Abstract
An enrichment process was employed by applying high ibuprofen concentration in an immobilized cell bioreactor in order to favor the ibuprofen-degrading community present in activated sludge. Experimental data showed the ability of the immobilized cell bioreactor to achieve high ibuprofen removal efficiencies (98.4 ± 0.3%), the tendency of the enriched biomass to acidify the treated liquor, and the inhibition of the nitrification process. Illumina sequencing revealed a massive increase in the relative abundance of Alphaproteobacteria and Gammaproteobacteria (from 29.1 to 80.8%) and a dramatic decrease in the proportion of Bacteroidetes, Planctomycetes, and Verrucomicrobia (from 42.7 to 2.1%) when pure ibuprofen served as the sole carbonaceous feeding substrate. This shift in the feeding conditions resulted in the predominance of Novosphingobium and Rhodanobacter (25.5 ± 10.8% and 25.2 ± 3.0%, respectively) and demonstrated a specialized ibuprofen-degrading bacterial community in activated sludge, which possessed the selective advantage to cope with its degradation. To the best of our knowledge, this bioreactor system was capable of effectively treating the highest ibuprofen concentration applied in wastewater treatment plants.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Ibuprofen, which is scientifically named 4-isobutylphenyl-2-propionic acid, is one of the widely used non-steroidal anti-inflammatory drugs (NSAIDs). The most common therapeutic properties of ibuprofen are its inflammatory action against arthritis and toothache and its anti-fever effects (Rainsford 2009). Nowadays, ibuprofen is detected in high concentrations in pharmaceutical industry and hospital wastewaters, which are considered as important point sources of NSAIDs (Li 2014; Davarnejad et al. 2018).
Pharmaceutical pollutants like ibuprofen pose negative effects on aquatic organisms at all stages of their life cycle (Zhou et al. 2009). Ibuprofen has been detected in pharmaceutical industry wastewater at concentrations even higher than 200 mg/L (Jallouli et al. 2018). Moreover, the presence of ibuprofen in pharmaceutical wastewaters can cause genotoxic effects under both acute and sub-chronic exposure conditions and induce free radical production and oxidative stress on various aquatic organisms (Ragugnetti et al. 2011; Jeffries et al. 2015; Candido et al. 2016; Islas-Flores et al. 2017). In addition, Moro et al. (2014), who investigated the negative impact of ibuprofen on Scenedesmus rubescens cultures, denoted ultrastructural and morphological alterations in cytoplasmic inclusions. Chlorophyll reduction and carotenoid increase were also among the negative consequences of ibuprofen (Moro et al. 2014).
Despite the importance of NSAID degradation, the examination of microbiota with proven capability of degrading ibuprofen is restricted solely to studies with single isolates. More specifically, Sphingobium sp. IbD51, which was isolated from activated sludge, could mineralize ibuprofen in R2A medium (Zhou et al. 2013). Variovorax Ibu-1 obtained from activated sludge was capable of utilizing ibuprofen as the sole carbon and energy source through 2,3-dioxygenation of the aromatic ring and dehydrogenation to catechol, reducing in batch cultures an initial ibuprofen concentration of 500 mg/L by 40% at the early stationary phase (Murdoch and Hay 2015). Sphingomonas Ibu-2 was also capable of utilizing ibuprofen as a sole carbon and energy source via 1,2-dioxygenation of the aromatic ring to isobutylcatechol and subsequent induction of the extradiol ring-cleavage pathway (Murdoch and Hay 2005). Fortunato et al. (2016) recently isolated a Comamonas aquatica and a Bacillus ibuprofen-degrading strain from river surface waters by using an ibuprofen-based minimal medium containing 0.2 g/L EDTA.
A limited number of studies have been performed in wastewater treatment systems in order to investigate the effects of ibuprofen on the composition of the activated sludge microbiota. In particular, Jiang et al. (2017) reported that Actinobacteria and Bacteroidetes population was increased when mixtures of ibuprofen with diclofenac or naproxen were applied at low concentration (5 μg/L) in sequencing batch reactors (SBRs) fed with synthetic wastewater. By contrast, Wang et al. (2016) denoted a relative decrease in the proportion of Bacteroidetes population during addition of five pharmaceutical and personal care products (PPCPs) (50 μg/L each), including ibuprofen, in a granular MBR system treating an acetate-based wastewater. Besides, Xia et al. (2014) mentioned that “PPCP drugs had relatively less effect on microbial diversity.” Kraigher et al. (2008) reported that greater divergence in activated sludge communities occurred when NSAIDs were added at 5 mg/L rather than 50 μg/L. Indeed, the complexity caused by the low concentration of PPCPs used in these studies, as compared to the high organic content of the wastewater treated, increases the uncertainty in the identification of the main NSAID degraders, and therefore, the investigation of the ibuprofen-degrading community present in the activated sludge is advantageous when such anti-inflammatory drug serves as the sole carbonaceous feeding substrate. However, no study of microbial diversity has been carried out in batch experiments or in bioreactor systems so far by using ibuprofen as the sole carbonaceous feeding substrate. Thus, the objectives of this research work were (a) to proliferate the ibuprofen-degrading community present in the activated sludge in order to study biodegradation aspects of ibuprofen through the performance of an enrichment process carried out in an aerobic immobilized cell bioreactor where high ibuprofen concentration was applied either by using commercially available tablets or the pure pharmaceutical substrate, (b) to evaluate biosystem’s performance by using high ibuprofen dose applications for a prolonged period of time, and (c) to identify the microbial community structure in the enriched ibuprofen-degrading biomass by employing high-throughput sequencing techniques, i.e., Illumina sequencing.
