Abstract
Background and aims
Global warming is predicted to alter the timing and magnitude of biogeochemical nitrogen cycling in paddy soils. However, little is known about its effect on active nitrifying populations. Here we investigated the responses of nitrification activity and active nitrifiers to elevated temperature in an acidic paddy soil.
Methods
13CO2-DNA-stable isotope probing (SIP), qPCR and high-throughput sequencing were used to determine active nitrifying phylotypes as well as difference in their abundance and community composition incubated at field temperature (15 °C) and elevated temperature (20 °C).
Results
Urea application led to significant production of nitrate and growth of ammonia-oxidizing bacteria (AOB) at both temperatures. Nitrification activity at elevated temperature was 148.3% and 18.5% higher than that of low temperature at day 28 and 56, respectively, accompanied by an increase in the extent of 13C-label incorporation by AOB. 13CO2-based SIP experiment indicated that both AOB and ammonia-oxidizing archaea (AOA) were involved in the nitrification activity and the active ammonia oxidizers changed from AOA to AOB with elevated temperature. Significant variation of AOA communities was observed under different temperatures. Dominant 13C-labeled nitrite-oxidizing bacteria (NOB) shifted from Nitrospira moscoviensis to Nitrospira japonica with higher temperature.
Conclusions
Our findings emphasized that elevated temperature had pronounced effects on autotrophic nitrification which was mediated by altering relative abundance of active AOB and AOA, as well as the community composition of AOA and NOB. AOB were more adaptable than AOA with increasing abundance but no alteration of composition at elevated temperature.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Nitrification, as a vital process of the global nitrogen cycle, can lead to significant nitrate loss and greenhouse gases emission. It is conventionally assumed to be catalyzed by chemolithoautotrophic ammonia-oxidizing archaea (AOA), bacteria (AOB) and nitrite-oxidizing bacteria (NOB) (Gruber-Dorninger et al. 2015; Wei et al. 2017; Yu et al. 2018). The discovery of complete ammonia oxidizers (comammox) further enriched the groups of nitrifiers (Hu and He 2017). Their relative contributions to nitrification have attracted the attention of soil scientists and a lot of research has been carried out around this issue in recent decades. Results showed that factors such as soil pH (Jia and Conrad 2009; Jiang et al. 2015; Luo et al. 2016), elevated CO2 (Long et al. 2012), water content (Hu et al. 2015), N concentration and forms (Maixner et al. 2006; Verhamme et al. 2011; Sun et al. 2015), stocking rates (Pan et al. 2018) and temperature (Taylor et al. 2017) differentially influenced the relative contributions of distinct nitrifying populations to nitrification.
Temperature is a key factor determining microbial growth and functional activity (Taylor et al. 2017; Ge et al. 2017). From 1880 to 2012, the global average land surface temperature has increased by 0.85 °C, and by the end of the twenty-first century, it is estimated to exceed 3.7 °C (IPCC 2013). Increased temperature may affect nitrification directly by stimulating enzymatic activity, and indirectly by changing the substrate availability, or by shifting the abundance and composition of the nitrifying populations (Schimel et al. 1994; Zhang et al. 2015; Hu et al. 2016; Osborne et al. 2016). The impacts of elevated temperature on nitrification and nitrifying populations are variable and soil specific (Hu et al. 2016). A few studies found that elevated temperature caused increase in nitrification rates (Grundmann et al. 1995; Larsen et al. 2011). However, other studies revealed that temperature was not correlated with nitrification rates (Shaw and Harte 2001; Niboyet et al. 2011; Baer et al. 2014; Osborne et al. 2016).
The optimum temperature for nitrifiers depends on the climate of the region (e.g. 35 °C and 20 °C for tropical and moderate soils, respectively) (Malhi and McGill 1982) and the common sense is that differences in the population structure lead to variations in temperature adaptability (Anderson et al. 1971). The large-scale distribution patterns of both AOA and AOB were found to be strongly affected by temperature (Fierer et al. 2009; Cao et al. 2013). Similarly, the community structure of NOB was also influenced by temperature (Alawi et al. 2009).
Rice production in waterlogged paddy soils feeds more than half of the world’s population with large amounts of N fertilizer application (Zhu et al. 2011, 2018; Luo et al. 2018). Paddy soils are characterized by constant alternation of drying and wetting, which will induce great changes in soil oxygen content and chemical properties, thus shape the indigenous microbial ecology (Hu et al. 2013; Atere et al. 2018; Liu et al. 2018). Competition between nitrifiers for oxygen in flooded paddy soils may also have great influence on their performance in nitrification (Wang et al. 2015). Therefore, the response of nitrifiers to elevated temperature in paddy soils will be quite different compared with upland soils.
Previous studies related to the effects of elevated temperature on nitrification mainly focused on the abundance and diversity of ammonia oxidizing populations in upland soils (Malchair et al. 2010; Szukics et al. 2010; Wang et al. 2016). However, the change in amoA gene abundance could not fully reflect ammonia oxidation activity (Jia and Conrad 2009), and little is known about the response of the active nitrifiers to elevated temperature, especially in paddy soils. Thus, DNA-based stable-isotope probing (SIP) combined with high-throughput sequencing were used to determine whether elevated temperature (+5 °C) had effects on the production of nitrate, as well as the richness and community composition of active nitrifiers in an acidic paddy soil (with mean annual temperature of 15.1 °C). We hypothesized that: elevated temperature will lead to higher soil nitrification rates by promoting the growth and activity of active nitrifiers. Additionally, we predicted that phylogenetically distinct groups of nitrifiers would respond differently to the elevated temperature, due to their distinct genetic features, energy use efficiency and substrate affinity.
