
The original soil (OS) was soil from the Three Gorges water level fluctuation zone. An experimental group and control group were set up. In the two groups, 20-g soil samples and 100-mL culture medium were put into 200-mL conical flasks. The culture medium contained 0.4-g MgCl2·6H2O, 0.5-g KCl, 0.2-g KH2PO4, 1.0-g NaCl, and 0.1-g CaCl2·2H2O. Potassium chlorate (10 mM) was added to each group to inhibit the oxidation of nitrite (Jung et al. 2014). The experimental group, named ATU, was treated with 100-mg/L ATU, and the untreated group was the control group (CON). All experiments were run in triplicate.
The culture experiment was carried out on the shaking table. Conical flasks were put on the shaking table and pre-cultured for 24 h at 30 °C and 150 rpm. Then, 1.0-mM ammonium chloride was added to each group at time zero of the culture experiment. The culture experiment was carried out for 7 days. Water was sampled during the course of the experiment (days 0, 1, 3, 5, and 7), whereas soil was taken only at the end of the experiment. The ammonia and nitrate concentrations of the water samples were measured by a spectrophotometer. The nitrite nitrogen concentrations of the water samples were measured by a spectrophotometer, and the detection limit was 0.003 mg/L. A schematic representation of the experiment is shown in Fig. S1.
Extracting DNA and RNA
DNA and RNA were extracted with an E.Z.N.A.® Soil DNA kit and RNA kit (Omega, USA), respectively, according to the manufacturers’ instructions. Extracted DNA and RNA were stored at − 20 °C and − 80 °C, respectively, until further use. The concentrations and purity of DNA and RNA were measured by a NanoDrop 2000. A PrimeScript™ RT reagent Kit with gDNA Eraser (Perfect Real Time) kit (TaKaRa) was used for reverse transcription of RNA.
Quantitative real-time PCR
Quantitative PCR was performed on the amoA of archaea and bacteria. The specific primers for amoA functional genes in AOA and AOB (amoA-26F/amoA-417R and amoA-1F/amoA-2R, respectively) were used to quantify the abundances of AOA and AOB in the samples (Rotthauwe et al. 1997; Park et al. 2008). Plasmid standard samples containing the AOA amoA (amoA-26F/amoA-417R) or AOB amoA (amoA-1F/amoA-2R) gene were diluted with ddH2O to yield a series of tenfold concentrations and subsequently used as standards for qPCR standard curves. The R2 value for each standard curve exceeded 0.99, indicating linear relationships over the concentration ranges used in this study. The efficiencies of AOA and AOB amoA were 0.995 and 0.998, respectively. The absolute quantification of the samples was performed by a MyiQ2 real-time PCR Detection System (Bio-Rad, USA). A 20-μL reaction system was used: 10.0-μL FastStart Universal SYBR Green Master (2 ×) (ROX, Roche), 0.5-μL forward primer (10 μM), 0.5-μL reverse primer (10 μM), 1.0-μL DNA template (standard/sample), and 8.0-μL sterile water. qPCR was performed in a three-step thermal cycling procedure, and the protocol and parameters for each target gene are presented in Table S1 (Supporting Information). Each qPCR amplification was performed in 40 cycles.
High-throughput sequencing
PCR amplification of the amoA functional genes of AOA and AOB was performed for DNA and cDNA. The specific primers for the amoA functional genes of AOA and AOB were the same as the primers used in quantitative real-time PCR. A 25-μL PCR system was prepared: 5 × reaction buffer 5 μL, 5 × GC buffer 5 μL, dNTP (2.5 mM) 2 μL, forward primer (10 μM) 1 μL, reverse primer (10 μM) 1 μL, DNA template 2 μL, ddH2O 8.75 μL, and Q5 DNA polymerase 0.25 μL. The following protocol was performed: an initial denaturation cycle (98 °C for 2 min), 25 cycles of denaturation (98 °C for 15 s), annealing (55 °C for 30 s), extension (72 °C for 30 s), and a final extension cycle (72 °C for 5 min). High-throughput sequencing was performed with the Illumina MiSeq platform at Shanghai Personal Biotechnology. The raw data in this study have been deposited in the GenBank Sequence Read Archive database with accession number SRP187919.
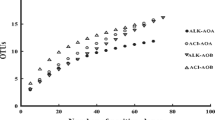
We used Quantitative Insights into Microbial Ecology (QIIME) software to identify unknown sequences (Caporaso et al. 2010). The obtained sequences were merged and classified into operational taxonomic units (OTUs) according to 97% sequence similarity by the sequence alignment tool UCLUST (Edgar et al. 2011). By comparing the representative OTU sequences with template sequences in the NCBI database, taxonomic information for the OTUs was obtained. OTUs whose relative abundances were less than 0.001% were removed (Bokulich et al. 2013). Bioinformatics information is provided in Table S2. The rarefaction curves of AOA and AOB indicated that the sequencing depth was sufficient, as shown in Fig. S2.
Statistical analysis
The co-occurrence networks were analyzed according to the relative OTU abundances of samples. There were two types of nodes in the network diagram: OTU nodes and sample nodes. If an OTU appeared in a sample, there would be an edge between the OTU node and the sample node. Gephi was used for network visualization.