Material and methods
Bioreactor design and enrichment of ibuprofen-degrading communities
An immobilized cell bioreactor of 550 mL working volume was installed. The unit consisted of a main reactor (biofilter) of 550 mL volume, which was filled with Siran porous beads as the immobilized carrier (Supplementary Material Fig. S1). The Siran beads formed a cell-immobilized column of 150 mL volume. A glass cylindrical tube of 120 mL, containing an air diffuser on the bottom that connected to an air pump, was used for aeration. Aeration was provided through recirculation of wastewater from the main biofilter to the aeration tube by the use of a high-speed peristaltic pump (550 mL/min). The immobilized bioreactor was fed automatically with a connected pump every 24 h by adding either synthetic wastewater containing commercially available ibuprofen tablets or minimal medium where ibuprofen served as the sole carbonaceous feeding substrate, as appropriate, resulting in a corresponding hydraulic retention time (HRT) of 3.5 days.
The immobilized cell bioreactor was initially seeded with 50 mL activated sludge receiving from a membrane reactor (MBR) treating municipal wastewater. Synthetic wastewater containing ibuprofen was initially prepared by adding one commercially available ibuprofen tablet of 600 mg in 1 L distilled water, although 43 days thereafter, ibuprofen concentration in the synthetic wastewater was decreased to 400 mg/L, supplementing also with 0.2 g/L yeast extract and 8 mg/L NH4Cl, in order to enhance microbial growth. Each commercial tablet used consisted of 297 mg sucrose as well as of trace amounts of silicon dioxide colloidal (aerosil), starch maize pregelatinized, stearic acid, starch maize, calcium sulfate dihydrate (terra alba), carmellose sodium, acacia (spray dried), opalux pink AS-1537, opaglos regular, and carnauba wax. After an extended operation period (more than 5 months), ibuprofen of high purity was used as the only carbonaceous feeding substrate in the bioreactor system. In particular, a minimal medium consisting of 400 mg/L of ibuprofen of high purity (99%, Alfa Aesar, Germany) as well as of trace elements and minerals, i.e., 10 mM NH4Cl, 0.2 mM NH4Fe(SO4)2·6H2O, 1 mM Na2HPO4, 5 mM KCl, 2 mM MgSO4·7H2O, 2 mM CaCl2·2H2O, 3 μM NiCl2·6H2O, 1 μM Na2SeO3, 3 μM CoCl2·6H2O, 3 μM NaMoO4·2H2O, 2 μM ZnSO4·7H2O, and 1 μM H3BO4 (Atlas 2010), was prepared to serve as the feeding substrate. The pH of the minimal medium was firstly adjusted to pH 8 and then to pH 7 in order to be converted to the ionic form of ibuprofen, a fact that increases the solubility of ibuprofen without the use of organic solvent, i.e., methanol, avoiding thus the addition of another carbon source, expect of ibuprofen. Moreover, no addition of yeast extract or any other organic compound was made when ibuprofen of high purity served as the sole carbonaceous feeding substrate. Both the synthetic wastewater consisting of commercial available ibuprofen tablets and the minimal wastewater containing ibuprofen of high purity were agitated in a magnetic stirrer prior to their addition to the immobilized cell bioreactor. After the acclimatization period where ibuprofen served as the sole carbonaceous feeding substrate, NH4Cl and Na2HPO4 concentrations were decreased to 4.67 mM and 0.35 mM, respectively, in order to reduce the residual ammonia and phosphorus concentrations in the effluent.
Physicochemical analyses
Dissolved oxygen (DO), pH, and electrical conductivity (EC) were determined by using the WTW Oxi 320, the Metrohm 632, and the Crison CM35 meters, respectively. Biochemical oxygen demand (BOD5), chemical oxygen demand (COD), and suspended solid (SS) concentrations were measured according to «Standard Methods for the Examination of Water and Wastewater» (Clesceri et al. 1998). Total Kjeldahl nitrogen (TKN) and ammonium nitrogen (NH4+-N) were estimated by following the Kjeldahl and ammonium distillation method, respectively. Nitrate and nitrite nitrogen (NO3−-Ν and ΝΟ2−-Ν) were measured colorimetrically after reducing nitrates to nitrites in a Cd-copperized column (Clesceri et al. 1998). PO43−-P was determined by reacting phosphates with ammonium molybdate reagent under acidic conditions and reducing the molybdophosphoric acid formed by stannous chloride to an intense molybdenum blue compound (Clesceri et al. 1998).
