Abstract
Ibuprofen is one of the most commonly detected pharmaceuticals in wastewater effluent; however, the effects of ibuprofen on aquatic organisms are poorly understood. This study presents the transcriptome-wide response of the inland silverside, Menidia beryllina, to chronic exposure to ibuprofen. At the lowest exposure concentration (0.0115 mg/L), we detected a downregulation of many genes involved in skeletal development, aerobic respiration, and immune function. At the highest exposure concentration (1.15 mg/L), we detected increased expression of regulatory genes in the arachidonic acid metabolism pathway and several immune genes involved in an inflammatory response. Additionally, there was differential expression of genes involved in oxidative stress responses and a downregulation of genes involved in osmoregulation. This study provides useful information for monitoring the effects of this common wastewater effluent contaminant in the environment and for the generation of biomarkers of exposure to ibuprofen that may be transferable to other fish species.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Municipal wastewater effluents are major point sources of numerous industrial contaminants, personal care products, hormones, and pharmaceuticals to aquatic systems (Kolpin et al. 2002; Metcalfe et al. 2003; Fernandez et al. 2007; Jeffries et al. 2010; reviewed in Brander 2013; Yu et al. 2013). Many of the compounds introduced to the environment via wastewater effluent have unknown or poorly understood consequences on aquatic organisms. Nonsteroidal anti-inflammatory drugs (NSAIDs) are a class of widely used pharmaceuticals that are routinely detected in wastewater effluent (ng/L to μg/L range; Metcalfe et al. 2003; reviewed in Fent et al. 2006; Yu et al. 2013) and receiving waters (pg/L to ng/L range; Kolpin et al. 2002; Brozinski et al. 2013), with detections reaching up to 5 μg/L at sites downstream from wastewater effluent discharge points (Ashton et al. 2004). Ibuprofen is one of the most commonly detected NSAIDs in the environment due to its high consumption (reviewed in Fent et al. 2006) and because much of the dose is unmetabolized (Pounds et al. 2008). Ibuprofen is not very persistent in the environment (Buser et al. 1999); however, its continual reintroduction into aquatic systems allows it to exist in a “pseudo-persistent” state. The concentrations of ibuprofen in water samples may be misleading as unmetabolized ibuprofen levels in wild fish plasma and bile samples can be 100 to 1000 times higher, respectively, than those found in surrounding water samples (Brozinski et al. 2013). Therefore, understanding how aquatic organisms respond to chronic exposure to low concentrations of ibuprofen in systems that receive wastewater effluent is critical for determining the consequences of ibuprofen in the environment.
Ibuprofen is a nonselective cyclooxygenase (COX) inhibitor that causes decreased production of prostaglandins, a group of compounds derived from arachidonic acid metabolism that contribute to inflammation, to produce the desired anti-inflammatory effect. The COX enzymes involved in prostaglandin synthesis, PTGS1 and PTGS2 (prostaglandin G H synthase 1 and 2, respectively; PTGS2 is in vertebrates only), also known as COX1 and COX2, are relatively evolutionarily conserved, and therefore, the effects of ibuprofen exposure have been detected in many diverse aquatic organisms, such as cladocerans (Heckmann et al. 2008; Han et al. 2010), molluscs (Pounds et al. 2008; Milan et al. 2013), and several fish species (David and Pancharatna 2009; Gravel et al. 2009; Bhandari and Venables 2011; Nallani et al. 2011; Morthorst et al. 2013; Ji et al. 2013; Islas-Flores et al. 2014). Consequently, understanding the effects of ibuprofen on aquatic life has gained significant research attention in recent years. Ibuprofen has been shown to reduce prostaglandin levels in fishes [e.g., bluntnose minnow (Pimephales notatus; Bhandari and Venables 2011) and zebrafish (Danio rerio; Morthorst et al. 2013)]. Prostaglandins also influence steroid synthesis in fishes and act as pheromones (reviewed in Munakata and Kobayashi 2010). Therefore, a reduction in prostaglandin production as a result of exposure to ibuprofen can affect the synthesis of hormones involved in reproduction (Ji et al. 2013); however, this may be dose- and species-dependent (Morthorst et al. 2013). An ibuprofen-induced reduction in reproductive hormones has been linked with embryonic developmental issues and delayed hatching, as well as reduced egg production in spawning adult zebrafish (David and Pancharatna 2009; Ji et al. 2013) and decreased numbers of spawning events in medaka (Oryzias latipes; Flippin et al. 2007). Ibuprofen exposure may also influence the synthesis of cortisol, a multipurpose hormone involved in many physiological processes that include stress responses and salinity tolerance. An ibuprofen-induced alteration in cortisol production was hypothesized to have contributed to an inhibition of N+/K+-ATPase activity in rainbow trout (Oncorhynchus mykiss; Gravel et al. 2009), an effect that could lead to reduced salinity tolerance in fishes. Although studies exist that demonstrate the effects of ibuprofen exposure on reproduction and osmoregulation in fishes, to our knowledge, a transcriptome-wide assessment on the effects of ibuprofen on fishes is lacking.
The goal of the present study was to examine the transcriptome-wide effects of exposure to ibuprofen on an environmentally relevant nonmodel fish species, the inland silverside (Menidia beryllina). Silverside species (Menidia spp.) are highly abundant in estuaries throughout North America (Moyle 2002) and have been suggested to be useful bioindicator species in estuaries and coastal systems (Doyle et al. 2011; Brander et al. 2013). The inland silverside is also a commercially available Menidia species used in whole effluent toxicity testing by the United States Environmental Protection Agency (http://water.epa.gov/). Although an invasive species in California, USA, the inland silverside habitat overlaps with many native pelagic fishes that have been listed as threatened or endangered (Moyle 2002), such as the critically endangered delta smelt (Hypomesus transpacificus). Therefore, the inland silverside may be a particularly useful, environmentally relevant bioindicator species of exposure to contaminants in California surface waters.
Genomics tools are valuable for identifying cellular responses in toxicological studies (Ankley et al. 2006; Villeneuve et al. 2011; Connon et al. 2012) and are increasingly being used in the study of nonmodel fishes (e.g., Connon et al. 2009, 2011; Jeffries et al. 2012, 2014). We developed a M. beryllina 4x44K probe oligonucleotide microarray and used it to examine the chronic effects of ibuprofen in an environmentally relevant fish species in North America. Ibuprofen is regularly detected in aquatic systems and has recently been identified as a “contaminant of emerging concern” in California (Maruya et al. 2014). We also examined the expression of specific genes involved in osmoregulatory function and prostaglandin synthesis, known to be affected by exposure to ibuprofen, to further identify some of the cellular responses in inland silversides. To the best of our knowledge, this is the first transcriptome-wide assessment of the effects of ibuprofen on a nonmodel fish.
Materials and methods
Ibuprofen exposures
Inland silversides were ordered from Aquatic Biosystems (Ft. Collins, CO, USA) and were allowed 4 days to acclimatize to holding conditions at the Aquatic Toxicology Laboratory at the University of California Davis. Fish were fed ad libitum with Artemia franciscana throughout the acclimatization and exposure period. Acute toxicity tests were conducted over 96-h exposures with nine nominal exposure concentrations ranging from 1.2 to 300 mg/L ibuprofen, along with a control. The actual exposure concentration was measured for two of the exposures, and the effective concentration was then estimated for the other treatment groups and used to determine the 10, 50, and 90 % lethal concentrations (LCs) for M. beryllina. Analytical chemistry resulted in a recovery of 56 %; thus, concentrations are presented as measured values. Reduced ibuprofen recovery, due to adsorption to glass, has previously been reported (Nallani et al. 2011). A nonlinear regression curve was fitted to the cumulative mortality data using a Weibull distribution, and the fit was confirmed by a goodness of fit test. A three-parameter regression model was fitted assuming a lower limit of 0.