Results
Ammonia oxidation activity
The variations in nitrogen concentration are shown in Fig. 1. Ammonia oxidation led to an accumulation of nitrite; thus, the addition of 10-mM potassium chlorate inhibited nitrite oxidation, and the concentration of nitrate did not increase (Fig. 1b). Therefore, the concentration of nitrite in the system reflected the activity of ammonia oxidation. When exposed to 100-mg/L ATU, the concentration of nitrite had no obvious changes from 0 to 7 days (Fig. 1a). The ammonia oxidation activity in the ATU group on days 1, 3, 5, and 7 was 1.4%, 2.6%, 2.5%, and 4.3%, respectively, of that in the control group. Under 100-mg/L ATU exposure, over 95.7% of the ammonia oxidation activity was inhibited.
Gene abundance of AOA and AOB
The amoA gene abundances of AOA and AOB in DNA and cDNA are shown in Fig. 2a. For DNA, the abundances of AOA amoA and AOB amoA in the ATU group were 6.23 times and 1.40 times as high as those in the control group, respectively. For cDNA, the abundances of AOA amoA and AOB amoA in the ATU group were 0.30 times and 0.93 times as high as those in the control group, respectively. The ATU group had more AOA and AOB amoA in DNA but less in cDNA compared with the control group. The T test results showed that there were significant differences in the abundance of AOA amoA in DNA (P < 0.05) and cDNA (P < 0.01) between the two groups.
The AOA/AOB ratios in DNA and cDNA were in the range of 101–102 and 10−1–100, respectively (Fig. 2b). The AOA/AOB ratios in DNA and cDNA in the ATU group were 4.45 times and 0.33 times as high as those in the control group, respectively. The ATU group had a higher AOA/AOB ratio in DNA but lower AOA/AOB ratio in cDNA compared with the control group. The cDNA/DNA ratios of AOA and AOB amoA genes were calculated as indicators of the potential transcriptional activity of the targeted microbial groups. The cDNA/DNA ratios in AOA and AOB were in the range of 10−4–10−1 and 10−1–100, respectively (Fig. 2b). The cDNA/DNA ratios in AOA and AOB in the ATU group were 0.05 times and 0.66 times as high as those in the control group, respectively. The ATU group had lower cDNA/DNA ratios in AOA and AOB compared with the control group.
Community structure of AOA and AOB
Influence of ATU on the community structure of AOA and AOB
The OTU sequences were compared with the NCBI database, and the OTUs of AOA and AOB were divided into 7 clusters and 12 clusters, respectively (Fig. 3). The initial communities of AOA and AOB are shown in Table S3. The phylogenetic trees for the clusters of AOA and AOB are shown in Fig. S3, indicating the taxonomic relationship among the clusters. As shown in Fig. 3a, exposure to ATU had no significant effect on the relative abundance of AOA clusters in DNA. AOA mainly consisted of Candidatus Nitrososphaera gargensis, and its relative abundance was more than 90%. Compared with the control group, the relative abundance of Candidatus Nitrososphaera gargensis remained almost unchanged when exposed to ATU (the control group was 90.7%; the ATU group was 90.1%). The relative abundance of Candidatus Nitrosocosmicus franklandus in the ATU group (5.1%) was higher than that in the control group (3.4%). Candidatus Nitrosocosmicus oleophilus had a lower relative abundance in the ATU group (3.9%) than in the control group (4.7%). For the AOA clusters, all clusters were detected in the control group and the ATU group. After exposure to ATU for 7 days, the community structure of AOA was relatively stable.
Figure 3b shows that ATU exposure altered the relative abundance of AOB clusters in DNA. The AOB were mainly composed of Nitrosomonas marina and Bacterium amoA.18.V-frei.kultur, and the total relative abundance of these two species was 96.1% ~ 99.4%. Nitrosomonas marina had a higher relative abundance in the ATU group (47.7%) than in the control group (39.0%), and the relative abundance of Bacterium amoA.18.V-frei.kultur in the ATU group (51.7%) was lower than that in the control group (57.8%). The relative abundances of Nitrosomonas sp. A2, Nitrosomonas sp. A7, Nitrosomonas sp. N2005, and Bacterium amoA.30.Sali.kultur in the ATU group were 0.5%, 0.2%, 0.2%, and 0.5%, respectively, whereas their relative abundances were below 0.01% in the control group. In the AOB clusters, Nitrosomonas europaea (9.67E-4) and Bacterium amoA.22.HaldeII.kultur (2.8E-5) were found in the control group, but their relative abundances were both zero in the ATU group. After exposure to ATU for 7 days, the community structure of AOB changed.
Community structure of cDNA and DNA when exposed to ATU
Comparing the relative abundance of AOA clusters in cDNA and DNA when exposed to ATU, it was found that Candidatus Nitrosocosmicus franklandus, Candidatus Nitrosocosmicus oleophilus, and Nitrososphaera sp. JG1 showed higher relative abundances at the transcriptional level, while Candidatus Nitrososphaera gargensis and Candidatus Nitrosocosmicus sp. G61 showed higher relative abundances at the genetic level (Fig. 3a). Candidatus Nitrososphaera gargensis was the main cluster of AOA, and its relative abundance in cDNA.ATU (72.4%) was lower than that in DNA.ATU (90.1%). The relative abundance of Candidatus Nitrosocosmicus franklandus in cDNA.ATU (22.0%) was significantly higher than that in DNA.ATU (5.1%). Candidatus Nitrosocosmicus oleophilus had a higher relative abundance in cDNA.ATU (3.7%) than in DNA.ATU (0.8%). The relative abundance of Nitrososphaera sp. JG1 in cDNA.ATU (1.2%) was significantly higher than that in DNA.ATU (0.02%). Candidatus Nitrosocosmicus sp. G61 had a lower relative abundance in cDNA.ATU (0.5%) than in DNA.ATU (3.9%). All clusters of AOA were detected in both DNA and cDNA.