Materials and methods
Soil sampling
Soils (Stagnic Anthrosol, pH 5.29) were sampled from Yunhe terrace, the largest terrace of Eastern China. It locates in Zhejiang province, a subtropical monsoon climate area, which has been cultivated with rice for more than 150 years. The mean annual temperature of this area is 15.1 °C. Soils were collected from three separate fields (>50 m apart, with an area of 30 m × 10 m). Three random soil cores (approximately 20 cm × 20 cm in area, and 15 cm depth) were collected and bulked into a single sample in each field. The collected samples were stored on ice and transported to the laboratory immediately. Obvious materials (stones, macro-fauna and plant residues) were removed before the moist soil samples were sieved through 2 mm sieve. One subsample of the sieved soil was stored at 4 °C prior to incubation and the other was air dried for physiochemical analysis. All results are the means of triplicate measurements and are expressed on an oven dry soil basis (24 h, 105 °C).
DNA-SIP microcosms
DNA-SIP microcosms were constructed as previously described (Zhang et al. 2019) in an attempt to simulate and predict the behavior of nitrifiers at 15 and 20 °C (T15 and T20), respectively. There were three treatments for each temperature: one labeled treatment (13CO2) and two control treatments (12CO2 and 13CO2 + C2H2) with three replicates of each treatment. For each treatment, 10 g sieved moist soil was placed in a 120 ml serum bottle, and was incubated at 60% of the soil maximum water-holding capacity in the dark for 56 days. The bottles were sealed with rubber stoppers and aluminum caps and 6 ml of 12C-CO2 or 13C-CO2 (99 at% carbon) was injected into the bottles through the rubber septum. The 13CO2 + C2H2 treatments were also applied with 100 Pa C2H2, which could completely inhibit ammonia oxidation. All microcosms received 100 μg urea-N g−1 soil, equivalent to the annual fertilizer N input in the field (approximately 300 kg urea-N ha−1 during the rice growing season). The 13CO2 and 13CO2 + C2H2 microcosms received 13C-urea to reduce the dilution of 13CO2 by CO2 released from the catalysis of urea, and 12C-urea was applied in 12CO2 treatments. The 13C-urea and 12C-urea were both applied weekly during the 8-week incubation period by dropwise addition of freshly prepared urea solution. In total, 90 (2 temperatures×3 treatments×3 replicates of each treatment×5 sampling points) bottles were prepared. Before the application of urea and CO2, a 14-day pre-incubation was conducted at 40% of soil maximum water-holding capacity to decrease the dilution of 13CO2 by soil-respired 12CO2, as previously described (Zhang et al. 2019).
Destructive sampling was performed for each treatment at days 0, 7, 14, 28 and 56. The concentrations of soil NO2−-N, NO3−-N and NH4+-N were determined colorimetrically by flow injection analysis (SAN++, Skalar, Holland) of 1 M KCl extracts. Soil DNA was extracted using a FastDNA Spin Kit for Soil (MP Biomedicals, Cleveland, OH, USA).
Isopycnic centrifugation and gradient fractionation
SIP fractionation was performed as previously described (Zhang et al. 2019). The mixture (with an initial CsCl buoyant density of 1.725 g ml−1) of 3.0 μg DNA extract in CsCl stock solution was placed in a 5.1 ml Quick-Seal polyallomer ultracentrifugation tube, and then ultracentrifuged at 20 °C for 44 h with a speed of 177,000 g in a Vti65.2 vertical rotor (Beckman Coulter Inc., Palo Alto, CA, USA). Fifteen DNA gradient fractions (~380 μl) were generated and their refractive index was measured with an AR200 digital hand-held refractometer (Reichert Inc., Buffalo, NY, USA) using a 65 μl aliquot of each fraction. The purified fractionated DNA was dissolved in 30 μl of TE buffer (Freitag et al. 2010).
PCR quantification of the amoA genes
The abundance of amoA genes in the total DNA and the fractionated DNA over the 56-day incubation were quantified with a LightCycler 480 II (Roche Applied Science). The qPCR primers and thermal-cycling conditions were described in Table S1. Each 20 μl PCR contained 1 μl of DNA template, 10 μl SYBR Premix Ex Taq (TaKaRa, Dalian, China), 0.4 mM of each primer, and 8.68 μl Milli-Q water. The qPCR standard was generated using plasmid DNA from one representative clone containing archaeal or bacterial amoA genes (Di et al. 2009). Specifically, AOA and AOB amoA genes were PCR amplified with the primers described in Table S1, and the resultant PCR products were purified using a PCR cleanup kit (Macherey-Nagel Inc., Germany) and cloned into the pGEM-T Easy Vector (Promega, Madison, WI, USA). Then the resulting ligation mix was transformed into Escherichia coli JM109 competent cells (Promega) according to the manufacturer’s recommendations. Plasmid DNA was extracted from the representative clones containing the target gene, quantified using a Nanodrop® ND-2000 UV-vis spectrophotometer (NanoDrop Technologies), and ten-fold serial dilutions of the plasmid DNA with a known copy number were used as the standard curve. That was 1.15 × 101 to 1.15 × 107 and 2.33 × 101 to 2.33 × 107 copies per assay for archaeal and bacterial amoA genes, respectively.
The PCR inhibition during the amplification was assessed using a serial dilution of the DNA template. All total DNA extracts were diluted at a ratio of 1:10 with nuclease-free water to reduce potential PCR inhibition, but no dilution was applied to the fractionated DNA. Amplification efficiencies were 97–101%, with R2 values of 0.99. The specificity of the amplification products was determined by standard agarose gel electrophoresis and melting-curve analysis.
Sequencing and processing of sequences
The 16S rRNA gene sequencing was conducted with primers 515F-907R targeting the V4-V5 regions (Table S1). Total DNA extracts at days 0, 28 and 56 and the fractionated heavy DNA (fractions 3–8) at days 28 and 56 in 13CO2 and 13CO2 + C2H2 microcosms were measured using an Illumina MiSeq platform. The raw data were submitted to the NCBI Sequence Read Archive database with assigned Study SRP102858.