Determination of ibuprofen through derivatization and GC-MS analysis
Samples from the influent and the effluent were dissolved in methanol (1:100 v/v or 1:5 v/v sample/methanol, respectively), evaporated to dryness with nitrogen gas, and derivatized with a mixture of 100 μL pyridine and 100 μL BSTF+ 1% (trimethylchorosilane) TMCS at 60 °C for 30 min. Ibuprofen concentrations were determined by using a GCMS-QP2010 Plus gas chromatograph mass spectrophotometer (Shimadzu, Japan) through the use of a 5MS MEGA (Italy) chromatographic capillary column (60-m length, 0.25-mm i.d., 0.25-μm film thickness, 330–350 °C max temperature). A calibration curve was constructed by derivatizing ibuprofen standard solution and detecting the respective mass spectrum of the target compound. Helium gas flowed through the column under a rate of 2 mL/min. The operating parameters of the GCMS-QP2010 Plus instrument were set as follows: purge flow of 3 mL/min and injector temperature of 250 °C for GC, as well as ion source of 200 °C, interface temperature of 300 °C, detector voltage of 1 kV, solvent cut time of 3 min, and scan range of 40–500 amu for MS, respectively. The derivatized ibuprofen was eluted by employing a ramp program of 1-min holding period at 100 °C, temperature rise to 300 °C at a temperature increase rate of 8 °C/min, and a final holding step of 4 min at 300 °C.
Genomic DNA extraction, Illumina sequencing, and ecological statistics
Genomic DNA from the immobilized cells was extracted by grinding the Siran beads with a small plastic pestle and then using the Wizard Genomic DNA extraction kit (Promega), following manufacturer’s instructions. The primer set 27F (5′-AGR GTT TGATCM TGG CTC AG 3′) and 519R (5′-GTN TTA CNG CGG CKG CTG-3′) was employed to amplify the V1–V3 16S rRNA gene region. PCR reactions were performed by using the Qiagen’s HotStarTaq Plus master mix kit and following a thermocycling program, which was comprised of an initial denaturation step of 3 min at 94 °C, 28 cycles consisting of 30-s denaturation at 94 °C, 40-s annealing at 53 °C and 1-min DNA polymerization at 72 °C, and a final 5-min elongation step at 72 °C. Amplicons were purified by using the AMPure beads (Agencourt Bioscience, USA), prior to their analysis in a MiSeq instrument (Illumina, at “Mr DNA,” USA).
Both demultiplexing and trimming of the amplicons were performed in RDP pipeline by employing the “Pipeline Initial Process” option (Cole et al. 2014). Reads, with inappropriate length size, Ν(s), or quality score (Q) lower than 20, were excluded by the PandaSeq program (Masella et al. 2012). Detection of chimeras was performed at RDP by employing the UCHIME algorithm of USEARCH 6.0 (Edgar et al. 2011). The alignment of the non-chimeric reads was carried out by using the aligner tool of Nawrocki et al. (2009), and their clustering was performed by using the “Complete Linkage Clustering” option in RDP pipeline. A total of 344,856 non-chimeric Illumina reads, which derived from two experimental setups of three independent samples each, corresponding to the first and the second experimental periods where the immobilized cell bioreactor was operated with wastewater containing commercial available ibuprofen tablets and pure ibuprofen as the sole carbonaceous feeding substrate, respectively, were deposited in the SRA database under the “Bioproject” PRJNA474987 (SRA accession SRX4178968–SRX4178973). The microbial composition of the inoculum used was also deposited in the SRA database under the Bioproject PRJNA521586 (SRA accession SRX5354718–SRX5354720).
Dissimilarity matrices were constructed based on Bray and Curtis distance calculation (1957), and subsequently, the transformed data was subjected to multidimensional scaling (MDS), where reliable stress value was obtained on Kruskal’s test (Kruskal’s stress-1 = 6.7%) (Cox and Cox 2008). Ecological indicators, i.e., the Shannon diversity index H′ and the Shannon evenness index (SEI), were calculated as previously reported in Magurran (1988). Statistically significant differences were identified between indices or phylogenetic abundances in Student’s t test for 95% and 99% confidence intervals (*p < 0.05 or **p < 0.01). MicrobiomeAnalyst web platform was used to visualize correlation matrix and dendrogram (Dhariwal et al. 2017).