Transcriptome profiling was used to assess the chronic effects of ibuprofen exposure. For this, another subset of fish was exposed for 14 days to nominal concentrations of 0.025, 0.25, and 2.5 mg/L (which will subsequently be referred to as the low, medium, and high concentrations, respectively), along with a control group handled identically to the other treatments. The highest concentration was analyzed for the actual ibuprofen level, and recovery was 46 % of the 2.5-mg/L nominal concentration. Therefore, the measured concentration was 1.15 mg/L for the high treatment, followed by effective concentrations for the low and medium treatments estimated to be 0.0115 and 0.115 mg/L, respectively. At the termination of the chronic 14-day exposure period, fish were sacrificed by a lethal dose of tricaine methanesulfonate (MS-222), and whole fish were immediately frozen in liquid nitrogen and stored at −80 °C until analysis.
For both the acute (n = 10 fish/container) and chronic exposures (n = 8 fish/container), there were four replicate 2-L containers for each treatment. All exposures were conducted in 1.5 L of freshwater on fish that were 66 days posthatch (dph) at the initiation of the experiment. Water was changed (75 % renewal), and the nominal exposure concentrations were renewed daily. Tanks were monitored for temperature (19.6 °C ± SD 0.9), electrical conductivity (729.4 μS/cm ± SD 21.5), dissolved oxygen (8.84 mg/L ± SD 0.4), total ammonia as nitrogen (0.23 mg/L ± SD 0.07), and pH (7.58 ± SD 0.3). There were no differences in these parameters between the treatment tanks. This research was approved by the University of California Davis Institutional Animal Care and Use Committee (IACUC #16845).
Microarray development
We generated the data to construct an oligonucleotide microarray (Agilent technologies) using RNA from tissues and whole body homogenates from laboratory-reared M. beryllina (Aquatic Biosystems) that were exposed to a number of physiological (temperature, salinity, handling) and chemical stressors (ibuprofen, bifenthrin), as well as wild-caught individuals, in order to capture a diverse suite of mRNA transcripts. Total RNA was extracted using an RNeasy kit (Qiagen) with on-column DNAse digestion following manufacturer’s guidelines. Total RNA integrity was verified using an Agilent Technologies 2100 Bioanalyzer that determined that the samples were high quality [RNA integrity numbers (RIN) values >9]. Samples were pooled to represent an equal proportion of RNA per tissue and whole organisms (larvae/juveniles) to normalize and maximize the identification of rare transcripts. Transcriptome sequencing was conducted at the University of California, Davis Genome Center DNA Technologies Core Facility. A library was constructed using a TruSeq RNA Sample Prep kit (Illumina) with poly-A selection. The library was sequenced on an Illumina HiSeq2000 sequencer, using a 100-bp paired-end read protocol, to generate 183 million pairs of reads.
Sequencing data were processed in collaboration with the University of California, Davis Genome Center Bioinformatics Core Facility. Raw reads were trimmed to remove adapter contamination and low-quality sequences using Scythe and Sickle (https://github.com/ucdavis-bioinformatics). The trimmed reads were used for a de novo transcriptome assembly with Trinity version r2013-02-16 (Grabherr et al. 2011; Haas et al. 2013), which included read normalization, open reading frame (ORF) predition, and isoform abundance filtering steps. Annotation was conducted by Basic Local Alignment Search Tool (BLAST) comparisons of all contigs against a custom subset of the nr database representing all teleost species using ncbi-blast-2.2.28+ (Altschul et al. 1997). The BLAST results were consolidated, and Gene Ontology (GO) terms were assigned using Blast2GO (Conesa et al. 2005). Additional BLASTs were made between the M. beryllina transcriptome assembly and the Ensembl protein sets for medaka and zebrafish. Custom microarrays GE 4x44K, designed utilizing Agilent specific software (https://earray.chem.agilent.com/earray/), consisted of 14,393 unique M. beryllina genes and nonspecific Agilent control probes in triplicate. Each gene was printed with three different overlapping probes.
RNA extraction and microarray analysis
Whole fish from the chronic 14-day exposures were homogenized using a pestle in 1 mL of Trizol reagent following manufacturer’s protocols. Concentration and purity (260/280, 260/230) were assessed using a Nanodrop spectrophotometer (Thermo Scientific), and RNA integrity was determined through electrophoresis. Two hundred nanograms of total RNA was amplified and labeled with Cy3 using the Low Input Quick Amp Labeling kit (Agilent Technologies). A total of 24 single color microarray hybridizations (6 fish per treatment) were performed with the custom-designed 4x44K oligonucleotide microarrays using Agilent Gene Expression Hybridization Kits (Agilent Technologies). To minimize technical artefacts, labeling reactions were performed simultaneously, and individuals from each treatment were randomized between slides. Prior to hybridization, 1.65 μg of dye-labeled complementary RNA for each sample was combined with 2.2 μL of 25× fragmentation buffer, 11 μL of 10× Gene Expression Blocking Agent, and nuclease-free water to bring the final volume to 55 μL. The fragmentation mix was incubated at 60 °C for 30 min, cooled on ice for 1 min, and the fragmentation was stopped by adding 55 μL of 2× Hi-RPM hybridization buffer. Samples were centrifuged for 1 min, placed on ice, and 100 μL of the mix was loaded onto arrays, and each microarray slide was placed in a single hybridization chamber. Samples were hybridized to the arrays for 17 h at 65 °C, followed by a wash with Gene Expression Wash Buffer 1 at room temperature and a wash with Gene Expression Wash Buffer 2 heated to 37 °C. Fluorescent images were scanned using an Axon GenePix 4000B Scanner and the analysis software GenePix Pro (Molecular Devices, LLC, Sunnyvale, CA). The images were quantified using Feature Extraction 11.5.1.1 (Agilent Technologies).
Normalization of the microarray data and statistical analysis was performed in Genespring v12.6 (Agilent Technologies). Data was quantile-normalized and log2-transformed prior to statistical analysis. Probes that had a raw signal less than 80 (~2 times the intensity of the background) were filtered out of the statistical analysis to eliminate sources of background noise, but still retain probes with low intensity (Sifakis et al. 2012). After the filtering step, there were 8789 probes remaining to be used for the statistical comparisons. Differences in the expression of probes between treatment groups were assessed by ANOVA followed by Student-Newman-Keuls (SNK) post hoc tests at α = 0.01. It is important to note that there are three different overlapping probes for the same gene that were treated as separate genes in the statistical analyses. Therefore, multiple copies of each gene that were significant in our analyses were used to validate the results and identify true patterns from background noise in the data. The microarray data have been deposited in the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) with the accession number GSE64967.
Functional analysis was performed using Blast2GO. Due to a relatively low number of significant genes detected in the ANOVA, functional enrichment analysis was performed on significant gene lists generated by separate t tests between each exposure treatment and the control group (α = 0.05). Redundancies in the GO term lists were reduced using ReVigo (Supek et al. 2011). Functional groups were considered significantly enriched in gene lists using Fisher exact tests at a FDR corrected α = 0.001.
cDNA synthesis and qPCR
We selected 28 genes to examine responses in greater scrutiny by qPCR (Table 1; n = 20 per treatment that included the same individuals from the microarray analysis), for microarray validation, and to examine the changes in osmoregulatory function and arachidonic acid metabolism expected to be affected by exposure to ibuprofen. Primers were designed using the Universal Probe Library Assay Design Center (Roche). Primer sequences and Roche probe numbers are available in Table 1.