Comparing the relative abundance of AOB clusters in cDNA and DNA when exposed to ATU, it was found that Nitrosomonas marina, Nitrosomonas sp. A2, Nitrosomonas sp. A7, Nitrosomonas aestuarii, and Nitrosomonas sp. N2005 at the transcriptional level were higher than those at the genetic level (Fig. 3b). Bacterium amoA.18.V-frei.kultur was the main cluster of AOB in DNA.ATU (51.7%), while its relative abundance was zero in cDNA.ATU, suggesting that it was not transcribed at all. Another major cluster of AOB, Nitrosomonas marina, had a higher relative abundance in cDNA.ATU (72.0%) than in DNA.ATU (47.7%). The relative abundances of Nitrosomonas sp. A2 and Nitrosomonas sp. A7 were 10.9% and 13.1% in cDNA.ATU, respectively, whereas their relative abundances were 0.006% and 0.003% in DNA.ATU. The relative abundances of Nitrosomonas aestuarii and Nitrosomonas sp. N2005 were 1.8% and 1.9% in cDNA.ATU and 0.04% and 0.006% in DNA.ATU, respectively. Specifically, the cDNA/DNA ratios of Bacterium amoA.18.V-frei.kultur, Bacterium amoA.2.V-frei.kultur, and Bacterium amoA.30.Sali.kultur were all zero when exposed to ATU.
Co-occurrence network of AOA and AOB
There were 53 OTUs in the co-occurrence network of AOA (Fig. 4a). Thirty-four OTUs occurred in both the control group and the ATU group. Six OTUs only occurred in the control group, and 13 OTUs only occurred in the ATU group. The co-occurrence network of AOA included Candidatus Nitrososphaera gargensis (44 OTUs), Candidatus Nitrosocosmicus sp. G61 (4 OTUs), Candidatus Nitrosocosmicus oleophilus (3 OTUs), and Candidatus Nitrosocosmicus franklandus (2 OTUs). Specifically, the 6 OTUs that only occurred in the control group were all Candidatus Nitrososphaera gargensis members. The 13 OTUs only occurred in the ATU group included Candidatus Nitrosocosmicus sp. G61 (1 OTU), Candidatus Nitrosocosmicus oleophilus (1 OTU), and Candidatus Nitrososphaera gargensis (11 OTUs). Candidatus Nitrosocosmicus franklandus (2 OTUs) occurred in both the control group and the ATU group.
There were 70 OTUs in the co-occurrence network of AOB (Fig. 4b). Twenty-three OTUs occurred in both the control group and the ATU group. Twenty-seven OTUs only occurred in the control group, and 20 OTUs only occurred in the ATU group. The co-occurrence network of AOB included Bacterium amoA.18.V-frei.kultur (43 OTUs), Nitrosomonas marina (15 OTUs), Nitrosomonas sp. A7 (5 OTUs), Nitrosomonas sp. A2 (4 OTUs), Bacterium amoA.2.V-frei.kultur (2 OTUs), and Nitrosomonas sp. N2005 (1 OTU). Bacterium amoA.2.V-frei.kultur (2 OTUs) occurred only in the control group, and Nitrosomonas sp. N2005 (1 OTU) occurred only in the ATU group.
Furthermore, the proportion of OTUs that occurred in both the control group and the ATU group in the co-occurrence network of AOA and AOB was 64.2% and 32.9%, respectively (Fig. 4). When exposed to ATU, AOA OTUs had a higher co-occurrence than AOB OTUs. In the co-occurrence network of AOA, 6 OTUs only occurred in the control group (Fig. 5). The relative abundances of the 6 OTUs in DNA.CON were 10−3 ~ 10−2. The relative abundance of one OTU in DNA.ATU was zero, and those of the other 5 OTUs were 10−4 ~ 10−3. In the co-occurrence network of AOB, 27 OTUs only occurred in the control group (Fig. 5). The relative abundances of the 27 OTUs in DNA.CON in AOB were 10−3 ~ 10−2. The relative abundances of 5 OTUs in DNA.ATU were zero. The relative abundances of 6 OTUs in DNA.ATU were 10−5 ~ 10−4, and those of the other 16 OTUs were 10−4 ~ 10−3. Among the 6 OTUs of AOA and the 27 OTUs of AOB, one OTU of AOA and 5 OTUs of AOB were not detected in the ATU group. The relative abundances of 21 OTUs (5 OTUs of AOA and 16 OTUs of AOB) in the ATU group were one order of magnitude lower than those in the control group. The relative abundances of 6 OTUs (6 OTUs of AOB) in the ATU group were two orders of magnitude lower than those in the control group. The relative abundances of AOB OTUs changed more than those of AOA OTUs. Accordingly, exposure to ATU had a greater influence on AOB OTUs than AOA OTUs.