Results and discussion
Performance and optimization of the immobilized cell bioreactor fed with commercial ibuprofen tablets and pure ibuprofen
The bioreactor system was initially fed with synthetic wastewater containing commercial ibuprofen tablet of 600 mg/L without the addition of ammonium ions and yeast extract. However, system instability after the first month of operation resulted in lowered ibuprofen removal efficiency. Thus, the influent ibuprofen concentration was reduced to 400 mg/L, and ammonium chloride and yeast extract were added to facilitate microbial growth. As a consequence, higher biosystem removal efficiencies were recorded. Despite the fact that mechanical pump malfunction occurred during the experimental period from day 65 to day 75, system ibuprofen removal efficiency was stable, reaching values of 97.7 ± 2.8% (p < 0.01) (Fig. 1a). Then, a shift in the operating conditions of the immobilized cell bioreactor was performed by feeding the system with 400 mg/L of pure ibuprofen. After system adaptation to the new feeding conditions established (day 20 and thereafter), ibuprofen removal efficiencies reached values equal to 98.4 ± 0.3% (p < 0.01) (Fig. 1b). This indicates that the microbial communities in the immobilized cell bioreactor were effectively adapted to these severe loading rates of ibuprofen. Moreover, the detection of trimethylsilyl catechol puryvate tris (trimethylsilyl) denotes the presence of catechol 2-hydroxypropionate, which is a possible degradation intermediate of ibuprofen. To the best of our knowledge, this is the highest ibuprofen concentration effectively treated in any bioreactor system employed for the degradation of such pharmaceutical compound. Langenhoff et al. (2013) reported almost complete removal of 250 mg/L ibuprofen in batch experiments within a period of 4 days only when adapted sludge was used, whereas the removal efficiency was lower than 60% in the case of an unadapted biomass. This period of 4 days was similar to the operating HRT of the immobilized cell bioreactor (HRT of 3.5 day). On the other hand, the system instabilities occurred in the immobilized cell bioreactor that was fed with 600 mg/L ibuprofen indicate either insufficient HRT to deal with the extremely high ibuprofen concentration or ibuprofen-induced toxicity at dose application above 400 mg/L. Xia et al. (2014) showed that prolonged sludge retention time (SRT) and HRT can improve the removal of pharmaceuticals in bioreactor systems. In addition, the application of ibuprofen as the sole carbonaceous feeding substrate demonstrates the existence of a specialized ibuprofen-degrading bacterial community, denoting the advantages of activated sludge versatility.
Adjustment of ibuprofen concentration from 600 to 400 mg/L resulted in a COD decrease, i.e., from influent total COD (tCOD) of 1095 ± 20 mg/L to effluent tCOD and soluble (sCOD) values of 286 ± 25 and 185 ± 23 mg/L, respectively, corresponding to tCOD removal efficiencies of 73.7 ± 2.3% (Fig. 2a). Regarding effluent sCOD, the removal efficiency was estimated to be 83.2 ± 2.1% (Fig. 2a). In addition, the effluent tCOD and sCOD values and their respective removal efficiencies were equal to 166 ± 10 and 100 ± 7 mg/L as well as to 80.2 ± 2.2 and 88.6 ± 0.9%, when ibuprofen served as the sole carbonaceous feeding substrate (Fig. 2b). Under both feeding conditions, the high COD removal efficiencies were in agreement with the high ibuprofen removal values detected through chromatographic analysis of ibuprofen (Figs. 1 and 2). The residual effluent COD concentration determined could be mainly attributed to the release of secondary metabolites and cell debris rather to the ibuprofen concentration remained in the effluent, which was steadily below 2 mg/L.
Considering other operating parameters, influent pH was set at 6.42 ± 0.07 and the respective effluent pH values obtained were equal to 6.75 ± 0.04 when the immobilized cell bioreactor was fed with commercial ibuprofen tablets (Fig. 3a). Under this feeding condition, the electrical conductivity (EC) in the influent and the effluent were estimated as 322 ± 5 and 327 ± 10 μS/cm, respectively, indicating no statistically significant differences (Fig. 3a). By contrast, the immobilized cell bioreactor exhibited the tendency to acidify the treated liquor when pure ibuprofen served as the sole carbonaceous feeding substrate, resulting in a pH reduction from an initial influent pH of 6.94 ± 0.06 to an effluent pH value of 5.69 ± 0.08. Interestingly, Kimura et al. (2010) reported effective degradation of ibuprofen at pH 6 rather at neutral pH. Higher ibuprofen removal efficiencies were also reported at pH 5, when the activated sludge of a submerged MBR was subjected to a broad pH range of operation (within pH 5 and 9) (Tadkaew et al. 2010). At these moderate acidic conditions, ibuprofen mainly exists in hydrophobic speciation (non-ionic form), a fact that facilitates its sorption to biosolids (Urase and Kikuta 2005). Besides, hydrophobicity may enhance ibuprofen availability to the negatively charged membranes of various bacteria, increasing thus degradation rates. In comparison to the operating conditions where commercial ibuprofen tablets were used, the electrical conductivity values in the influent and effluent of the immobilized cell bioreactor that was fed with pure ibuprofen were much greater and equal to 3.04 ± 0.06 and 2.92 ± 0.06 mS/cm as the consequence of metal microelements’ addition.
The organic nitrogen was partially mineralized as indicated by the remaining effluent TKN concentration, while the ammonium nitrogen remained almost unoxidized (Table 1) since nitrates and nitrites were detected in negligible amounts in the effluent of the immobilized cell bioreactor under both feeding conditions examined. This could be attributed either to the acidic pH of the treated wastewater in the immobilized cell bioreactor (Zhang and Bishop 1996) and/or to the possible inhibitory effects of the high ibuprofen concentration to nitrifiers. Interestingly, Katsou et al. (2016) reported that nitrification process was inhibited by 80% when biomass spiked with ibuprofen concentrations above 250 mg/L in ex situ bioassays. The reduction in NH4+-N concentration, i.e., from 15.47 ± 3.32 to 7.69 ± 5.25 mg/L and from 84.58 ± 10.60 to 55.00 ± 14.19 mg/L, when the immobilized cell bioreactor was fed with commercial ibuprofen tablets and pure ibuprofen, respectively, appeared to be the consequence of ammonia assimilation in the immobilized microbial cells (Table 1). The concentration of the soluble phosphorus determined in the effluent under both feeding conditions indicated that adequate phosphorus concentration was provided to the immobilized microbial cells (Table 1).