One microgram of total RNA was used for cDNA synthesis. For each sample, nuclease-free water was added to bring the total volume up to 12 μL followed by the addition of 2 μL of gDNA wipeout buffer (Qiagen) and an incubation at 42 °C for 2 min. Each cDNA synthesis reaction consisted of 4-μL 5× Quantiscript RT Buffer, 1-μL RT Primer Mix, 1-μL Quantiscript RT (Qiagen), and 14 μL of diluted sample for a total reaction volume of 20 μL. Reactions were carried out at 42 °C for 30 min, followed by 95 °C for 3 min, and an end hold at 4 °C. The cDNA was stored at −20 °C until qPCR analysis.
A 1:6 dilution of cDNA was used as a template in the qPCR reactions. Assays were performed on an automated fluorometer (ABI HT 7900 A FAST Sequence Detection System, Applied Biosystems) in 384-well plates using 12-μL reaction volumes composed of 6 μL of PCR Master Mix (Qiagen), 0.6-μL primer probe mix (forward primer, reverse primer, the appropriate fluorescent probe, and nuclease-free water), 2.4 μL of nuclease-free water, and 3 μL of diluted cDNA. Cycling conditions were 2 min at 50 °C, 10 min at 95 °C, 40 cycles of 15 s at 95 °C, and 60 s at 60 °C, and amplifications were analyzed using SDS 2.4 software (Applied Biosystems). Relative expression of target genes was determined using the 2−ΔΔCt method and are presented as relative fold change (Livak and Schmittgen 2001). Target gene expression was normalized to four reference genes: beta-actin (actin, cytoplasmic; ACTB), elongation factor 1-alpha-like (EEF1A), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), and 60s ribosomal protein l7 (RPL7). Stability of the reference genes across experimental treatments was assessed using the program geNorm (Vandesompele et al. 2002). Statistical differences were determined via two different approaches. First, individual treatments were compared using one-factor ANOVAs on log2-scaled ΔΔCt’s, followed by SNK post hoc tests at α = 0.05. We then fit dose-response curves using maximum likelihood estimates, similar to previous toxicological studies (Brander et al. 2009; Brander et al. 2012) that fit two different curves to each gene’s dataset: a sigmoidal curve (four parameter logistic model) and a nonmonotonic curve (a modified four parameter logistic model; Brain and Cousens 1989). We used a likelihood ratio test (LRT) with a significance level of α = 0.05 (Bolker 2008) to test whether each model fit significantly better than a flat line (i.e., no dose-response) and whether the nonmonotonic curve afforded a significantly better fit than the sigmoidal curve. Curves were fit by maximum likelihood using the mle2 algorithm (Bolker 2008) in R 3.1.1 (R Core Development Team 2010). All curves were fit to log10(x + 0.025) transformed concentration data to avoid taking the log of concentration 0 (control).
Results
The LC10, LC50, and LC90 of ibuprofen over 96-h exposures were estimated to be 17.0 (SE ± 0.59), 34.35 (SE ± 0.40), and 53.77 mg/L (SE ± 1.06), respectively, for M. beryllina. There was no mortality observed at any of the ibuprofen concentrations in the 14-day chronic exposures used in the transcriptomic assessments.
Microarray and functional analysis results
At the highest exposure concentration, genes associated with functional categories involved in proteolysis (e.g., catalytic activity), metabolism (e.g., metabolic process), and cell structure (e.g., intracellular part) were differentially expressed in the 387 significant gene list (665 microarray probes; t test, P < 0.05; Table 2). Many genes associated with aerobic respiration and protein synthesis [e.g., oxidative phosphorylation, respiratory electron transport chain, and NADH dehydrogenase (ubiquinone) activity; ribonucleoprotein complex biogenesis, ribosome, and translation] were differentially expressed in the 481 and 525 (828 and 913 microarray probes) significant gene lists (t test, P < 0.05) for the medium and low concentration treatments, respectively. Similar to the high treatment, genes associated with cell structure (e.g., structural molecule activity) were differentially expressed at the lowest exposure concentration.
Overall responses to the ibuprofen exposures were relatively subtle, as only 57 different genes (96 microarray probes; see Table 3) were statistically significant across all treatments (ANOVA, significant at P < 0.01; Fig. 1). The relative fold change differences between each treatment group and the control for the 96 probes are presented in Table S1. There were two distinct responses occurring as 59.4 % of the genes significantly differing between the high exposure concentration, and control groups were upregulated, compared with only 6.7 and 8.0 % significantly upregulated in the medium and low exposure concentration groups, respectively.
Venn diagram of a upregulated and b downregulated genes that differed significantly from the control group in M. beryllina exposed to different concentrations of ibuprofen as determined by a one-factor ANOVA (P < 0.01). High, medium, low treatments refer to the 1.15, 0.115, and 0.0115 mg/L ibuprofen exposure concentrations, respectively
There were ten genes significantly downregulated compared with the control group that were common between all three treatment groups (Fig. 2). These included genes involved in apoptosis [caspase-3 precursor (CASP3)], an immune response [c-c motif chemokine 28 precursor (CCL28)], and cell proliferation [proline-rich protein 15-like protein a-like (PRR15LA). There was a downregulation of transcriptional enhancer factor tef-5-like (TEAD3), also involved in cell proliferation; however, this was not significant in the medium exposure group. There was also a downregulation of genes associated with cellular structural components [complement c1q tumor necrosis factor-related protein 5-like (C1QTNF5), type i enveloping like (CYT1L), type i cytoskeletal 13-like (KRT13), and muscle m-line assembly protein unc-89-like (UNC-89)], lipid metabolism [sphingomyelin phosphodiesterase 3-like (SMPD3)] and cell adhesion [fermitin family homolog 1-like (FERMT1)] at all exposure concentrations. Last, there was a downregulation of a gene involved in ion transport [zinc transporter zip14-like (SLC39A14)].
Heat map showing the 96 microarray probes that were statistically different from the control group (57 different genes; ANOVA, P < 0.01) in M. beryllina following 14-day exposures to ibuprofen. Microarray probes are organized by differences in expression from the control group as determined by post hoc analysis of the ANOVA: a common at all exposure concentrations (note that TEAD3 was only significant in the low and high concentration treatments); b differentially expressed in the high concentration treatment; c differentially expressed in the two lower exposure concentration treatments; d differentially expressed in the medium concentration treatment; e differentially expressed in the low concentration treatment. High, medium, and low treatments refer to the 1.15, 0.115, and 0.0115 mg/L ibuprofen exposure concentrations, respectively. Expression levels are presented as normalized log2 values with yellow indicating upregulation and blue indicating downregulation. Gene symbols are presented along the right side of the heat map
Differentially expressed genes at the high exposure concentration
There were several genes that were upregulated at the highest exposure concentration (i.e., 1.15 mg/L) involved in extracellular matrix and protein breakdown [matrix metalloproteinase 9 (MMP9; also involved in an inflammatory response), proteasome subunit alpha type-7 (PSMA7; also associated with an apoptotic response), and 26S protease regulatory subunit 6B (PSMC4)]. There was also an upregulation of several genes involved in cell structure [pdz and lim domain protein 1 (PDLIM1), pleckstrin homology-like domain family a member 2-like (PHLDA2), and von willebrand factor type egf and pentraxin domain-containing protein 1-like (SVEP1)]. This was in contrast to the downregulated genes associated with the cytoskeleton and cell structure (e.g., KRT13, CYT1L, FERMT1, and UNC-89) that were common at all treatment concentrations.