Pyrosequencing of AOB and AOA amoA genes in the total DNA extracted from the 13CO2 microcosms at days 0, 28 and 56 was also performed on a Roche 454 GS FLX+ Titanium platform (Roche 454 Life Sciences, Branford, CT, U.S.). Primers amoA-1F/amoA-2R and Arch-amoAF/Arch-amoAR were used for AOB and AOA, respectively (Rotthauwe et al. 1997; Francis et al. 2005) (Table S1). The sequence data were deposited in the NCBI Sequence Read Archive (SRA) database (Accession Number: SRP102865 for AOB and SRP102863 for AOA).
Quality control and analysis of the resulting sequences were performed by the Quantitative Insights into Microbial Ecology (QIIME) software (Caporaso et al. 2010). Sequences with mismatched primers, ambiguous characters, < 20 average quality score, or shorter than 200 bp were removed. Operational Units (OTUs) were clustered using UPARSE (version 7.1) with 97% similarity cutoff. The taxonomy of 16S rRNA gene sequences and amoA gene sequences were analyzed by RDP Classifier against the Silva 16S rRNA database (Release 123) and FGR (Release 7.3) respectively, with a confidence threshold of 70%. Neighbour-joining phylogenetic trees of AOB, AOA and NOB were constructed using representative sequences of the 16S rRNA and amoA genes with the Kimura 2-parameter distance in MEGA (version 4.0) using 1000 bootstrap replicates (Tamura et al. 2007). The relative abundance of specific lineages was calculated from: (Reads attributed to this lineage)/(the total 16S rRNA reads).
Statistical analysis
The differences in nitrate concentration, ammonia concentration and amoA gene abundance between different treatments were assessed using one-way analysis of variance. Multiple comparisons were performed by Tukey’s post hoc tests. All analyses were realized in SPSS 17.0 (IBM, Armonk, NY, USA) and values at P < 0.05 were considered statistically significant. Non-metric multidimensional scaling (NMDS) plots were conducted with the vegan package in R.
Results
Changes of nitrification activity
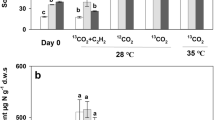
The nitrite concentration was below the detection limit (0.05 mg/L) and stepwise accumulation of nitrate occurred during the incubation period (Fig. S1). The production of nitrate at T20 was always higher than T15 throughout the incubation, in accordance with lower ammonium contents (Fig. S1). The differences between nitrate production at different temperatures after incubation for 28 and 56 days were statistically significant (Fig. 1a). There were no significant differences in the production of soil nitrate between the 12CO2 and 13CO2 microcosms (Fig. S1), indicating that 13CO2 was not discriminated by ammonia oxidizers. Therefore, soil nitrification activity was calculated as the rate of soil nitrate production in 13CO2 microcosms throughout the 56-day incubation. The nitrification activity from day 0 to day 28 was similar to that between day 28 and 56 at T20, with 4.66 μg NO3−-N g−1 soil per day during the 56-day incubation (Table 1). In contrast, biphasic kinetics was observed for the nitrate production rate at T15. Specifically, nitrate was produced at a rate of 0.51 μg NO3−-N g−1 soil per day from 0 to 28 days, while the rate between day 28 and 56 was much higher, resulting in a rate of 3.79 μg NO3−-N g−1 soil per day during the 56-day incubation (Table 1). The increase in soil nitrate content was completely stopped by acetylene at both temperatures (Fig. S1c).
Soil nitrate content (a) and abundance of ammonia oxidizers in SIP microcosms before and after the 56-day incubation. The AOB (b) and AOA (c) amoA gene copy numbers were determined using qPCR. Relative abundance of AOB (d) is calculated as the ratio: (the AOB 16S rRNA gene reads)/(the total microbial 16S rRNA gene reads) in each microcosm. The standard deviations of the triplicate microcosms were represented by the error bars. A significant difference (P < 0.05) based on the analysis of variance was indicated by the different letters above the columns
Changes of nitrifying communities
The amoA gene abundances of bacteria at both temperatures were significantly lower than that of archaea at day 0 (Fig. 1). Urea fertilization led to significant increase in the copy numbers of AOB amoA genes during the incubation at both temperatures (Fig. 1b). In T15 microcosms, it gradually increased from 1.89 × 105 copies g−1 soil at day 0 to 2.89 × 106 and 3.34 × 107 copies g−1 soil at day 28 and 56, respectively. With T20 13C-labeled treatments, it significantly increased from 3.86 × 105 at day 0 to 4.88 × 107 and 6.14 × 107 copies g−1 soil at day 28 and 56, representing 125- and 158-fold increases, respectively (Fig. 1b). Increase in bacterial amoA gene abundance was completely abolished by the addition of C2H2 at both temperatures (Fig. 1b). After incubation, the AOA amoA gene copy number slightly declined at T15, from 4.12 × 107 copies g−1 soil at day 0 to 3.31 × 107 copies g−1 soil at day 56 (Fig. 1c). While in T20 microcosms, their abundance decreased significantly from 5.40 × 107 copies g−1 soil at day 0 to 2.06 × 107 and 1.99 × 107 copies g−1 soil at day 28 and 56, respectively.