Bacterial community structure in the immobilized cell bioreactor fed with commercial ibuprofen tablets and pure ibuprofen
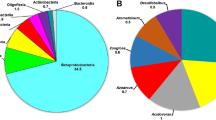
The dominant bacterial population in the Siran beads of the immobilized cell bioreactor was assessed by Illumina sequencing during treatment of synthetic wastewater containing commercial ibuprofen tablets. At genus level, the predominant bacterial taxon was placed within the genus Novosphingobium, which belongs to the same family as the members of the genus Sphigomonas, i.e., the family Sphigomonadaceae, representing 14.40 ± 3.98% of the relative abundance. Other major bacterial genera identified were those of Runella and Terrimonas, showing relative phylogenetic abundances of 7.21 ± 2.24 and 5.84 ± 0.41%, respectively. The prevalent bacterial taxa at phylum/class level in the immobilized cell bioreactor fed with commercial ibuprofen tablet were placed in Alphaproteobacteria and Bacteroidetes (Fig. 4), as a consequence of the high phylogenetic abundances of the genus Novosphingobium and the genera Runella and Terimonas, respectively.
The shift in the feeding conditions, when ibuprofen served as the sole carbonaceous feeding substrate, resulted in the predominance of the genera Novosphingobium and Rhodanobacter, which represented at this stage the 25.52 ± 10.77 and 25.15 ± 3.04% of the total reads, respectively, followed by strains of the genera Sphingomonas, Rhodopseudomonas, Pandoraea, and Dokdonella. On the other hand, Runella and Terrimonas, as well as members of the genera Crocinitomix, Ohtaekwangia, Alicycliphilus, Flavobacterium, Paracoccus, Bacillus, and Flavihumibacter, which were among the major microbiota when the bioreactor was fed with wastewater containing commercial ibuprofen tablets, were not further detected when ibuprofen served as the sole carbonaceous feeding substrate. This indicates a massive increase (from 29.06 to 80.80% of the total reads) in the relative abundances of Alphaproteobacteria and Gammaproteobacteria and a respective decrease in the proportion of Bacteroidetes, Planctomycetes, and Verrucomicrobia (from 42.65 to 2.05% of the total reads) (Fig. 4). In particular, the relative abundance of 29 taxa (each represented more than 0.1% of the total bacterial reads) reduced significantly (p < 0.01 or p < 0.05, in Student’s t test), while the respective number of taxa with increased abundance was 19 (Fig. 5). Estimation of ecological indicators showed a significant decrease (p < 0.05 in Student’s t test) in Shannon diversity index H′ when the shift from commercial ibuprofen tablets to pure ibuprofen occurred, resulting in the reduction of H′ indicator from 3.85 ± 0.07 to 2.92 ± 0.28. This shift also affected Shannon evenness index (SEI), reducing community evenness after feeding shift from 0.66 ± 0.01 to 0.50 ± 0.05 (p < 0.05 in Student’s t test). In addition, multidimensional scaling of Illumina data showed that the district bacterial community structures favored in the immobilized cell bioreactor under the two feeding conditions examined (commercial ibuprofen tablets vs pure ibuprofen) (Supplementary Material Fig. S2). The predominance of Novosphingobium under both feeding conditions could be attributed to its ability to catabolize ibuprofen as the sole carbon and energy source, although the growth of the remaining dominant taxa identified in the immobilized cell bioreactor that was fed with wastewater consisting of ibuprofen tablets appeared to be relied mainly on excipient consumption (e.g., each ibuprofen tablet of 400 mg contains 297 mg sucrose as well as starch, stearate, etc.) since these microbita disappeared or were minor constituents of microbial community structure when pure ibuprofen served as the feeding substrate. Based on the current and previous studies, specialized microbiota of the family Sphingomonadaceae like Novosphingobium and Sphingomonas spp., which are capable of degrading NSAIDs, appeared to gain an adaptive advantage (Murdoch and Hay 2005; Iasur-Kruh et al. 2011). A positive relationship among the families Sphingobacteriaceae, Pseudomonadaceae, and Rhodobacteraceae was found (Fig. 6), which is also indicative of the degradation ability of these taxa (Nojiri et al. 2014). Moreover, Rhodobacteraceae spp., which are effective degraders of recalcitrant compounds (Nalin et al. 1999), positively correlated with Nakamurellaceae representatives.