There was a significant upregulation of genes involved in an inflammatory response [signal transducer and activator of transcription 1 (STAT1; however, STAT1 has also been identified as potentially important in bone development in fish; Vieira et al. 2013) and signal transducer and activator of transcription 2 (STAT2)], apoptosis [dna damage-inducible transcript 3 (DDIT3)], RNA processing [thioredoxin-like protein 4a (TXNL4A)], and DNA repair [protein bccip homolog (BCCIP)]. There was also an upregulation of genes involved in fatty acid beta-oxidation [acyl- dehydrogenase family member 11 (ACAD11)], regulating cell signaling pathways [14-3-3 protein beta alpha (YWHAB)], the secretory pathway [transmembrane emp24 domain-containing protein 10 precursor (TMED10)], protein folding [calreticulin precursor (CALR)], and two genes with unknown function [plac8-like protein 1 (PLAC8L1) and SI:DKEY-6 N6.2]. Interestingly, tributyltin binding protein type 1 (TBT-BP1), discovered because of its ability to bind to the marine pollutant tributyltin, may also play a role in osteoblast activity and development in fishes in addition to aiding in detoxification (Satone et al. 2011) and was upregulated at the highest exposure concentration. There was variable expression of genes involved in an oxidative stress response as cytochrome b-245 light chain-like (CYBA), and glutathione reductase (GR) were upregulated while manganese superoxide dismutase (SOD2) was downregulated. There was also decreased expression of thyroid hormone receptor beta (THRb; ANOVA P < 0.05]; however, this was not significant at the P < 0.01 threshold.
Differentially expressed genes at the two lower exposure concentrations
There was a downregulation of genes involved in lipid metabolism [fatty acid-binding protein 6 (FABP6; that binds to bile salts and bilirubins) and 5-aminolevulinate mitochondrial-like (ALAS1)], proteolysis [serine protease htra3-like (HTRA3)], translation [40s ribosomal protein s13 (RPS13)], cell proliferation [beta-galactoside-binding lectin (GALECTIN) and nonspecific cytotoxic cell receptor protein 1 homolog (NCCRP1)], and with an unknown function [upf0139 membrane protein c19 or f56 homolog (CS056)], which was common at the two lowest exposure concentrations. There was also a downregulation of a nonenzymatic cofactor of the multisynthase complex, aminoacyl trna synthase complex-interacting multifunctional protein 1-like (AIMP1), which can act as a cytokine in an immune response, however can also interact with components of the cytoskeleton (Jackson et al. 2011). The only gene that was upregulated in both the medium and low concentration exposures was ras-related protein rab-21-like (RAB21).
Differentially expressed genes at the medium exposure concentration
Unique to the medium exposure treatment was the upregulation of f-box lrr-repeat protein 18-like (FBXL18). However, there was a downregulation of several other genes associated with ubiquitin [cop9 signalosome complex subunit 6 (COPS6), ubiquitin-like protein fubi (FAU), and ubiquitin carboxyl-terminal hydrolase 42-like (USP42)]. Collectively, these results suggest that proteasomal breakdown was affected by exposure to ibuprofen. There was also a downregulation of genes involved in transcription and translation [ccr4-not transcription complex subunit 6-like (CNOT6), elongation factor 1-delta (EEF1D), 40s ribosomal protein s16 (RPS16)], metabolism [arginase- mitochondrial-like (ARG) and glucosamine-6-phosphate isomerase (NAG1)], immune function [gtpaseimap family member 4-like (GIMAP4)], and cell proliferation [pancreatic progenitor cell differentiation and proliferation factor b (PPDPF-B)].
Differentially expressed genes at the low exposure concentration
Unique to the low exposure concentration was the upregulation of the immune response gene c4b-binding protein alpha chain precursor (C4BPA). As in the responses detected at the other concentrations, there was a downregulation of genes involved in proteolysis [alpha-aspartyl dipeptidase (AAD-A)], transcription and translation [nitric oxide-associated protein 1-like (NOA1, which is also associated with mitochondria-mediated cell death) and nuclear receptor subfamily 0 group b member 2 (NR0B2) that inhibits receptor-dependent signaling pathways], and the cytoskeleton [protein app1-like (APP1)].
Quantitative PCR results
Several qPCR assays were designed to examine responses to ibuprofen exposure of genes specifically associated with general stress and metabolism, arachidonic acid metabolism (prostaglandin and leukotriene synthesis), and osmoregulatory function. Among the genes assayed, most were significant only at the highest exposure concentration in the ANOVA tests; however, several additional genes had a significant sigmoidal dose-response when curves were fit using an LRT indicating that these genes respond in a dose-dependent manner. Although both sigmoidal and nonmonotonic dose-response curves were fit for each gene, the sigmoidal curve was the best fit for all (Supplemental Table 2). We report P values from the ANOVA and also from the LRTs used to fit dose-response curves.
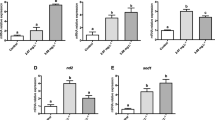
There was a significant upregulation of MMP9 (ANOVA, P < 0.00001; LRT, P < 0.0001), a pattern detected in both the qPCR and microarray results (Fig. 3). Also associated with an immune response was a downregulation of CCL28 at the highest exposure concentration (ANOVA, P < 0.01; LRT, P < 0.01), similar to the pattern detected in the microarray. We also detected an upregulation of STAT1 in the microarray results; however, this pattern was not evident in the qPCR assay. We confirmed a significant downregulation of C1QTNF5 (detected in both the qPCR and microarray analysis) at the highest exposure concentration and a nearly significant negative sigmoidal dose-response in the qPCR results (ANOVA, P < 0.05; LRT, P = 0.056). Similar to the microarray analysis, decreased expression of KRT13 was present in the qPCR results; however, it was not statistically significant.
Changes in the expression of M. beryllina genes involved in 1 apoptosis, 2 immune function, 3 metabolism, 4 oxidative stress, and 5 structural components due to ibuprofen exposure as determined by quantitative PCR. These genes were selected to verify expression patterns observed in the microarray analysis. High, medium, and low treatments refer to the 1.15, 0.115, and 0.0115 mg/L ibuprofen exposure concentrations, respectively. There is a reference line at one because a negative mean relative fold change is impossible using the 2−ΔΔCt method for qPCR analysis; therefore, values greater than one indicate an upregulation and values below one indicate a downregulation of a gene. For each gene, differences in expression from the control, as determined by ANOVA (P < 0.05), are indicated by an asterisk (*). The sigmoidal dose-response curves for each gene fit using a likelihood ratio test (P < 0.05) are indicated using a solid line. Nonstatistically significant dose-response trends are indicated with a dashed line
The upregulation of GR and CYBA detected in the microarray results was confirmed in the qPCR assessment (ANOVA, P < 0.05; LRT, P < 0.05, 0.0001, respectively). Additionally, the iron and copper transport protein ceruloplasmin precursor (CP), which may be associated with an antioxidant and inflammatory response, was significant in the microarray (at P < 0.05) and was also significantly upregulated in the qPCR assay (ANOVA, P < 0.01; LRT, P < 0.01). There was a decrease in the expression of genes involved in aerobic metabolism as cytochrome b-c1 complex subunit 7 (UQCRB), and cytochrome c oxidase subunit mitochondrial-like (COI) were downregulated (P < 0.05) in the microarray, and this response pattern was confirmed by the qPCR results as we detected a significant decrease in UQCRB levels (ANOVA, P < 0.05; LRT, P < 0.05). The decrease in COI was not statistically significant (ANOVA, P > 0.05). The expression of ARG was also significantly downregulated in the qPCR (ANOVA, P < 0.05; LRT, P < 0.01), similar to the patterns detected in the microarray. Additionally, ACAD11 was upregulated at the lower exposure concentrations in the microarray, a trend that was present in the qPCR assessment; however, this was not statistically significant (P > 0.05). The response for PSMA7, which was significantly upregulated in the microarray, was not confirmed in the qPCR assessment. A downregulation trend was observed in the qPCR assays for genes associated with apoptosis [bcl-2-modifying factor-like (BMF) and CASP3]; however, these were not statistically significant.