Discussion
Effect of ATU on ammonia oxidation activity
The ammonia oxidation activity in the 100-mg/L ATU group was 4.3% of that in the control group after 7 days, indicating that nitrification was significantly inhibited by 100-mg/L ATU. Previous studies have reported the inhibition of ammonia oxidation by ATU. A 10-mg/L concentration of ATU selectively inhibited ammonia oxidation without affecting the activity of other microorganisms, completely inhibited the ammonia oxidation of activated sludge and enriched nitrifying bacteria (Ginestet et al. 1998; Ali et al. 2013). In our study, although 100-mg/L ATU was higher than the ATU concentration (10 mg/L) that completely inhibited nitrification, there was still 4.3% ammonia oxidation activity in the ATU group. It was obvious that the ammonia-oxidizing microorganisms in the Three Gorges water level fluctuation zone were more tolerant to ATU. From the perspective of agriculture, 100-mg/L ATU could not completely inhibit the oxidation of ammonia nitrogen in the soil of the water level fluctuation zone.
Santoro’s research showed that 100-mg/L ATU could inhibit nitrification completely (Santoro and Casciotti 2011). However, exposure to 100-mg/L ATU failed to completely inhibit ammonia oxidation activity in the present study, probably because AOA were more abundant in the water level fluctuation zone. ATU had different inhibitory effects on AOA and AOB, which has been reported previously. ATU only partially inhibited ammonia oxidation by AOA at the concentration that completely inhibited ammonia oxidation by AOB. Some studies have reported that 11.6-mg/L ATU completely inhibited the activity of AOB but only partially inhibited AOA (Hatzenpichler et al. 2008; Santoro and Casciotti 2011). The effective concentration of ATU (EC50) in AOA culture was 1000 times higher than that in AOB (Shen et al. 2013). Compared with AOB, AOA were less sensitive to ATU and more difficult to affect (Taylor et al. 2010; Lehtovirta-Morley et al. 2013). There were more AOA than AOB in the water level fluctuation zone, because AOA might be more adaptive to anoxic environments than AOB (Pett-Ridge et al. 2013; Liu et al. 2015). The copy number of AOA amoA was 1–2 orders of magnitude higher than that of AOB amoA (Fig. 2a), which might be the reason for the existence of ammonia oxidation activity.
Exposure to 100-mg/L ATU failed to completely inhibit ammonia oxidation activity, possibly due to the protection by the soil structure. Inhibitors diffuse through soil, resulting in a weakened effect. Lehtovirta-Morley’s research showed that ATU in liquid culture could completely inhibit ammonia oxidation but ATU in soil did not inhibit ammonia oxidation (Lehtovirta-Morley et al. 2013). Therefore, it might also be the presence of soil that weakened the effectiveness of ATU.
Effects of ATU on AOA and AOB
When exposed to ATU, the gene abundance of AOA was affected more than that of AOB. There were statistically significant differences in the abundance of AOA amoA between the two groups in DNA and cDNA, but not in the abundance of AOB amoA in DNA and cDNA (Fig. 2a). Furthermore, AOA transcriptional activity decreased significantly in the allylthiourea-containing microcosms compared with the control, whereas no significant difference was observed for AOB transcriptional activity (Fig. 2b). Consequently, the ammonia oxidation potential of the soil of water level fluctuation areas, based on ATU inhibition, was associated mainly with AOA amoA gene abundance shifts in DNA and cDNA. ATU at 100 mg/L could inhibit the growth of AOA and AOB within 5–10 days, and ATU had a more obvious inhibition on the growth of AOA (Wang and Gu 2014). Our results indicated that exposure to ATU stimulated the growth of AOA (Fig. 2a). Similarly, it has been found that ATU stimulated rather than inhibited Nitrosotalea devanaterra, which is an ammonia-oxidizing archaea (Lehtovirta-Morley et al. 2013).
Exposure to ATU had a greater influence on AOB cluster composition than AOA cluster composition. Figure 3a shows that all AOA clusters were detected in DNA.CON, DNA.ATU, and cDNA.ATU. However, for AOB clusters in DNA, Nitrosomonas europaea and Bacterium amoA.22.HaldeII.kultur were detected in the control group but not in the ATU group (Fig. 3b). Moreover, for AOB clusters in the ATU group, three AOB clusters (Bacterium amoA.18.V-frei.kultur, Bacterium amoA.2.V-frei.kultur, and Bacterium amoA.30.Sali.kultur) were detected in DNA but not in cDNA (Fig. 3b). When exposed to ATU, the community structure of AOB in DNA and cDNA was affected more than that of AOA. In addition, AOA OTUs had a higher co-occurrence than AOB OTUs under ATU exposure (Fig. 4). In the OTUs that only occurred in the control group in the co-occurrence network of AOA and AOB, the relative abundances of AOB OTUs changed more than those of AOA OTU (Fig. 5). This observation was also confirmed by the results from PCA analysis (Fig. S4). Figure S4 illustrates that the distance between the OTUs of AOB under different treatments was greater than that of AOA. That is, the community structure of AOB was affected more than that of AOA when exposed to ATU.