Illustration of bacterial taxa that their abundances (denoted inside the grey squares) were significantly increased or decreased (*p < 0.05 or **p < 0.01, in Student’s t test), when the shift from commercial ibuprofen tablets to pure ibuprofen as the feeding substrate occurred. Only bacterial taxa, which their relative abundances were greater than 0.1%, at least in one of the two feeding conditions examined, and statistically differed, are included in this figure. A shift from pale to dark grey squares corresponds to an increase in the relative abundance of a taxon and vice versa. CIT commercial ibuprofen tablets, PIS pure ibuprofen solution
Under both feeding conditions examined, the microbial community structure identified was distinct from the dominant diversity detected in treatment systems where low doses of ibuprofen had been applied. In these cases, despite the occurred differences after addition of low ibuprofen concentrations (up to 0.5 mg/L), the major bacterial diversity was the typical diversity detected in wastewater treatment plants, showing the dominance of Betaproteobacteria, consisting mainly of Thauera, Sphaerotilus, and Acidovorax spp. (Kraigher et al. 2008; Muter et al. 2017). It appears that the ibuprofen degraders comprise of a minor part of the microbial diversity in these systems, while ibuprofen may exert toxic effects only on certain taxa. However, the high ibuprofen concentration applied in this study was adequate to sustain sufficient growth of ibuprofen-degrading bacteria and permitted to identify the ibuprofen-degrading microbiota existing in the activated sludge. The increase in cyanobacterial reads may be attributed to the increased electrical conductivity that was caused by the addition of micronutrient solution when ibuprofen served as the sole carbonaceous feeding substrate. On the other hand, increased pharmaceutical concentrations, including that of ibuprofen, have been reported to sustain or even increase cyanobacterial population (Proia et al. 2013; Corcoll et al. 2014; Bácsi et al. 2016). Based on Illumina sequencing, the restricted nitrifying population identified in the immobilized cell bioreactor that was fed with wastewater consisting of commercial ibuprofen tablets was comprised of Nitrosovibrio and Nitrospira spp., as the main ammonia and nitrite oxidizers, respectively. On the other hand, Nitrosospira rather than Nitrosovibrio was the main ammonia-oxidizing taxon, while Nitrospira still remained as the predominant nitrite oxidizer when ibuprofen served as the sole carbonaceous feeding substrate. Negligible proportion of the nitrite-oxidizing genus Nitrobacter was also detected under both feeding conditions. The limited proportion of ammonia-oxidizing bacteria supports the hypothesis of Tran et al. (2013) that biodegradation of ibuprofen can be almost exclusively carried out by heterotrophs.
Apart from Sphingomonadaceae-affiliated taxa and the autotrophic Nitrospira, which appears to have a persistent presence in activated sludge systems under elevated NSAID concentrations (Wang et al. 2016; Vuono et al. 2016), Nakamurella population was also detected at low abundances when a mixture of diclofenac, ibuprofen, and naproxen was added in the activated sludge of a SBR system (Jiang et al. 2017). Lipases, which were related with those of Thermus spp. and were capable of hydrolyzing ibuprofen-phenyl ester, have been also detected in a metagenome library (Chow et al. 2012). Moreover, Propionicimonas has been identified in a MBR performing the post-treatment of the secondary effluent of a full-scale WWTP (Arriaga et al. 2016). Recently, a Pseudomonas strain isolated from a sewage plant in China was found to be capable of degrading ibuprofen (Li et al. 2015). No previous report on ibuprofen degradation for the remaining major taxa detected under both feeding conditions examined exists in the international literature regarding any natural and artificial ibuprofen-polluted environment.
Conclusions
The application of ibuprofen as the sole carbonaceous feeding substrate indicated that a specific part of activated sludge consisted of specialized ibuprofen degraders, which possessed the selective advantage to effectively degrade NSAIDs. Such specialized ibuprofen degraders were mainly members of the family Sphingomonadaceae, like Sphingomonas and Novosphingobium spp., which were capable of catabolizing ibuprofen. Inhibition of the nitrification process occurred, due to the tendency of the immobilized biomass to acidify the effluent and/or the potent toxic effects of the high ibuprofen concentration onto the nitrifying bacteria.