There was a significant upregulation of PTGS2 and arachidonate 5-lipoxygenase-activating protein (ALOX5AP) assays detected in the qPCR (ANOVA, P < 0.001; LRT, P < 1.0E−08, 0.0001, respectively; Fig. 4). Involved in ion regulation, sodium potassium-transporting atpase subunit alpha-2-like (ATP1A2) was significantly downregulated (ANOVA, P < 0.05; LRT, P < 0.05), and sodium potassium-transporting atpase subunit alpha-3-like (ATP1A3) was near significantly downregulated in the ANOVA, but had a highly significant negative sigmoidal dose response (ANOVA, P = 0.076; LRT, P < 1.0E−11; Fig. 5) in the qPCR analyses. Similarly, sodium potassium-transporting atpase alpha-1 subunit (ATP1A1) had a significant negative sigmoidal dose response (LRT, P < 1.0E−07). There appeared to be slight downregulation of sodium potassium-transporting atpase subunit beta-233 (ATNB233); however, this pattern was not statistically significant.
Changes in the expression of M. beryllina genes involved in arachidonic acid metabolism, specifically 1 prostaglandin and 2 leukotriene synthesis, due to ibuprofen exposure as determined by quantitative PCR. High, medium, and low treatments refer to the 1.15, 0.115, and 0.0115 mg/L ibuprofen exposure concentrations, respectively. There is a reference line at one because a negative mean relative fold change is impossible using the 2−ΔΔCt method for qPCR analysis; therefore, values greater than one indicate an upregulation and values below one indicate a downregulation of a gene. For each gene, differences in expression from the control, as determined by ANOVA (P < 0.05), are indicated by an asterisk (*). The sigmoidal dose-response curves for each gene fit using a likelihood ratio test (P < 0.05) are indicated using a solid line. Nonstatistically significant dose-response trends are indicated with a dashed line
Changes in the expression of M. beryllina genes involved in osmoregulation due to ibuprofen exposure as determined by quantitative PCR. High, medium, and low treatments refer to the 1.15, 0.115, and 0.0115 mg/L ibuprofen exposure concentrations, respectively. There is a reference line at one because a negative mean relative fold change is impossible using the 2−ΔΔCt method for qPCR analysis; therefore, values greater than one indicate an upregulation and values below one indicate a downregulation of a gene. For each gene, differences in expression from the control, as determined by ANOVA (P < 0.05), are indicated by an asterisk (*). The sigmoidal dose-response curves for each gene fit using a likelihood ratio test (P < 0.05) are indicated using a solid line. Nonstatistically significant dose-response trends are indicated with a dashed line
Discussion
The goal of the present study was to determine the transcriptome-wide effects of ibuprofen exposure on the inland silverside, a nonmodel fish species with specific ecological relevance to the assessment of contaminants in California surface waters (e.g., Brander et al. 2013). The 96 h LC50 of ibuprofen was 34.35 mg/L for the inland silverside. While the acutely toxic concentration is quite high relative to environmental concentrations, inland silversides are more sensitive to ibuprofen than the Indian major carp (Cirrhinus mrigala) with a 24 h LC50 of 142 mg/L (Saravanan et al. 2012) or the common carp (Cyprinus carpio) with a 96 h LC50 of 175.6 mg/L (Islas-Flores et al. 2014). We also chronically exposed inland silversides to ibuprofen and detected different transcriptomic responses based on exposure concentration. At the lowest exposure concentration (0.0115 mg/L), consistent with those detected in undiluted wastewater effluent (e.g., Metcalfe et al. 2003; reviewed in Fent et al. 2006; Yu et al. 2013) and ocassionally downstream of effluent discharge points (Ashton et al. 2004), we detected a downregulation of many processes including those involved in skeletal development. At the highest exposure concentration (1.15 mg/L), we detected an inflammatory response that included increased expression of regulatory genes in the arachidonic acid metabolism pathway. There are numerous examples in the literature that address nonlinear relationships between exposure concentration and measured endpoint (reviewed in Vandenberg et al. 2012), where functional responses to exposure at higher concentrations, upon which modes of action are generally determined, differ to responses observed at lower, more environmentally realistic concentrations. The microarray analysis in the present study showed that at the low concentration, there was a downregulation of several biological processes, whereas at the higher exposure concentration, there was an upregulation of several biological processes, patterns that were not detected at the lower exposure concentrations. This difference in responses, and the subtle response patterns described herein, highlight the importance of measuring transcriptome-wide cellular responses to a range of concentrations to identify the diversity of cellular mechanisms that underlie responses to this common wastewater contaminant.
Arachidonic acid metabolism is vital for many physiological processes in fishes. There are two main branches of arachidonic acid metabolism: one branch involves the production of prostaglandins and thromboxanes that are used in many physiological functions including inflammation and bone formation; the second branch involves the production of leukotrienes that are used in inflammatory responses and paradoxically, regulating prostaglandin and thromboxane activity (Lutz and Cornett 2013). Both PTGS2 and ALOX5AP are highly inducible and are key enzymes in regulating inflammatory responses in vertebrates. Consequently, they are highly regulated through a variety of cellular mechanisms to maintain tight control of arachidonic acid metabolism (Lutz and Cornett 2013). We observed an upregulation of PTGS2 and ALOX5AP after 14-day exposure to the highest concentration of ibuprofen. This pattern was not observed at the lower exposure concentrations, although both genes followed a sigmoidal dose-response pattern. The activation of the leukotriene branch of arachidonic acid metabolism is consistent with responses observed in Daphnia magna exposed to ≥20 mg/L of ibuprofen (Heckmann et al. 2008). Because the mode of action of ibuprofen is to inhibit PTGS1 and PTGS2 activity, the increased abundance of transcripts for the inducible PTGS2 and ALOX5AP may indicate a compensatory mechanism to counteract PTGS1 and PTGS2 enzyme inhibition at the protein level to maintain arachidonic acid metabolism-dependent processes. Therefore, future research should examine PTGS2 activity along with mRNA patterns to determine whether increased PTGS2 transcript abundance maintains sufficient PTGS2 enzyme activity and successfully counteracts the effects of exposure to ibuprofen.