We obtained 1,616,364 high-quality reads after the high-throughput sequencing of 16S rRNA genes (Table S2), and the rarefaction curves were shown in Fig. S2. Reads belonging to AOB, AOA and NOB were screened out according to Zhang et al. (2019). The application of urea led to significant increase in the relative abundance of AOB after incubation (Fig. 1d). It increased dramatically only between the second half of the incubation at T15, from 0.01% at day 28 to 0.47% at day 56, representing 48-fold increase (Fig. 1d). With T20, the relative abundance of AOB increased gradually from lower than 0.01% at day 0 to 0.21% and 0.27% after incubated for 28 and 56 days, representing 62- and 76-fold increases, respectively (Fig. 1d). There were no significant changes of the AOA relative abundance during incubation and the relative abundance of NOB decreased significantly after incubation at both temperatures (Fig. S3).
In total, 93,728 and 133,380 high quality amoA reads were obtained for archaea and bacteria, respectively, after the pyrosequencing of amoA genes (Table S2). The cumulative number of species reached an asymptote at 2000 reads in all samples (Fig. S4). There was no significant difference between the amoA gene phylogeny and their 16S rRNA gene phylogeny for both AOA and AOB (Fig. S5 and S6). Approximately 90% of the AOA reads were classified into the marine group 1.1a-associated lineage (Fig. S5) and almost all AOB reads had high sequence similarities to members within the Nitrosospira cluster 3 lineage (Fig. S6). With NOB, more than 99% of the reads fell within the genera Nitrospira, which dominated the nitrite oxidizer populations (Fig. S7).
13C-labeled active nitrifying communities
Strong labeling of AOB was revealed by qPCR of the amoA genes in the DNA fractions in all 13CO2-labeled treatments (Fig. 2). In the control (12CO2 and 13CO2 + C2H2) microcosms, the highest copy number of AOB amoA genes was observed in the “light” fraction with a buoyant density of 1.720 g ml−1 at both temperatures (Fig. 2a and c). However, in the T15 13CO2 microcosms, the AOB amoA gene copies peaked in the “heavy” DNA fractions with buoyant densities of 1.732 and 1.739 g ml−1 at days 28 and 56, respectively (Fig. 2a). Similarly, in the T20 13CO2 microcosms, the genome of AOB populations was also gradually labeled with the highest AOB amoA gene abundance occurring in the “heavy” fractions with buoyant densities of 1.739 and 1.742 g ml−1 at days 28 and 56, respectively (Fig. 2c). AOB was labeled more heavily at T20 than T15 (Fig. 2a and c), which was further supported by higher ratios: (bacterial amoA gene copy numbers in heavy DNA)/(bacterial amoA gene copy numbers in all DNA fractions) at T20 than T15 at both days 28 and 56 (Table 1). For instance, at day 28, the ratios were 86.2% and 94.7% at T15 and T20, respectively.
The relative frequency of the bacterial (a, c) and archaeal (b, d) amoA genes based on qPCR across the entire buoyant density gradient of the DNA fractions from the 12CO2, 13CO2 or 13CO2 + C2H2 microcosms at day 28 and 56. The standard errors of the triplicate microcosms were represented by the error bars. All other designations are as in Fig. 1
Similarly, the increasing labeling of AOA during incubation was also observed in 13C-labeled microcosms and they were also labeled to a greater extent at T20 than T15 (Fig. 2b and d). In T20 13CO2 microcosms, the archaeal amoA gene abundance peaked in heavy fractions with buoyant density of 1.732 g ml −1 at both day 28 and 56 (Fig. 2d), and ratios: (archaeal amoA gene copy numbers in heavy DNA)/(archaeal amoA gene copy numbers in all DNA fractions) reached 76.4% and 69.7%, respectively (Table 1). With T15, the archaeal amoA gene abundance peaked in lighter fractions, at around a buoyant density of 1.720 g ml−1 at day 28 (Fig. 2b). After the 56-day incubation, the peak of AOA abundance clearly shifted towards the heavy fractions with buoyant densities of 1.732 g ml −1 (Fig. 2b). The assimilation of 13CO2 by both AOA and AOB was completely inhibited by C2H2 (Fig. 2).
Significant enrichment of nitrifying community in the ‘heavy’ DNA fractions was also revealed by the 16S rRNA gene sequencing. For instance, in T15 13CO2 microcosms, the highest relative abundance of AOB were 12.3% and 33.7% at day 28 and 56, respectively, in the ‘heavy’ DNA fractions (Fig. 3 and Table S3). The maximum percentages increased to 43.4% and 41.6% in T20 13CO2 microcosms at day 28 and 56, respectively, much higher than in their control treatments (less than 0.30%; Fig. 3 and Table S3).
Similarly, the relative abundance of AOA in the ‘heavy’ DNA fractions reached as high as 57.6% and 39.8% at T15 and T20, respectively at day 28, then decreased to 45.5% and 18.7% at day 56, respectively, which were still much higher than those in the control (13CO2 + C2H2) treatments (lower than 0.48%; Fig. 3 and Table S3). With NOB, their relative abundance in the ‘heavy’ DNA fractions were 29.9- and 4.42-fold higher than those in the control microcosms after the 56-day incubation in T15 and T20, respectively (Fig. 3 and Table S3).
Phylogenetic analyses indicated that there was no significant difference between proportions of active nitrifying populations at day 28 and 56 (data not shown), so we choose data at day 56 for the construction of phylogenetic trees of AOA (Fig. S8), AOB (Fig. S9) and NOB (Fig. 4). Active ammonia-oxidizing communities were dominated by members within the 1.1a-associated cluster AOA and Nitrosospira cluster 3 AOB at both temperatures (Table 2). The 13C-labeled nitrite oxidizers were dominated by Nitrospira-like NOB (Fig. 4). Nitrospira-like bacteria comprised ecologically versatile sublineages, for instance, members within Nitrospira japonica lineage occupied 24.4% and 57.7% of the 13C-16S rRNA genes at T15 and T20, respectively. Approximately 55.9% and 32.2% of the 13C-labeled NOB could be assigned to Nitrospira moscoviensis at T15 and T20, respectively. The remained NOB belonged to three other Nitrospira lineages and one Nitrobacter lineage (Table 2).