At the genetic level, exposure to ATU altered the community structure of AOB and had a slight impact on the community structure of AOA in the Three Gorges water level fluctuation zone soil, which was consistent with results for mangrove soil (Wang and Gu 2014). The community structure of AOA was relatively stable (Fig. 3a), and ATU had poor selectivity to AOA. However, exposure to ATU had a greater impact on the community structure of AOB and completely inhibited Nitrosomonas europaea and Bacterium amoA.22.HaldeII.kultur (Fig. 3b). Nitrosomonas europaea is one of the most studied AOB, and many reports have confirmed that ATU completely inhibits the ammonia oxidation activity of Nitrosomonas europaea (Ginestet et al. 1998; Ali et al. 2013; Martens-Habbena et al. 2015). A low concentration of ATU significantly inhibited the ammonia-oxidizing activity of Nitrosomonas europaea (Martens-Habbena et al. 2015), indicating that Nitrosomonas europaea is sensitive to ATU. The results showed that different AOB responded differently to ATU and that ATU affected AOB selectively.
Responses at the genetic and transcriptional levels
The copy number of AOA amoA was 1–2 orders of magnitude higher than that of AOB amoA at the genetic level. Nevertheless, the copy number of AOA amoA was 0–1 orders of magnitude lower than that of AOB amoA at the transcriptional level. It has been reported that AOA are abundant at the genetic level and active at the transcriptional level in soil, while AOB are abundant in sediment (Zhang et al. 2018). The gene abundances and transcriptional activity of AOA and AOB were closely related to the source and environment of the samples. The results indicated that AOA had higher gene abundance than AOB in the water level fluctuation zone, but AOB transcribed more than AOA. In addition, the cDNA/DNA ratios of AOB were significantly higher than those of AOA. That is, the transcriptional activity of AOB was higher than that of AOA in the water level fluctuation zone. Other studies have also shown that the cDNA/DNA ratios of AOA were one order of magnitude lower than those of AOB (Lipsewers et al. 2014; Ling et al. 2018), consistent with our results, supporting that AOB had higher transcriptional activity. Moreover, the gene abundance of AOA amoA in cDNA and the cDNA/DNA ratio of AOA in the ATU group significantly decreased compared with the control group, suggesting that exposure to ATU significantly reduced the transcription and transcriptional ratio of AOA.
Candidatus Nitrososphaera gargensis was the main AOA in the Three Gorges water level fluctuation zone, although the relative abundance varied at the genetic and transcriptional levels (Fig. 3a). Candidatus Nitrososphaera gargensis belongs to Nitrososphaera, and it is a main type of AOA in cattle manure and municipal sludge compost (Yamamoto et al. 2011; Oishi et al. 2012; Yan et al. 2015). When exposed to ATU, the relative abundances of Candidatus Nitrosocosmicus franklandus and Candidatus Nitrosocosmicus oleophilus at the transcriptional level were significantly higher than those at the genetic level. Candidatus Nitrosocosmicus franklandus and Candidatus Nitrosocosmicus oleophilus had higher transcriptional activity when exposed to ATU.
When exposed to ATU, the AOB community in DNA consisted of Nitrosomonas and unclassified bacteria, while the AOB community in cDNA was mainly composed of Nitrosomonas. Six clusters of AOB belonged to Nitrosomonas. Nitrosomonas europaea was completely inhibited by ATU, and the relative abundances of the other five Nitrosomonas clusters at the transcriptional level were higher than those at the genetic level. Previous studies have reported that Nitrosomonas might be the main AOB at the genetic level (Burrell et al. 2001; Layton et al. 2005; Pan et al. 2018). The results of the present study confirmed that Nitrosomonas might play a dominant role in AOB from the perspective of transcription. Nitrosomonas marina was one of the main AOB in the Three Gorges water level fluctuation zone (Fig. 3b). Nitrosomonas marina is a marine species (PommereningRoser et al. 1996), and it is the main type of AOB in wastewater treatment plants and marine aquaculture biofilms (Foesel et al. 2008; Zhang et al. 2011; Pan et al. 2018). Nitrosomonas marina was also one of the main AOB in terms of ammonia oxidation at the transcriptional level (Fig. 3b), and it was an active AOB (Pan et al. 2018). When exposed to ATU, the relative abundances of Nitrosomonas sp. A2 and Nitrosomonas sp. A7 at the transcriptional level were significantly higher than those at the genetic level. Furthermore, Nitrosomonas sp. A2 and Nitrosomonas sp. A7 had higher relative abundances (> 10%) than other Nitrosomonas except for Nitrosomonas marina at the transcriptional level. Therefore, Nitrosomonas sp. A2 and Nitrosomonas sp. A7 had higher transcriptional activity than other Nitrosomonas except for Nitrosomonas marina when exposed to ATU.
In conclusion, the ammonia oxidation activity in the 100-mg/L ATU group was 4.3% of that in the control group after 7 days. From the perspective of agriculture, 100-mg/L ATU could not completely inhibit the oxidation of ammonia nitrogen in the soil of the water level fluctuation zone. Exposure to 100-mg/L ATU failed to completely inhibit ammonia oxidation activity, possibly due to abundant AOA in the water level fluctuation zone and the protection by the soil structure. When exposed to ATU, the gene abundance of AOA was favored compared with that of AOB, and there were no statistically significant differences in the abundance of AOB amoA in DNA and cDNA between the two groups. The gene abundance of AOA significantly increased, while the transcription and transcriptional ratio of AOA significantly decreased compared with the control group. Our results indicated that the ammonia oxidation potential of the soil of water level fluctuation areas, based on ATU inhibition, was associated mainly with AOA amoA gene abundance and AOB community shifts in DNA and cDNA.