Exposure to ibuprofen led to the differential expression of several genes associated with an immune response, mostly involved in an inflammatory response. Differential regulation of genes involved in inflammation could be expected based on ibuprofen’s intended anti-inflammatory mode of action in humans. The inducible gelatinase MMP9 can be regulated by PTGS2 and was significantly upregulated at the highest exposure concentration of ibuprofen, directly linking exposure to an inflammatory response. Interpretation of the MMP9 induction, however, can be complicated because of its crucial role in bone matrix degradation and skeletal development (Vieira et al. 2013), in addition to its association with a response to, and recovery from, inflammation (Chadzinska et al. 2008). Similarly, CP is generally associated with an inflammation response (Gitlin 1988); however, it has also been linked to antioxidant responses in fishes (Liu et al. 2011) and was significantly upregulated due to exposure to ibuprofen. Because these and many other genes involved in an inflammatory response (e.g., STAT1 and STAT2), along with PTGS2 and ALOX5AP, were still upregulated after 14 days of exposure to the highest concentration of ibuprofen, it suggests maintenance of an inflammatory response due to chronic exposure to high concentrations of ibuprofen.
There was differential regulation of several genes involved in oxidative stress, which include the downregulation of SOD2 and the upregulation of GR, CYBA, and CP. The downregulation of SOD2 is consistent with a decrease in SOD activity detected in clams (Milan et al. 2013), however is in contrast to an increase in SOD activity in common carp acutely exposed to ibuprofen (Islas-Flores et al. 2014). Additionally, the upregulation of GR and downregulation of SOD2 observed in the present study is contradictory to the patterns observed in Mediterranean mussel (Mytilus galloprovincialis) that were chronically exposed to 250 ng/L of ibuprofen for 14 days (Gonzalez-Rey and Bebianno 2011). Because SOD2 is located in the mitochondrion and responds to superoxides produced through aerobic respiration (Robb et al. 2014), the decrease in SOD2 transcript abundance detected in the present study may be associated with the overall decrease in the expression of genes involved in aerobic metabolism. However, the upregulation of other genes involved in an oxidative stress response suggests that the fish in the present study were responding to other oxidative stress conditions related to ibuprofen exposure. Collectively, these studies indicate that exposure to ibuprofen affects the oxidative stress response of aquatic organisms; however, the patterns of the response appear to be dependent on species, exposure duration, and concentration.
There was evidence of differential expression of genes involved in bone development and structure due to exposure to ibuprofen. Previous work has confirmed a downregulation of numerous hormone receptors in response to ibuprofen exposure in zebrafish, which was linked to reduced hormone levels due to an inhibition of prostaglandin production (Ji et al. 2013). There was a decrease in THRb in the present study (although not significant at the P < 0.01 threshold), which is critical to growth and development, and may be associated with an ibuprofen-induced decrease in hormone levels. Decreases in sex steroids can lead to bone loss in humans (reviewed in Nelson et al. 2013), and a potential endocrine regulation of bone has been identified in gilthead seabream (Sparus auratus; Vieira et al. 2013). In the present study, at all exposure concentrations, there was an upregulation of genes involved in extracellular matrix degradation and calcium binding, variable regulation of genes associated with cell adhesion, and a downregulation of genes involved in collagen and cytoskeleton structure, along with calcium metabolism, which collectively, may be associated with ibuprofen effects on skeletal development. The fish used in the present study were ~66 dph and consequently were in a critical stage of skeletal growth and development. Disruption in bone developmental processes may have implications on overall fish growth throughout the life cycle. Due to the importance of fish size in predator avoidance, a decrease in skeletal growth may result in reduced survival in fish chronically exposed to ibuprofen in aquatic systems. Future research should examine the effects of exposure to low concentrations of ibuprofen across the entire life cycle of fishes (e.g., Han et al. 2010) to examine potential long-term decreases in growth and development.
Various genes involved in aerobic metabolism were downregulated in the inland silverside in the present study. This is in contrast to previous studies that detected increased expression of genes involved in glycolysis, the TCA cycle and oxidative phosphorylation in clams (Milan et al. 2013), and increased gill glycolytic capacity and reduced glycogen content in the livers of rainbow trout, which was suggested to be related to maintaining energetically costly ion pumps (Gravel et al. 2009). Both studies reported these responses to ibuprofen following exposures conducted in salt water. Interestingly, there was a trend of higher glycogen content in the livers of rainbow trout exposed to ibuprofen in freshwater; however, this was not statistically significant (Gravel et al. 2009). There was a downregulation of CYBA in inland silverside exposed to ibuprofen in freshwater, and functional categories associated with metabolism were significantly enriched in the functional analysis. Additionally, there was a downregulation of genes involved in proteolysis and protein synthesis, and several functional categories associated with protein synthesis were enriched in the significant gene lists that collectively suggest a reduction in protein turnover.
A downregulation of genes involved in osmoregulation at the highest exposure concentration was suggestive of an ibuprofen-dependent decrease in osmoregulatory ability. This is consistent with previous work on rainbow trout that showed an inhibition of the required elevated Na+/K+-ATPase activity in salt water (Gravel et al. 2009). We found decreased expression of Na+/K+-ATPase genes (ATP1A1, ATP1A2, ATP1A3) at the highest exposure concentration, genes required for ion regulation in fishes (Liao et al. 2009). Downregulation of Na+/K+-ATPases may be associated with the decreased expression of genes involved in aerobic metabolism observed in the present study, as this physiological process can use a significant portion of the cellular energy budget (Staples and Buck 2009). These results suggest an overall disturbance in osmoregulatory mechanisms and reduced metabolism in inland silversides. An ibuprofen-dependent decrease in these genes may affect the osmoregulatory capability of fishes in salt water, which could have significant consequences in fishes that are chronically exposed to ibuprofen in an estuary or marine environment, such as amphidromous fishes like the inland silverside.
Many of the cellular responses in the present study occurred at ibuprofen exposure concentrations that exceed those detected in the aquatic environment. While the responses detected at 1.15 mg/L provide valuable insight into the mode of action of ibuprofen in fish, it may be difficult to directly extrapolate these results to the environment. At the most environmentally relevant concentration (0.0115 mg/L) used in this study, the fish were still responding to the ibuprofen exposure at the level of the transcriptome. There was evidence of differential expression of genes involved in immune function and a downregulation of genes associated with cell structure, and many functional categories associated with aerobic respiration were significantly enriched. Disturbances in these physiological processes over exposures longer than 14 days may have significant consequences on the health of wild fishes, especially those with limited home ranges (e.g., inland silverside; Brander et al. 2013) at locations where they may be continually exposed to wastewater effluents. Additionally, the exposure concentration in the environment likely fluctuates seasonally and may spike randomly due to changes in water usage or ibuprofen consumption that lead to variable ibuprofen concentrations over time; therefore, it is difficult to predict true environmental exposure concentrations. Given that ibuprofen levels can often be orders of magnitude higher in bile and plasma samples (Brozinski et al. 2013), dissolved ibuprofen concentrations measured in individual water samples may not accurately reflect the concentrations relevant to fishes in a system. Last, it is critical to note that ibuprofen is often detected along with many other NSAIDs and pharmaceuticals in systems that receive wastewater effluent (e.g., Ashton et al. 2004), and therefore, it is the response to the entire suite of compounds that is ecologically relevant and not necessarily the response to a single compound.
This study presents the transcriptome-wide response of an environmentally relevant fish species to chronic exposure to ibuprofen. Interestingly, some of the response patterns are similar to those of previous transcriptome-wide assessments of the effects of ibuprofen exposure on evolutionarily distant invertebrate species (Heckmann et al. 2008; Milan et al. 2013). The similarity of the responses across taxa suggests that these results collectively may be useful for generating biomarkers of exposure to ibuprofen for various aquatic species for monitoring the effects of this common wastewater effluent contaminant. Additionally, this study significantly expands on the current suite of molecular tools available for studying inland silversides, a highly abundant and widely distributed fish species that can potentially be used as a bioindicator of contaminant exposure in coastal ecosystems in North America.