Phylogenetic analysis of NOB 16S rRNA genes in 13C-labeled DNA at day 56. The designation ‘T15-NOB-16S rRNA-HF-OTU-1-772-(55.9%)’ indicates that OTU-1 accounts for 55.9% of the total NOB 16S rRNA gene reads in the 13C-DNA from the heavy fractions at T15, containing 772 reads with 97% sequence similarity
Discussion
The application of urea stimulated the growth of AOB but not AOA, in accordance with other studies which found that only AOB abundance increased after N fertilizer applications (Di et al. 2009; Jia and Conrad 2009). Among numerous climatic and environmental factors, temperature is recognized to be one of the most important driving factors of soil nitrification, which is a microbially-mediated process predominantly controlled by nitrifiers (Hu et al. 2016). The contributions of nitrifiers to nitrification are determined by their enzyme activity, which is temperature-sensitive and can directly respond to warming through acclimation (physiological responses) and thermal adaptation (genetic changes) (Tourna et al. 2008; Karhu et al. 2014).
Effects of elevated temperature on active ammonia oxidizers
Mounting evidence has shown the effect of elevated temperature on ammonia oxidizers, but there was no consistent conclusion (He et al. 2007; Xu et al. 2017). The absence of the middle peak for T20 AOB at day 28 indicated that their growth rate was higher than at T15 (Fig. 2a, c), which was also supported by amoA gene copies at day 28 at T15 compared with T20 (Fig. 1b). We concluded that the application of urea stimulated the growth of AOB, but the stimulation was greater at T20 compared with T15, implying that the elevated temperature might provide beneficial conditions for nitrifiers by promoting cell survival and maintenance, as well as reducing the lag time for cell growth (Avrahami and Bohannan 2007).
The ratio: (amoA gene copy numbers in heavy DNA)/(amoA gene copy numbers in all DNA fractions) of both AOA and AOB was higher under higher temperature after incubation (Table 1). Besides, the abundance of AOB amoA genes in heavy DNA was stimulated to a much greater extent at T20 than T15, in accordance with another study which found an increasing abundance of AOB, but not AOA, with increasing soil temperature from 5 to 25 °C (Szukics et al. 2010).
The changes in the relative abundance of active AOA and AOB resulted in the change in AOA/AOB ratio at elevated temperature. In T15 labeled treatment, the AOA/AOB ratios in heavy fractions were 29.1 and 2.01 at day 28 and 56, respectively (Table 1). While in T20 treatments, the 13CO2 was assimilated to a significantly greater extent in bacterial amoA genes compared with the archaeal amoA genes, and the ratio of AOA/AOB decreased to 0.85 and 0.60 at day 28 and 56, respectively (Table 1). The above results were further supported by the max ratios of AOA or AOB 16S rRNA genes to the total microbial communities in the 13C-DNA, which showed similar patterns of AOA 16S rRNA gene ratios/AOB 16S rRNA gene ratios of 4.70 and 1.35 for T15 while 0.90 and 0.45 for T20 at day 28 and 56, respectively (Fig. 3 and Table S3).
The AOA/AOB ratio in 13C-DNA could largely reflect the relative importance of archaeal and bacterial ammonia oxidation in the DNA-SIP based nitrification experiment (Wang et al. 2015). Therefore, in the tested acidic soil, dominant ammonia-oxidizing microorganisms changed from AOA to AOB with the increase of temperature. This could be explained by the presumption that the increased temperature accelerated the diffusion rates of ammonia into microbial cells (Avrahami and Bohannan 2007), and ammonia concentration determined differential growth of AOA and AOB (Verhamme et al. 2011). Decrease in soil pH leads to exponentially declines of ammonia concentration due to the ionization of ammonia to ammonium (Allison and Prosser 1993; Burton and Prosser 2001; Jiang et al. 2015). In the tested acidic soil (pH 5.29), the ammonia concentrations (<10 μM) may not always be sufficient for ammonia-oxidizing microorganisms. Besides, the reaction from ammonium to ammonia is endothermic and high temperature is beneficial to the production of ammonia. AOA were reported to play important roles in acidic and ammonia-poor environments benefited from their high affinity for ammonia (Martens-Habbena et al. 2009; He et al. 2012), while AOB preferred environments with relatively higher ammonia concentration (Di et al. 2009, 2010; Höfferle et al. 2010; Verhamme et al. 2011; Hu et al. 2012). Therefore, the elevated temperature was more suitable for AOB in the tested soil due to promotion of the production of ammonia and accelerating its diffusion rates into microbial cells.
The amoA gene-based community analyses revealed that fertilization treatments induced community shifts of both AOA and AOB after 8 weeks of incubation (Fig. 5), indicating the crucial role of ammonia in selecting the community responses of ammonia oxidizers (Szukics et al. 2010). In line with previous study, the structure of AOA community shifted in response to temperature variation (Fig. 5a), but AOB appeared to be more stable and there was no significant differentiation of AOB community at different temperatures (Fig. 5b), implying that AOA were more sensitive than AOB communities at elevated temperature (Tourna et al. 2008).
Effects of elevated temperature on active nitrite oxidizers
There was significant difference in the population structures of nitrite oxidizing populations at these two temperatures especially in 13C-labeled DNA (Table 2 and Fig. S10), implying the different temperature adaptability of NOB groups (Anderson et al. 1971). NOB community was dominated by a widely distributed genus Nitrospira, a monophyletic but diverse group of organisms, which are the dominant NOB in many soil environments (Wang et al. 2015; Ushiki et al. 2017). Phylogenetic analysis demonstrated marked dissimilarities in Nitrospira composition under elevated temperature, specifically, dominant NOB shifted from Nitrospira moscoviensis at T15 to Nitrospira japonica at T20 (Table 2 and Fig. 4, S10), implying that the biochemical processes associated with nitrite oxidation by the two species differ thermodynamically and the existence of their ecological niche partitioning.