References
Arriaga S, Jonge N, Nielsen ML, Andersen HR, Borregaard V, Jewel K, Ternes TA, Nielsen JL (2016) Evaluation of a membrane bioreactor system as post-treatment in waste water treatment for better removal of micropollutants. Water Res 107:37–46
Atlas RM (2010) Handbook of microbiological media, 4th edn. CRC Press, Taylor & Francis group, Boca Raton, Florida, p 63
Bácsi I, B-Béres V, Kókai Z, Gonda S, Novák Z, Nagy SA, Vasas G (2016) Effects of non-steroidal anti-inflammatory drugs on cyanobacteria and algae in laboratory strains and in natural algal assemblages. Environ Poll 212:508–518
Bray RJ, Curtis JT (1957) An ordination of the upland forest communities of southern Wisconsin. Ecol Monogr 27(4):325–349
Candido JP, Andrade SJ, Fonseca AL, Silva FS, Silva MRA, Kondo MM (2016) Ibuprofen removal by heterogeneous photocatalysis and ecotoxicological evaluation of the treated solutions. Environ Sci Poll Res 23(19):19911–19920
Chow J, Kovacic F, Dall Antonia Y, Krauss U, Fersini F, Schmeisser C, Lauinger B, Bongen P, Pietruszka J, Schmidt M, Menyes I, Bornscheuer UT, Eckstein M, Thum O, Liese A, Mueller-Dieckmann J, Jaeger KE, Streit WR (2012) The metagenome-derived enzymes LipS and LipT increase the diversity of known lipases. PLoS One 7(10):e47665
Clesceri LS, Greenberg AE, Eaton AD (1998) Standard methods for the examination of water and wastewater, 20th edn. American Public Health Association (APHA), Washington DC
Cole JR, Wang Q, Fish JA, Chai B, McGarrell DM, Sun Y, Brown CT, Porras-Alfaro A, Kuske CR, Tiedje JM (2014) Ribosomal database project: data and tools for high throughput rRNA analysis. Nucleic Acids Res 42(D1):D633–D642
Corcoll N, Acuña V, Barceló D, Casellas M, Guasch H, Huerta B, Petrovic M, Ponsatí L, Rodríguez-Mozaz S, Sabater S (2014) Pollution-induced community tolerance to non-steroidal anti-inflammatory drugs (NSAIDs) in fluvial biofilm communities affected by WWTP effluents. Chemosphere 112:185–193
Cox MAA, Cox TF (2008) Multidimensional scaling. In: Chen C-H, Hardle W, Unwin A (eds) Handbook of data visualization. Springer, New York, pp 315–347
Davarnejad R, Soofi B, Farghadani F, Behfar R (2018) Ibuprofen removal from a medicinal effluent: a review on the various techniques for medicinal effluents treatment. Environ Technol Innov 11:308–320
Dhariwal A, Chong J, Habib S, King I, Agellon LB, Xia J (2017) MicrobiomeAnalyst—a web-based tool for comprehensive statistical, visual and meta-analysis of microbiome data. Nucleic Acids Res 45(W1):W180–W188
Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R (2011) UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27(16):2194–2200
Fortunato MS, Fuentes Abril NP, Martinefski M, Trípodi V, Papalia M, Rádice M, Gutkind G, Gallego A, Korola SE (2016) Aerobic degradation of ibuprofen in batch and continuous reactors by an indigenous bacterial community. Environ Technol 37(20):2617–2626
Iasur-Kruh L, Hadar Y, Minz D (2011) Isolation and bioaugmentation of an estradiol-degrading bacterium and its integration into a mature biofilm. Appl Environ Microbiol 77(11):3734–3740
Islas-Flores H, Manuel Gómez-Oliván L, Galar-Martínez M, Michelle Sánchez-Ocampo E, SanJuan-Reyes N, Ortíz-Reynoso M, Dublán-García O (2017) Cyto-genotoxicity and oxidative stress in common carp (Cyprinus carpio) exposed to a mixture of ibuprofen and diclofenac. Environ Toxicol 32(5):1637–1650
Jallouli N, Pastrana-Martínez LM, Ribeiro AR, Moreira NFF, Faria JL, Hentati O, Silva AMT, Ksibi M (2018) Heterogeneous photocatalytic degradation of ibuprofen in ultrapure water, municipal and pharmaceutical industry wastewaters using a TiO2/UV-LED system. Chem Eng J 334:976–984
Jeffries KM, Brander SM, Britton MT, Fangue NA, Connon RE (2015) Chronic exposures to low and high concentrations of ibuprofen elicit different gene response patterns in a euryhaline fish. Environ Sci Poll Res 22(22):17397–17413
Jiang C, Geng J, Hu H, Ma H, Gao X, Ren H (2017) Impact of selected non-steroidal anti-inflammatory pharmaceuticals on microbial community assembly and activity in sequencing batch reactors. PLoS One 12(6):e0179236
Katsou E, Alvarino T, Malamis S, Suarez S, Frison N, Omil F, Fatone F (2016) Effects of selected pharmaceuticals on nitrogen and phosphorus removal bioprocesses. Chem Eng J 295:509–517
Kimura K, Hara H, Watanabe Y (2010) Elimination of selected pharmaceuticals by biosolids from municipal wastewater treatment plants: importance of modest pH change and degree of mineralization. Water Sci Technol 62(5):1084–1089
Kraigher B, Kosjek T, Heath E, Kompare B, Mandic-Mulec I (2008) Influence of pharmaceutical residues on the structure of activated sludge bacterial communities in wastewater treatment bioreactors. Water Res 42(17):4578–4588