References
Altschul S, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Ankley GT, Daston GP, Degitz SJ, Denslow ND, Hoke RA, Kennedy SW, Miracle AL, Perkins EJ, Snape J, Tillitt DE, Tyler CR, Versteeg D (2006) Toxicogenomics in regulatory ecotoxicology. Environ Sci Technol 40:4055–4065
Ashton D, Hilton M, Thomas KV (2004) Investigating the environmental transport of human pharmaceuticals to streams in the United Kingdom. Sci Total Environ 333:167–184
Bhandari K, Venables B (2011) Ibuprofen bioconcentration and prostaglandin E2 levels in the bluntnose minnow Pimephales notatus. Comp Biochem Physiol C Toxicol Pharmacol 153:251–257
Bolker BM (2008) Ecological models and data in R. Princeton University Press
Brain P, Cousens R (1989) An equation to describe dose responses when there is stimulation of growth at low doses. Weed Res 29:93–96
Brander SM (2013) Thinking outside the box: Assessing endocrine disruption in aquatic life.In:Ajuha S (ed) Monitoring water quality: Pollution assessment, analysis, and remediation. Elsevier, pp 103-147
Brander SM, Werner I, White JW, Deanovic LA (2009) Toxicity of a dissolved pyrethroid mixture to Hyalellaazteca at environmentally relevant concentrations. Environ Toxicol Chem 28:1493–1499
Brander SM, He G, Smalling KL, Denison MS, Cherr GN (2012) The in vivo estrogenic and in vitro anti-estrogenic activity of permethrin and bifenthrin. Environ Toxicol Chem 31:2848–2855
Brander SM, Connon RE, He G, Hobbs JA, Smalling KL, Teh SJ, White JW, Werner I, Denison MS, Cherr GN (2013) From ’omics to otoliths: responses of an estuarine fish to endocrine disrupting compounds across biological scales. PLoS One 8:e74251
Brozinski J-M, Lahti M, Meierjohann A, Oikari A, Kronberg L (2013) The anti-inflammatory drugs diclofenac, naproxen and ibuprofen are found in the bile of wild fish caught downstream of a wastewater treatment plant. Environ Sci Technol 47:342–348
Buser H, Poiger T, Muller MD (1999) Occurrence and environmental behavior of the chiral pharmaceutical drug ibuprofen in surface waters and in wastewater. Environ Sci Technol 33:2529–2535
Chadzinska M, Baginski P, Kolaczkowska E, Savelkoul HFJ, Kemenade BMLV (2008) Expression profiles of matrix metalloproteinase 9 in teleost fish provide evidence for its active role in initiation and resolution of inflammation. Immunology 125:601–610
Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21:3674–3676
Connon RE, Geist J, Pfeiff J, Loguinov AV, D’Abronzo LS, Wintz H, Vulpe CD, Werner I (2009) Linking mechanistic and behavioral responses to sublethal esfenvalerate exposure in the endangered delta smelt; Hypomesus transpacificus (Fam. Osmeridae). BMC Genomics 10:608
Connon RE, Beggel S, D’Abronzo LS, Geist JP, Pfeiff J, Loguinov AV, Vulpe CD, Werner I (2011) Linking molecular biomarkers with higher level condition indicators to identify effects of copper exposures on the endangered delta smelt (Hypomesus transpacificus). Environ Toxicol Chem 30:290–300
Connon RE, Geist J, Werner I (2012) Effect-based tools for monitoring and predicting the ecotoxicological effects of chemicals in the aquatic environment. Sensors 12:12741–12771
Core Development Team R (2010) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria
David A, Pancharatna K (2009) Developmental anomalies induced by a non-selective COX inhibitor (ibuprofen) in zebrafish (Danio rerio). Environ Toxicol Pharmacol 27:390–395
Doyle MA, Bosker T, Munkittrick KR (2011) The potential use of Atlantic silverside (Menidia menidia) for monitoring estuarine pollution. J Environ Monit 13:3168–3177
Fent K, Weston AA, Caminada D (2006) Ecotoxicology of human pharmaceuticals. Aquat Toxicol 76:122–159
Fernandez MP, Ikonomou MG, Buchanan I (2007) An assessment of estrogenic organic contaminants in Canadian wastewaters. Sci Total Environ 373:250–269
Flippin JL, Huggett D, Foran CM (2007) Changes in the timing of reproduction following chronic exposure to ibuprofen in Japanese medaka, Oryzias latipes. Aquat Toxicol 81:73–78
Gitlin JD (1988) Transcriptional regulation of ceruloplasmin gene expression during inflammation. J Biol Chem 263:6281–6287
Gonzalez-Rey M, Bebianno MJ (2011) Non-steroidal anti-inflammatory drug (NSAID) ibuprofen distresses antioxidant defense system in mussel Mytilus galloprovincialis gills. Aquat Toxicol 105:264–269
Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, Chen Z, Mauceli E, Hacohen N, Gnirke A, Rhind N, di Palma F, Birren BW, Nusbaum C, Lindblad-Toh K, Friedman N, Regev A (2011) Full-length transcriptome assembly from RNA-seq data without a reference genome. Nat Biotechnol 29:644–652
Gravel A, Wilson JM, Pedro DFN, Vijayan MM (2009) Non-steroidal anti-inflammatory drugs disturb the osmoregulatory, metabolic and cortisol responses associated with seawater exposure in rainbow trout. Comp Biochem Physiol C Toxicol Pharmacol 149:481–490
Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, Couger MB, Eccles D, Li B, Lieber M, Macmanes MD, Ott M, Orvis J, Pochet N, Strozzi F, Weeks N, Westerman R, William T, Dewey CN, Henschel R, Leduc RD, Friedman N, Regev A (2013) De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc 8:1494–512
Han S, Choi K, Kim J, Ji K, Kim S, Ahn B, Yun J, Choi K, Khim JS, Zhang X, Giesy JP (2010) Endocrine disruption and consequences of chronic exposure to ibuprofen in Japanese medaka (Oryzias latipes) and freshwater cladocerans Daphnia magna and Moina macrocopa. Aquat Toxicol 98:256–264
Heckmann L-H, Sibly RM, Connon R, Hooper HL, Hutchinson TH, Maund SJ, Hill CJ, Bouetard A, Callaghan A (2008) Systems biology meets stress ecology: linking molecular and organismal stress responses in Daphnia magna. Genome Biol 9:R40
Islas-Flores H, Gómez-Oliván LM, Galar-Martínez M, García-Medina S, Neri-Cruz N, Dublán-García O (2014) Effect of ibuprofen exposure on blood, gill, liver, and brain on common carp (Cyprinus carpio) using oxidative stress biomarkers. Environ Sci Pollut R 21:5157–5166
Jackson VC, Dewilde S, Albo AG, Lis K, Corpillo D, Canepa B (2011) The activity of aminoacyl-tRNA synthetase-interacting multi-functional protein 1 (AIMP1) on endothelial cells is mediated by the assembly of a cytoskeletal protein complex. J Cell Biochem 112:1857–1868
Jeffries KM, Jackson LJ, Ikonomou MG, Habibi HR (2010) Presence of natural and anthropogenic organic contaminants and potential fish health impacts along two river gradients in Alberta, Canada. Environ Toxicol Chem 29:2379–2387