Because of the evident differences between ammonium and nitrate concentrations at the two temperatures during the incubation, we speculate that the differences in nitrite contents existed under different temperatures, which will influence the population composition of Nitrospira-like bacteria (Maixner et al. 2006). According to previous studies, two members of Nitrospira sublineage II, Nitrospira japonica and Nitrospira moscoviensis, were distantly related to each other, with 95.6% similarity (Ehrich et al. 1995; Ushiki et al. 2013). There were difference in their optimal temperature, tendency to aggregate, and utilization of organic substrate of the two species. For example, Nitrospira japonica could grow with formate and was mixotrophic, but the utilization of organic substrate of Nitrospira moscoviensis was not observed (Ushiki et al. 2013). Besides, the abundance and distribution of different Nitrospira clades could be co-affected by protozoan grazing, the affinity for oxygen, the availability of organic substrates and the presence of specific phages under complex soil conditions, which could be influenced by temperature (Daims et al. 2001; Maixner et al. 2006). There was no direct evidence for the factors driving the niche partitioning between these two closely related species and the difference in their nitrite oxidation kinetics in situ was also unclear. Due to the huge phylogenetic diversity of nitrite oxidizers, our insights into the physiology of NOB remain the tip of an iceberg.
In conclusion, with the combination of DNA-SIP and high-throughput sequencing, we found different response of AOB and AOA to elevated temperature, with increasing dominance of AOB in ammonia oxidation and significant shift of AOA community composition. Elevated temperature also resulted in niche differentiation of two Nitrospira sublineage II species. The above results revealed pronounced direct and indirect effects of elevated temperature on soil nitrification by selecting phylogenetically distinct dominant nitrifiers in the tested acidic paddy soil. Our results provide novel evidence for the response of nitrification and active nitrifiers in paddy soils to elevated temperature, and future research should focus on the mechanism of temperature selections for particular active nitrifying assemblages.
Abbreviations
- AOA:
-
Ammonia-oxidizing archaea
- AOB:
-
Ammonia-oxidizing bacteria
- NOB:
-
Nitrite-oxidizing bacteria
- DNA-SIP:
-
DNA-stable isotope probing
References
Alawi M, Off S, Kaya M, Spieck E (2009) Temperature influences the population structure of nitrite-oxidizing bacteria in activated sludge. Environ Microbiol Rep 1:184–190
Allison SM, Prosser JI (1993) Ammonia oxidation at low pH by attached populations of nitrifying bacteria. Soil Biol Biochem 25:935–941
Anderson OE, Boswell FC, Harrison RM (1971) Variations in low temperature adaptability of nitrifiers in acid soils. Soil Sci Soc Amer Proc 35:68–71
Atere CT, Ge TD, Zhu ZK, Liu SL, Huang XZ, Shibsitova O, Guggenberger G, Wu JS (2018) Assimilate allocation by rice and carbon stabilisation in soil: effect of water management and phosphorus fertilization. Plant Soil. https://doi.org/10.1007/s11104-018-03905-x
Avrahami S, Bohannan BJM (2007) Response of Nitrosospira sp. strain AF-like ammonia oxidizers to changes in temperature, soil moisture content, and fertilizer concentration. Appl Environ Microbiol 73:1166–1173
Baer SE, Connelly TL, Sipler RE, Yager PL, Bronk DA (2014) Effect of temperature on rates of ammonium uptake and nitrification in the western coastal Arctic during winter, spring, and summer. Glob Biogeochem Cycles 28:1455–1466
Burton SA, Prosser JI (2001) Autotrophic ammonia oxidation at low pH through urea hydrolysis. Appl Environ Microbiol 67:2952–2957
Cao H, Auguet JC, Gu JD (2013) Global ecological pattern of ammonia-oxidizing archaea. PLoS One 8:e52853
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R (2010) QIIME allows analysis of highthroughput community sequencing data. Nat Methods 7:335–336
Daims H, Nielsen JL, Nielsen PH, Schleifer KH, Wagner M (2001) In situ characterization of Nitrospira-like nitrite-oxidizing bacteria active in wastewater treatment plants. Appl Environ Microbiol 71:5273–5284
Di HJ, Cameron KC, Shen JP, Winefield CS, O'Callaghan M, Bowatte S, He JZ (2009) Nitrification driven by bacteria and not archaea in nitrogen-rich grassland soils. Nat Geosci 2:621–624
Di HJ, Cameron KC, Shen JP, Winefield CS, O'Callaghan M, Bowatte S, He JZ (2010) Ammonia-oxidizing bacteria and archaea grow under contrasting soil nitrogen conditions. FEMS Microbiol Ecol 72:386–394
Ehrich S, Behrens D, Lebedeva E, Ludwig W, Bock E (1995) A new obligately chemolithoautotrophic, nitrite-oxidizing bacterium, Nitrospira moscoviensis sp. nov. and its phylogenetic relationship. Arch Microbiol 164:16–23
Fierer N, Carney KM, Horner-Devine MC, Megonigal JP (2009) The biogeography of ammonia-oxidizing bacterial communities in soil. Microb Ecol 58:435–445
Francis CA, Roberts KJ, Beman JM, Santoro AE, Oakley BB (2005) Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. Proc Natl Acad Sci U S A 102:14683–14688