References
Abbasi MK, Adams WA (1998) Loss of nitrogen in compacted grassland soil by simultaneous nitrification and denitrification. Plant Soil 200:265–277. https://doi.org/10.1023/A:1004398520150
Ali TU, Kim M, Kim D (2013) Selective inhibition of ammonia oxidation and nitrite oxidation linked to N2O emission with activated sludge and enriched Nitrifiers. J Microbiol Biotechnol 23:719–723. https://doi.org/10.4014/jmb.1302.02017
Aoi Y, Shiramasa Y, Masaki Y, Tsuneda S, Hirata A, Kitayama A, Nagamune T (2004) Expression of amoA mRNA in wastewater treatment processes examined by competitive RT-PCR. J Biotechnol 111:111–120. https://doi.org/10.1016/j.jbiotec.2004.02.017
Beman JM, Francis CA (2006) Diversity of ammonia-oxidizing archaea and bacteria in the sediments of a hypernutrified subtropical estuary: Bahia del Tobari, Mexico. Appl Environ Microbiol 72:7767–7777. https://doi.org/10.1128/AEM.00946-06
Beman JM, Popp BN, Alford SE (2012) Quantification of ammonia oxidation rates and ammonia-oxidizing archaea and bacteria at high resolution in the Gulf of California and eastern tropical North Pacific Ocean. Limnol Oceanogr 57:711–726. https://doi.org/10.4319/lo.2012.57.3.0711
Bokulich NA, Subramanian S, Faith JJ, Gevers D, Gordon JI, Knight R, Mills DA, Caporaso JG (2013) Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat Methods 10:11–57. https://doi.org/10.1038/NMETH.2276
Burrell PC, Phalen CM, Hovanec TA (2001) Identification of bacteria responsible for ammonia oxidation in freshwater aquaria. Appl Environ Microbiol 67:5791–5800. https://doi.org/10.1128/AEM.67.12.5791-5800.2001
Caffrey JM, Bano N, Kalanetra K, Hollibaugh JT (2007) Ammonia oxidation and ammonia-oxidizing bacteria and archaea from estuaries with differing histories of hypoxia. ISME J 1:660–662. https://doi.org/10.1038/ismej.2007.79
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Tumbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. https://doi.org/10.1038/nmeth.f.303
Clough TJ, Di HJ, Cameron KC, Sherlock RR, Metherell AK, Clark H, Rys G (2007) Accounting for the utilization of a N2O mitigation tool in the IPCC inventory methodology for agricultural soils. Nutr Cycl Agroecosyst 78:1–14. https://doi.org/10.1007/s10705-006-9069-z
De Corte D, Yokokawa T, Varela MM, Agogue H, Herndl GJ (2009) Spatial distribution of bacteria and archaea and amoA gene copy numbers throughout the water column of the eastern Mediterranean Sea. ISME J 3:147–158. https://doi.org/10.1038/ismej.2008.94
Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R (2011) UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200. https://doi.org/10.1093/bioinformatics/btr381
Foesel BU, Gieseke A, Schwermer C, Stief P, Koch L, Cytryn E, de la Torre JR, van Rijn J, Minz D, Drake HL, Schramm A (2008) Nitrosomonas Nm143-like ammonia oxidizers and Nitrospira marina-like nitrite oxidizers dominate the nitrifier community in a marine aquaculture biofilm. FEMS Microbiol Ecol 63:192–204. https://doi.org/10.1111/j.1574-6941.2007.00418.x
Ginestet P, Audic JM, Urbain V, Block JC (1998) Estimation of nitrifying bacterial activities by measuring oxygen uptake in the presence of the metabolic inhibitors allylthiourea and azide. Appl Environ Microbiol 64:2266–2268
Hatzenpichler R, Lebedeva EV, Spieck E, Stoecker K, Richter A, Daims H, Wagner M (2008) A moderately thermophilic ammonia-oxidizing crenarchaeote from a hot spring. Proc Natl Acad Sci U S A 105:2134–2139. https://doi.org/10.1073/pnas.0708857105
He J, Shen J, Zhang L, Zhu Y, Zheng Y, Xu M, Di H (2007) Quantitative analyses of the abundance and composition of ammonia-oxidizing bacteria and ammonia-oxidizing archaea of a Chinese upland red soil under long-term fertilization practices. Environ Microbiol 9:2364–2374. https://doi.org/10.1111/j.1462-2920.2007.01358.x
Jung M, Park S, Min D, Kim J, Rijpstra WIC, Damste JSS, Kim G, Madsen EL, Rhee S (2011) Enrichment and characterization of an autotrophic ammonia-oxidizing archaeon of mesophilic crenarchaeal group I.1a from an agricultural soil. Appl Environ Microbiol 77:8635–8647. https://doi.org/10.1128/AEM.05787-11
Jung M, Park S, Kim S, Kim J, Damste JSS, Jeon CO, Rhee S (2014) A mesophilic, autotrophic, ammonia-oxidizing Archaeon of thaumarchaeal group I.1a cultivated from a deep oligotrophic soil horizon. Appl Environ Microbiol 80:3645–3655. https://doi.org/10.1128/AEM.03730-13
Layton AC, Dionisi H, Kuo HW, Robinson KG, Garrett VM, Meyers A, Sayler GS (2005) Emergence of competitive dominant ammonia-oxidizing bacterial populations in a full-scale industrial wastewater treatment plant. Appl Environ Microbiol 71:1105–1108. https://doi.org/10.1128/AEM.71.2.1105-1108.2005