Jeffries KM, Hinch SG, Sierocinski T, Clark TD, Eliason EJ, Donaldson MR, Li S, Pavlidis P, Miller KM (2012) Consequences of high temperatures and premature mortality on the transcriptome and blood physiology of wild adult sockeye salmon (Oncorhynchus nerka). Ecol Evol 2:1747–1764
Jeffries KM, Hinch SG, Sierocinski T, Pavlidis P, Miller KM (2014) Transcriptomic responses to high water temperature in two species of Pacific salmon. Evol Appl 7:286–300
Ji K, Liu X, Lee S, Kang S, Kho Y, Giesy JP, Choi K (2013) Effects of non-steroidal anti-inflammatory drugs on hormones and genes of the hypothalamic-pituitary-gonad axis, and reproduction of zebrafish. J Hazard Mater 254–255:242–251
Kolpin DW, Furlong ET, Meyer MT, Thurman EM, Zaugg SD, Barber LB, Buxton HT (2002) Pharmaceuticals, hormones, and other organic wastewater contaminants in U.S. streams, 1999-2000: a national reconnaissance. Environ Sci Technol 36:1202–1211
Liao B, Chen R, Hwang P (2009) Expression regulation of Na + -K + -ATPaseα1-subunit subtypes in zebrafish gill ionocytes. Am J Physiol Regul Integr Comp Physiol 296:R1897–R1906
Liu H, Peatman E, Wang W, Abernathy J, Liu S, Kucuktas H, Terhune J, Xu D-H, Klesius P, Liu Z (2011) Molecular responses of ceruloplasmin to Edwardsiella ictaluri infection and iron overload in channel catfish (Ictalurus punctatus). Fish Shellfish Immun 30:992–997
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25:402–408
Lutz CS, Cornett AL (2013) Regulation of genes in the arachidonic acid metabolic pathway by RNA processing and RNA-mediated mechanisms. WIREs RNA 4:593–605
Maruya KA, Schlenk D, Anderson PD, Denslow ND, Drewes JE, Olivieri AW, Scott GI, Snyder SA (2014) An adaptive, comprehensive monitoring strategy for chemicals of emerging concern (CECs) in California’s aquatic ecosystems. Integr Environ Assess Manag 10:69–77
Metcalfe CD, Koenig BG, Bennie DT, Servos M, Ternes TA, Hirsch R (2003) Occurrence of neutral and acidic drugs in the effluents of Canadian sewage treatment plants. Environ Toxicol Chem 22:2872–2880
Milan M, Pauletto M, Patarnello T, Bargelloni L, Marin MG, Matozzo V (2013) Gene transcription and biomarker responses in the clam Ruditapes philippinarum after exposure to ibuprofen. Aquat Toxicol 126:17–29
Morthorst JE, Lister A, Bjerregaard P, Van Der Kraak G (2013) Ibuprofen reduces zebrafish PGE2 levels but steroid hormone levels and reproductive parameters are not affected. Comp Biochem Physiol C Toxicol Pharmacol 157:251–257
Moyle PB (2002) Inland Fishes of California. University of California Press, Berkley and Los Angeles
Munakata A, Kobayashi M (2010) Endocrine control of sexual behavior in teleost fish. Gen Comp Endocr 165:456–468
Nallani GC, Paulos PM, Constantine LA, Venables BJ, Huggett DB (2011) Bioconcentration of ibuprofen in fathead minnow (Pimephalespromelas) and channel catfish (Ictaluruspunctatus). Chemosphere 84:1371–1377
Nelson ER, Wardell SE, McDonnell DP (2013) The molecular mechanisms underlying the pharmacological actions of estrogens, SERMs and oxysterols: Implications for the treatment and prevention of osteoporosis. Bone 53:42–50
Pounds N, Maclean S, Webley M, Pascoe D, Hutchinson T (2008) Acute and chronic effects of ibuprofen in the mollusc Planorbis carinatus (Gastropoda: Planorbidae). Ecotoxicol Environ Saf 70:47–52
Robb EL, Christoff CA, Maddalena LA, Stuart JA (2014) Mitochondrial reactive oxygen species (ROS) in animal cells: Relevance to aging and normal physiology. Can J Zool 613:603–613
Saravanan M, Devi KU, Malarvizhi A, Ramesh M (2012) Effects of Ibuprofen on hematological, biochemical and enzymological parameters of blood in an Indian major carp, Cirrhinus mrigala. Environ Toxicol Pharmacol 34:14–22
Satone H, Lee JM, Oba Y, Kusakabe T, Akahoshi E, Miki S, Suzuki N, Sasayama Y, Nassef M, Shimasaki Y, Kawabata S-I, Honjo T, Oshima Y (2011) Tributyltin-binding protein type 1, a lipocalin, prevents inhibition of osteoblastic activity by tributyltin in fish scales. Aquat Toxicol 103:79–84
Sifakis EG, Prentza A, Koutsouris D, Chatziioannou AA (2012) Evaluating the effect of various background correction methods regarding noise reduction, in two-channel microarray data. ComputBiol Med 42:19–29
Staples JF, Buck LT (2009) Matching cellular metabolic supply and demand in energy-stressed animals. Comp Biochem Physiol A 153:95–105
Supek F, Bošnjak M, Škunca N, Šmuc T (2011) REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One 6:e21800
Vandenberg LN, Colborn T, Hayes TB, Heindel JJ, Jacobs DR Jr, Lee DH, Shioda T, Soto AM, vom Saal FS, Welshons WV, Zoeller T, Myers JP (2012) Hormones and endocrine-disrupting chemicals: low-dose effects and nonmonotonic dose responses. Endocr Rev 33:378–455
Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F (2002)Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3:RESEARCH0034
Vieira FA, Thorne MAS, Stueber K, Darias M, Reinhardt R, Clark MS, Gisbert E, Power DM (2013) Comparative analysis of a teleost skeleton transcriptome provides insight into its regulation. Gen Comp Endocr 191:45–58
Villeneuve DL, Garcia-Reyero N, Escalon BL, Jensen KM, Cavallin JE, Makynen EA, Durhan EJ, Kahl MD, Thomas LM, Perkins EJ, Ankley GT (2011) Ecotoxicogenomics to support ecological risk assessment: a case study with bisphenol A in fish. Environ Sci Technol 46:51–59
Yu Y, Wu L, Chang AC (2013) Seasonal variation of endocrine disrupting compounds, pharmaceuticals and personal care products in wastewater treatment plants. Sci Total Environ 442:310–316
Acknowledgments
The authors thank Linda Deanovic and Marie Stillway of the University of California Davis Aquatic Toxicology Laboratory for maintaining the fish and conducting the exposures, Jennifer Truong and Bethany DeCourten for assisting with the processing of the samples, and Will White and Simone Hasenbein for statistical support. This research was funded by the California Department of Fish and Wildlife (contract #E1183010, to REC and SMB), and the State and Federal Contractors Water Agency (contract #201301957, to REC). A Delta Science Postdoctoral Fellowship from the Delta Stewardship Council provided funding for KMJ.
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible editor: Henner Hollert
Rights and permissions
About this article
Cite this article
Jeffries, K.M., Brander, S.M., Britton, M.T. et al. Chronic exposures to low and high concentrations of ibuprofen elicit different gene response patterns in a euryhaline fish. Environ Sci Pollut Res 22, 17397–17413 (2015). https://doi.org/10.1007/s11356-015-4227-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11356-015-4227-y