Freitag TE, Chang L, Prosser JI (2010) Changes in the community structure and activity of betaproteobacterial ammonia-oxidizing sediment bacteria along a freshwater-marine gradient. Environ Microbiol 8:684–696
Ge TD, Wei XM, Razavi BS, Zhu ZK, Hu YJ, Kuzyakov Y, Jones DL, Wu JS (2017) Stability and dynamics of enzyme activity patterns in the rice rhizosphere: effects of plant growth and temperature. Soil Biol Biochem 113:108–115
Gruber-Dorninger C, Pester M, Kitzinger K, Savio DF, Loy A, Rattei T (2015) Functionally relevant diversity of closely related Nitrospira in activated sludge. ISME J 9:643–655
Grundmann GL, Renault P, Rosso L, Bardin R (1995) Differential effects of soil water content and temperature on nitrification and aeration. Soil Sci Soc Am J 59:1342–1349
He JZ, Shen JP, Zhang LM, Zhu YG, Zheng YM, Xu MG, Di HJ (2007) Quantitative analyses of the abundance and composition of ammonia-oxidizing bacteria and ammonia-oxidizing archaea of a Chinese upland red soil under long-term fertilization practices. Environ Microbiol 9:2364–2374
He JZ, Hu HW, Zhang LM (2012) Current insights into the autotrophic thaumarchaeal ammonia oxidation in acidic soils. Soil Biol Biochem 55:146–154
Höfferle Š, Nicol GW, Pal L, Hacin J, Prosser JI, Mandić-Mulec I (2010) Ammonium supply rate influences archaeal and bacterial ammonia oxidizers in a wetland soil vertical profile. FEMS Microbiol Ecol 74:302–315
Hu HW, He JZ (2017) Comammox-a newly discovered nitrification process in the terrestrial nitrogen cycle. J Soils Sediments 17:2709–2717
Hu BL, Liu S, Shen LD, Zheng P, Xu XY, Lou LP (2012) Effects of different ammonia concentrations on community succession of ammonia-oxidizing microorganisms in a simulated paddy soil column. PLoS One 7:e44122
Hu HW, Zhang LM, Yuan CL, He JZ (2013) Contrasting Euryarchaeota communities between upland and paddy soils exhibited similar pH-impacted biogeographic patterns. Soil Biol Biochem 64:18–27
Hu HW, Zhang LM, Yuan CL, Zheng Y, Wang JT, Chen D, He JZ (2015) The large-scale distribution of ammonia oxidizers in paddy soils is driven by soil pH, geographic distance, and climatic factors. Front Microbiol 6:938
Hu HW, Macdonald CA, Trivedi P, Anderson IC, Zheng Y, Holmes B (2016) Effects of climate warming and elevated CO2 on autotrophic nitrification and nitrifiers in dryland ecosystems. Soil Biol Biochem 92:1–15
IPCC (2013) Climate change 2013: the physical science basis. Contribution of working group I to the fifth assessment reports of the intergovernmental panel on climate change. Cambridge University Press, Cambridge, UK
Jia Z, Conrad R (2009) Bacteria rather than archaea dominate microbial ammonia oxidation in an agricultural soil. Environ Microbiol 11:1658–1671
Jiang XJ, Hou XY, Zhou X, Xin XP, Wright A, Jia ZJ (2015) pH regulates key players of nitrification in paddy soils. Soil Biol Biochem 81:9–16
Karhu K, Auffret MD, Dungait JAJ, Hopkins DW, Prosser JI, Singh BK, Subke JA, Wookey PA, Ågren GI, Sebastià MT, Gouriveau F, Bergkvist G, Meir P, Nottingham AT, Salinas N, Hartley IP (2014) Temperature sensitivity of soil respiration rates enhanced by microbial community response. Nature 513:81–84
Larsen KS, Andresen LC, Beier C, Jonasson S, Albert KR, Ambus PER (2011) Reduced N cycling in response to elevated CO2, warming, and drought in a Danish heathland: synthesizing results of the CLIMAITE project after two years of treatments. Glob Chang Biol 17:1884–1899
Liu H, Ding Y, Zhang Q, Liu X, Xu J, Li Y, Di H (2018) Heterotrophic nitrification and denitrification are the main sources of nitrous oxide in two paddy soils. Plant Soil. https://doi.org/10.1007/s11104-018-3860-x
Long X, Chen C, Xu Z, Oren R, He JZ (2012) Abundance and community structure of ammonia-oxidizing bacteria and archaea in a temperate forest ecosystem under ten-years elevated CO2. Soil Biol Biochem 46:163–171
Luo Y, Yu Z, Zhang K, Xu J, Brookes PC (2016) The properties and functions of biochars in forest ecosystems. J Soils Sediments 16:2005–2020
Luo Y, Zhu Z, Liu S, Peng P, Xu J, Brookes P, Ge T, Wu J (2018) Nitrogen fertilization increases rice rhizodeposition and its stabilization in soil aggregates and the humus fraction. Plant Soil. https://doi.org/10.1007/s11104-018-3833-0
Maixner F, Noguera DR, Anneser B, Stoecker K, Wegl G, Wagner M, Daims H (2006) Nitrite concentration influences the population structure of Nitrospira-like bacteria. Environ Microbiol 8:1487–1495
Malchair S, De Boeck HJ, Lemmens CMHM, Ceulemans R, Merckx R, Nijs I, Carnol M (2010) Diversity-function relationship of ammonia-oxidizing bacteria in soils among functional groups of grassland species under climate warming. Appl Soil Ecol 44:15–23
Malhi SS, McGill WB (1982) Nitrification in three Alberta soils: effect of temperature, moisture and substrate concentration. Soil Biol Biochem 14:393–399
Martens-Habbena W, Berube PM, Urakawa H, José R, Stahl DA (2009) Ammonia oxidation kinetics determine niche separation of nitrifying archaea and Bacteria. Nature 461:976–979