Lehtovirta-Morley LE, Verhamme DT, Nicol GW, Prosser JI (2013) Effect of nitrification inhibitors on the growth and activity of Nitrosotalea devanaterra in culture and soil. Soil Biol Biochem 62:129–133. https://doi.org/10.1016/j.soilbio.2013.01.020
Leininger S, Urich T, Schloter M, Schwark L, Qi J, Nicol GW, Prosser JI, Schuster SC, Schleper C (2006) Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature 442:806–809. https://doi.org/10.1038/nature04983
Ling J, Lin X, Zhang Y, Zhou W, Yang Q, Lin L, Zeng S, Zhang Y, Wang C, Ahmad M, Long L, Dong J (2018) Community composition and transcriptional activity of Ammonia-oxidizing prokaryotes of Seagrass Thalassia hemprichii in coral reef ecosystems. Front Microbiol 9. https://doi.org/10.3389/fmicb.2018.00007
Lipsewers YA, Bale NJ, Hopmans EC, Stefan S, Damsté JSS, Laura V (2014) Seasonality and depth distribution of the abundance and activity of ammonia oxidizing microorganisms in marine coastal sediments (North Sea). 5:472
Liu S, Hu B, He Z, Zhang B, Tian G, Zheng P, Fang F (2015) Ammonia-oxidizing archaea have better adaptability in oxygenated/hypoxic alternant conditions compared to ammonia-oxidizing bacteria. Appl Microbiol Biotechnol 99:8587–8596. https://doi.org/10.1007/s00253-015-6750-7
Liu J, Zeng B, Lin F, Ayi Q (2017) Effects of water level regulation on the seed germination and production of annual plant Xanthium sibiricum in the water-level-fluctuating-zone of Three Gorges Reservoir. Sci Rep 7:5056. https://doi.org/10.1038/s41598-017-04599-4
Martens-Habbena W, Qin W, Horak REA, Urakawa H, Schauer AJ, Moffett JW, Armbrust EV, Ingalls AE, Devol AH, Stahl DA (2015) The production of nitric oxide by marine ammonia-oxidizing archaea and inhibition of archaeal ammonia oxidation by a nitric oxide scavenger. Environ Microbiol 17:2261–2274. https://doi.org/10.1111/1462-2920.12677
Moir JL, Cameron KC, Di HJ (2007) Effects of the nitrification inhibitor dicyandiamide on soil mineral N, pasture yield, nutrient uptake and pasture quality in a grazed pasture system. Soil Use Manag 23:111–120. https://doi.org/10.1111/j.1475-2743.2006.00078.x
Molina V, Dorador C, Fernández C, Bristow L, Eissler Y, Hengst M, Hernandez K, Olsen LM, Harrod C, Marchant F, Anguita C, Cornejo M (2018) The activity of nitrifying microorganisms in a high-altitude Andean wetland. FEMS Microbiol Ecol 94. https://doi.org/10.1093/femsec/fiy062
Newell SE, Babbin AR, Jayakumar A, Ward BB (2011) Ammonia oxidation rates and nitrification in the Arabian Sea. Glob Biogeochem Cycles 25. https://doi.org/10.1029/2010GB003940
Oishi R, Tada C, Asano R, Yamamoto N, Suyama Y, Nakai Y (2012) Growth of ammonia-oxidizing archaea and bacteria in cattle manure compost under various temperatures and ammonia concentrations. Microb Ecol 63:787–793. https://doi.org/10.1007/s00248-011-9971-z
Pan K, Gao J, Li H, Fan X, Li D, Jiang H (2018) Ammonia-oxidizing bacteria dominate ammonia oxidation in a full-scale wastewater treatment plant revealed by DNA-based stable isotope probing. Bioresour Technol 256:152–159. https://doi.org/10.1016/j.biortech.2018.02.012
Park S, Park B, Rhee S (2008) Comparative analysis of archaeal 16S rRNA and amoA genes to estimate the abundance and diversity of ammonia-oxidizing archaea in marine sediments. Extremophiles 12:605–615. https://doi.org/10.1007/s00792-008-0165-7
Pett-Ridge J, Petersen DG, Nuccio E, Firestone MK (2013) Influence of oxic/anoxic fluctuations on ammonia oxidizers and nitrification potential in a wet tropical soil. FEMS Microbiol Ecol 85:179–194. https://doi.org/10.1111/1574-6941.12111
PommereningRoser A, Rath G, Koops HP (1996) Phylogenetic diversity within the genus Nitrosomonas. Syst Appl Microbiol 19:344–351. https://doi.org/10.1016/S0723-2020(96)80061-0
Prosser JI, Nicol GW (2008) Relative contributions of archaea and bacteria to aerobic ammonia oxidation in the environment. Environ Microbiol 10:2931–2941. https://doi.org/10.1111/j.1462-2920.2008.01775.x
Rotthauwe JH, Witzel KP, Liesack W (1997) The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl Environ Microbiol 63:4704–4712
Santoro AE, Casciotti KL (2011) Enrichment and characterization of ammonia-oxidizing archaea from the open ocean: phylogeny, physiology and stable isotope fractionation. ISME J 5:1796–1808. https://doi.org/10.1038/ismej.2011.58
Shen J, Zhang L, Di HJ, He J (2012) A review of ammonia-oxidizing bacteria and archaea in Chinese soils. Front Microbiol 3. https://doi.org/10.3389/fmicb.2012.00296