Langenhoff A, Inderfurth N, Veuskens T, Schraa G, Blokland M, Kujawa-Roeleveld K, Rijnaarts H (2013) Microbial removal of the pharmaceutical compounds ibuprofen and diclofenac from wastewater. BioMed Res Int. https://doi.org/10.1155/2013/325806
Li WC (2014) Occurrence, sources, and fate of pharmaceuticals in aquatic environment and soil. Environ Poll 187:193–201
Li Z, Cheng K, Teng X, Ma X, Hou Y, Wei Y (2015) Degradation spectrum of ibuprofen degrading strains. Acta Sci Circumst 35(1):177–183
Magurran AE (1988) Ecological diversity and its measurement. Princeton University Press, Princeton
Masella AP, Bartram AK, Truszkowski JM, Brown DG, Neufeld JD (2012) PANDAseq: paired-end assembler for illumina sequences. BMC Bioinformatics 13(31). https://doi.org/10.1186/1471-2105-13-31
Moro I, Matozzo V, Piovan A, Moschin E, Dalla Vecchia F (2014) Morpho-physiological effects of ibuprofen on Scenedesmus rubescens. Environ Toxicol Pharmacol 38(2):379–387
Murdoch RW, Hay AG (2005) Formation of catechols via removal of acid side chains from ibuprofen and related aromatic acids. Appl Environ Microbiol 71(10):6121–6125
Murdoch RW, Hay AG (2015) The biotransformation of ibuprofen to trihydroxyibuprofen in activated sludge and by Variovorax Ibu-1. Biodegradation 26(2):105–113
Muter O, Perkons I, Selga T, Berzins A, Gudra D, Radovica-Spalvina I, Fridmanis D, Bartkevics V (2017) Removal of pharmaceuticals from municipal wastewaters at laboratory scale by treatment with activated sludge and biostimulation. Sci Total Environ 584-585:402–413
Nalin R, Simonet P, Vogel TM, Normand P (1999) Rhodanobacter lindaniclasticus gen. nov., sp. nov., a lindane-degrading bacterium. Int J Syst Evol Microbiol 49(1):19–23
Nawrocki EP, Kolbe DL, Eddy SR (2009) Infernal 1.0: inference of RNA alignments. Bioinformatics 25(10):1335–1337
Nojiri H, Tsuda M, Fukuda M, Kamagata Y (2014) Biodegradative bacteria—how bacteria degrade, survive, adapt, and evolve. Springer, Tokyo
Proia L, Osorio V, Soley S, Köck-Schulmeyer M, Pérez S, Barceló D, Romaní AM, Sabater S (2013) Effects of pesticides and pharmaceuticals on biofilms in a highly impacted river. Environ Poll 178:220–228
Ragugnetti M, Adams ML, Guimarães ATB, Sponchiado G, Carvalho de Vasconcelos E, Ribas de Oliveira CM (2011) Ibuprofen genotoxicity in aquatic environment: an experimental model using Oreochromis niloticus. Water Air Soil Poll 218(1–4):361–364
Rainsford KD (2009) Ibuprofen: pharmacology, efficacy and safety. Inflammopharmacology 17(6):275–342
Tadkaew N, Sivakumar M, Khan SJ, McDonald JA, Nghiem LD (2010) Effect of mixed liquor pH on the removal of trace organic contaminants in a membrane bioreactor. Bioresour Technol 101(5):1494–1500
Tran NH, Urase T, Ngo HH, Hu J, Ong SL (2013) Insight into metabolic and cometabolic activities of autotrophic and heterotrophic microorganisms in the biodegradation of emerging trace organic contaminants. Bioresour Technol 146:721–731
Urasa T, Kikuta T (2005) Separate estimation of adsorption and degradation of pharmaceutical substances and estrogens in the activated sludge process. Water Res 39(7):1289–1300
Vuono DC, Regnery J, Li D, Jones ZL, Holloway RW, Drewes JE (2016) rRNA gene expression of abundant and rare activated-sludge microorganisms and growth rate induced micropollutant removal. Environ Sci Technol 50(12):6299–6309
Wang X-C, Shen J-M, Chen Z-L, Zhao X, Xu H (2016) Removal of pharmaceuticals from synthetic wastewater in an aerobic granular sludge membrane bioreactor and determination of the bioreactor microbial diversity. Appl Microbiol Biotechnol 100(18):8213–8223
Xia Z, Xiao-chun W, Zhong-lin C, Hao X, Qing-fang Z (2014) Microbial community structure and pharmaceuticals and personal care products removal in a membrane bioreactor seeded with aerobic granular sludge. Appl Microbiol Biotechnol 99(1):425–433
Zhang TC, Bishop PL (1996) Evaluation of substrate and pH effects in a nitrifying biofilm. Water Environ Res 68(7):1107–1115
Zhou JL, Zhang ZL, Banks E, Grover D, Jiang JQ (2009) Pharmaceutical residues in wastewater treatment works effluents and their impact on receiving river water. J Hazard Mater 166(2–3):655–661
Zhou NA, Lutovsky AC, Andaker GL, Gough HL, Ferguson JF (2013) Cultivation and characterization of bacterial isolates capable of degrading pharmaceutical and personal care products for improved removal in activated sludge wastewater treatment. Biodegradation 24(6):813–827
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Responsible editor: Gerald Thouand
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Navrozidou, E., Melidis, P. & Ntougias, S. Biodegradation aspects of ibuprofen and identification of ibuprofen-degrading microbiota in an immobilized cell bioreactor. Environ Sci Pollut Res 26, 14238–14249 (2019). https://doi.org/10.1007/s11356-019-04771-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11356-019-04771-5