Niboyet A, Le Roux X, Dijkstra P, Hungate BA, Barthes L, Blankinship JC (2011) Testing interactive effects of global environmental changes on soil nitrogen cycling. Ecosphere 2:1–24
Osborne BB, Baron JS, Wallenstein MD (2016) Moisture and temperature controls on nitrification differ among ammonia oxidizer communities from three alpine soil habitats. Front Earth Sci 10:1–12
Pan H, Xie K, Zhang Q, Jia Z, Xu J, Di H, Li Y (2018) Archaea and bacteria respectively dominate nitrification in lightly and heavily grazed soil in a grassland system. Biol Fertil Soils 54:41–54
Rotthauwe JH, Witzel KP, Liesack W (1997) The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl Environ Microbiol 63:4704–4712
Schimel DS, Braswell BH, Holland EA, McKeown R, Ojima DS, Painter TH (1994) Climatic, edaphic, and biotic controls over storage and turnover of carbon in soils. Glob Biogeochem Cycles 8:279–293
Shaw MR, Harte J (2001) Response of nitrogen cycling to simulated climate change: differential responses along a subalpine ecotone. Glob Chang Biol 7:193–210
Sun R, Guo X, Wang D, Chu H (2015) Effects of long-term application of chemical and organic fertilizers on the abundance of microbial communities involved in the nitrogen cycle. Appl Soil Ecol 95:171–178
Szukics U, Abell GC, Hödl V, Mitter B, Sessitsch A, Hackl E, Zechmeister-Boltenstern S (2010) Nitrifiers and denitrifiers respond rapidly to changed moisture and increasing temperature in a pristine forest soil. FEMS Microbiol Ecol 72:395–406
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Taylor AE, Giguere AT, Zoebelein CM, Myrold DD, Bottomley PJ (2017) Modeling of soil nitrification responses to temperature reveals thermodynamic differences between ammonia-oxidizing activity of archaea and bacteria. ISME J 11:896–908
Tourna M, Freitag TE, Nicol GW, Prosser JI (2008) Growth, activity and temperature responses of ammonia-oxidizing archaea and bacteria in soil microcosms. Environ Microbiol 7:1985–1995
Ushiki N, Fujitani H, Aoi Y, Tsuneda S (2013) Isolation of Nitrospira belonging to sublineage II from a wastewater treatment plant. Microbes Environ 28:346–353
Ushiki N, Jinno M, Fujitani H, Suenaga T, Terada A, Tsuneda S (2017) Nitrite oxidation kinetics of two Nitrospira strains: the quest for competition and ecological niche differentiation. J Biosci Bioeng 123:581–589
Verhamme DT, Prosser JI, Nicol GW (2011) Ammonia concentration determines differential growth of ammonia-oxidising archaea and bacteria in soil microcosms. ISME J 5:1067–1071
Wang BZ, Zhao J, Guo ZY, Ma J, Xu H, Jia ZJ (2015) Differential contributions of ammonia oxidizers and nitrite oxidizers to nitrification in four paddy soils. ISME J 9:1062–1075
Wang C, Chen Z, Unteregelsbacher S, Lu H, Gschwendtner S, Gasche R (2016) Climate change amplifies gross nitrogen turnover in montane grasslands of Central Europe in both summer and winter seasons. Glob Chang Biol 22:2963–2978
Wei XM, Hu YJ, Peng PQ, Zhu ZK, Atere CT, O'Donnell AG, Wu JS, Ge TD (2017) Effect of P stoichiometry on the abundance of nitrogen-cycle genes in phosphorus-limited paddy soil. Biol Fertil Soils 53:767–776
Xu X, Liu X, Li Y, Ran Y, Liu Y, Zhang Q (2017) High temperatures inhibited the growth of soil bacteria and archaea but not that of fungi and altered nitrous oxide production mechanisms from different nitrogen sources in an acidic soil. Soil Biol Biochem 107:168–179
Yu MJ, Yu L, Su W, Afzal M, Li Y, Brookes PC, Redmile-Gordon M, Luo Y, Xu J (2018) Changes in nitrogen related functional genes along soil pH, C and nutrient gradients in the charosphere. Sci Total Environ 650:626–632
Zhang JJ, Li YF, Chang SX, Qin H, Fu SL, Jiang PK (2015) Understory management and fertilization affected soil greenhouse gas emissions and labile organic carbon pools in a Chinese chestnut plantation. For Ecol Manag 337:126–134
Zhang Q, Li Y, He Y, Liu H, Dumont MG, Brookes PC, Xu J (2019) Nitrosospira cluster 3-like bacterial ammonia oxidizers and Nitrospira-like nitrite oxidizers dominate nitrification activity in acidic terrace paddy soils. Soil Biol Biochem 131:229–237. https://doi.org/10.1016/j.soilbio.2019.01.006
Zhu G, Wang S, Wang Y, Wang C, Risgaard-Petersen N, Jetten MS, Yin C (2011) Anaerobic ammonia oxidation in a fertilized paddy soil. ISME J 5:1905–1912
Zhu ZK, Ge TD, Liu SL, Hu YJ, Ye RZ, Xiao ML, Tong CL, Kuzyakov Y, Wu JS (2018) Rice rhizodeposits affect organic matter priming in paddy soil: the role of N fertilization and plant growth for enzyme activities, CO2 and CH4 emissions. Soil Biol Biochem 116:369–377
Acknowledgements
This research was financially supported by the National Natural Science Foundation of China (41721001 and 41671249).
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible Editor: Zucong Cai.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOCX 1523 kb)
Rights and permissions
About this article
Cite this article
Zhang, Q., Li, Y., He, Y. et al. Elevated temperature increased nitrification activity by stimulating AOB growth and activity in an acidic paddy soil. Plant Soil 445, 71–83 (2019). https://doi.org/10.1007/s11104-019-04052-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11104-019-04052-7