Shen T, Stieglmeier M, Dai J, Urich T, Schleper C (2013) Responses of the terrestrial ammonia-oxidizing archaeon Ca. Nitrososphaera viennensis and the ammonia-oxidizing bacterium Nitrosospira multiformis to nitrification inhibitors. FEMS Microbiol Lett 344:121–129. https://doi.org/10.1111/1574-6968.12164
Srithep P, Pornkulwat P, Limpiyakorn T (2018) Contribution of ammonia-oxidizing archaea and ammonia-oxidizing bacteria to ammonia oxidation in two nitrifying reactors. Environ Sci Pollut Res 25:8676–8687. https://doi.org/10.1007/s11356-017-1155-z
Taylor AE, Zeglin LH, Dooley S, Myrold DD, Bottomley PJ (2010) Evidence for different contributions of archaea and bacteria to the ammonia-oxidizing potential of diverse Oregon soils. Appl Environ Microbiol 76:7691–7698. https://doi.org/10.1128/AEM.01324-10
Wang Y, Gu J (2014) Effects of allylthiourea, salinity, and pH on ammonia/ammonium-oxidizing prokaryotes in mangrove sediment incubated in laboratory microcosms. Appl Microbiol Biotechnol 98:3257–3274. https://doi.org/10.1007/s00253-013-5399-3
Wang J, He Y, Zhu J, Guan H, Huang M (2017) Screening and optimizing of inhibitors for ammonia-oxidizing bacteria in sediments of malodorous river. Appl Microbiol Biotechnol 101:6193–6203. https://doi.org/10.1007/s00253-017-8318-1
Wuchter C, Abbas B, Coolen MJL, Herfort L, van Bleijswijk J, Timmers P, Strous M, Teira E, Herndl GJ, Middelburg JJ, Schouten S, Damste JSS (2006) Archaeal nitrification in the ocean. Proc Natl Acad Sci U S A 103:12317–12322. https://doi.org/10.1073/pnas.0600756103
Yamamoto N, Asano R, Yoshii H, Otawa K, Nakai Y (2011) Archaeal community dynamics and detection of ammonia-oxidizing archaea during composting of cattle manure using culture-independent DNA analysis. Appl Microbiol Biotechnol 90:1501–1510. https://doi.org/10.1007/s00253-011-3153-2
Yan L, Li Z, Bao J, Wang G, Wang C, Wang W (2015) Diversity of ammonia-oxidizing bacteria and ammonia-oxidizing archaea during composting of municipal sludge. Ann Microbiol 65:1729–1739. https://doi.org/10.1007/s13213-014-1012-y
Yang Y, Zhang J, Zhao Q, Zhou Q, Li N, Wang Y, Xie S, Liu Y (2016) Sediment ammonia-oxidizing microorganisms in two plateau freshwater lakes at different trophic states. Microb Ecol 71:257–265. https://doi.org/10.1007/s00248-015-0642-3
You J, Das A, Dolan EM, Hu Z (2009) Ammonia-oxidizing archaea involved in nitrogen removal. Water Res 43:1801–1809. https://doi.org/10.1016/j.watres.2009.01.016
Yu B, Wang X, Yu S, Li Q, Zhou Q (2014) Effects of roxithromycin on ammonia-oxidizing bacteria and nitrite-oxidizing bacteria in the rhizosphere of wheat. Appl Microbiol Biotechnol 98:263–272. https://doi.org/10.1007/s00253-013-5311-1
Zhang T, Ye L, Tong AHY, Shao M, Lok S (2011) Ammonia-oxidizing archaea and ammonia-oxidizing bacteria in six full-scale wastewater treatment bioreactors. Appl Microbiol Biotechnol 91:1215–1225. https://doi.org/10.1007/s00253-011-3408-y
Zhang L, Hu H, Shen J, He J (2012) Ammonia-oxidizing archaea have more important role than ammonia-oxidizing bacteria in ammonia oxidation of strongly acidic soils. ISME J 6:1032–1045. https://doi.org/10.1038/ismej.2011.168
Zhang Y, Huang L, Zhang Z, Wei L, Sun C, Chen D, Wu W (2016) Phosphorus fractions and phosphorus adsorption characteristics of soils from the water-level fluctuating zone of Nansi Lake, China. Pol J Environ Stud 25:865–872. https://doi.org/10.15244/pjoes/61007
Zhang L, Duff AM, Smith CJ (2018) Community and functional shifts in ammonia oxidizers across terrestrial and marine (soil/sediment) boundaries in two coastal bay ecosystems. Environ Microbiol 20:2834–2853. https://doi.org/10.1111/1462-2920.14238
Funding
This study was funded by the Water Pollution Control and Control of Major National Science and Technology Projects in China (2018ZX07110005), the National Key Research and Development Project of China (No.2019YFC0409202), and the National Natural Science Foundation of China (No. 51679001).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Statement of informed consent
Additional informed consent was obtained from all individual participants for whom identifying information is included in this article.
Additional information
Responsible editor: Diane Purchase
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOCX 558 kb)
Rights and permissions
About this article
Cite this article
He, X., Ji, G. Responses of AOA and AOB activity and DNA/cDNA community structure to allylthiourea exposure in the water level fluctuation zone soil. Environ Sci Pollut Res 27, 15233–15244 (2020). https://doi.org/10.1007/s11356-020-07952-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11356-020-07952-9