Abstract
Decabromodiphenyl ether (DBDE), which has been identified as an endocrine disrupting compound, is used as brominated flame retardant, and this can result in serious bioaccumulation within ecological systems. The objective of this study was to explore DBDE bioremediation (25 mg/kg) using laboratory scale soil slurry microcosms. It was found that effective biodegradation of DBDE occurred in all microcosms. Various biometabolites were identified, namely polybrominated diphenyl ethers congeners and hydroxylated brominated diphenyl ether. Reductive debrominated products such as tri-BDE to hepta-BDE congeners were also detected, and their total concentrations ranged from 77.83 to 91.07 ng/g. The mechanism of DBDE biodegradation in soil slurry microcosms is proposed to consist of a series of biological reactions involving hydroxylation and debromination. Catechol 2,3-oxygenase genes, which are able to bring about meta-cleavage at specific unbrominated locations in carbon backbones, were identified as present during the DBDE biodegradation. No obvious effect on the ecological functional potential based on community-level physiological profiling was observed during DBDE biodegradation, and one major facultative Pseudomonas sp. (99 % similarity) was identified in the various soil slurry microcosms. These findings provide an important basis that should help environmental engineers to design future DBDE bioremediation systems that use a practical microcosm system. A bacterial-mixed culture can be selected as part of the bioaugmentation process for in situ DBDE bioremediation. A soil/water microcosm system can be successfully applied to carry out ex situ DBDE bioremediation.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Decabromodiphenyl ether (DBDE) makes up a major proportion of all polybrominated diphenyl ethers (PBDE) congeners and is commonly used as a brominated flame retardant in numerous commercial products, including electronic equipment, furniture upholstery, building materials, textiles, carpets, and plastics. DBDE-contaminated environments occur due to the inadequate disposal of waste containing the above products. DBDE has a low water solubility and a high lipophilicity due to its structure (Mackay et al. 2006). As a result, these compounds undergo strong and almost complete sorption onto many environmental matrices and such accumulation can occur over a long period of time (Fangstrom et al. 2005). These PBDE congeners are widespread in their distribution and are frequently detected in the environment (Toms et al. 2008; Yogui and Sericano 2009; Chen et al. 2012a; Cincinelli et al. 2012; Gevao et al. 2011; Li et al. 2012; Wang et al. 2012; Wei et al. 2012; Moon et al. 2012). DBDE has been found to exist at concentrations that range from microgram per kilogram to milligram per kilogram across various soil slurry systems; these include aqueous sediments from reservoirs/lakes (Yogui and Sericano 2009; Moon et al. 2012), ocean sediments (Li et al. 2012; Wei et al. 2012), river sediments (Toms et al. 2008; Chen et al. 2012a), municipal wastewater treatment plant sludge (Cincinelli et al. 2012), soils (Gevao et al. 2011; Wang et al. 2012), and other similar situations. PBDE congeners have a high resistance to biodegradation. They are listed as persistent organic pollutants (POPs) by the Stockholm Convention and belong to “emerging contaminants” group of compounds.
According to the material safety data sheet of the U.S. EPA, acute toxicity due to DBDE does not occur with living organisms (U.S. EPA 2008). Similarly, EU risk assessments have found no human health risk related to DBDE (Bromide Science and Environment Forum, http://www.bsef.com). Nevertheless, more and more evidence has appeared suggesting that DBDE subchronic/chronic toxicity is possible; this is related to thyroid hormone disruption and the development of neurotoxic diseases. For example, significant dose-related disruption in mice occurs after exposure to a high-dose of DBDE on postnatal day 3; this involves changes in locomotion, rearing activity, and total activity (Viberg et al. 2003). Furthermore, DBDE may exert a direct effect on neuronal development in the rat brain, and hypothyroidism may occur when DBDE is given at high doses (Saegusa et al. 2012). The nervous system of mice has been found to be permanently damaged by DBDE via the chemical action on the cholinergic enzyme system (Liang et al. 2010). Furthermore, in developing zebra fish larvae, the hypothalamic-pituitary-thyroid axis is altered by DBDE via thyroid endocrine disruption (Chen et al. 2012b). As a result of the above findings, a practical and effective method for the elimination of DBDE seems to be necessary and has become an urgent ecological issue.
Bioremediation, which is often less expensive than other alternatives, can be used as eco-friendly treatment process that allows the elimination of POPs permanently from contaminated soil. Anaerobic biodegradation of high bromine number (HBN) PBDE (including DBDE) is possible, and during this process, the chemicals serve as electron acceptors during anaerobic respiratory; this approach has been a popular methodology up to the present (Tokarz et al. 2008; Nyholm et al. 2010; Qiu et al. 2012). Low bromine number (LBN) PBDE congeners are generated by biological reductive debromination of DBDE. Significant DBDE removal efficiency has been obtained by adding an extra carbon source like starch and yeast under anaerobic conditions (Gerecke et al. 2006). However, incomplete debromination of DBDE and HBN PBDE congeners usually occurs under micro-aerobic conditions. This results in LBN PBDE congener metabolites being released into the biota. Some LBN PBDE congeners may affect hormone homeostasis, and this may result in irreversible damage to cognitive performance and motor skills, as well as causing altered behavior (Frederiksen et al. 2010). For instance, dominant PBDE congeners, such as tetrabromodiphenyl ether (BDE-47) and pentabromodiphenyl ether (BDE-99), are commonly found in ecological systems and are thought to cause subchronic/chronic toxicity when fish, other wildlife, and humans are exposed to them (Shao et al. 2008; Albina et al. 2010).
Aerobic biodegradation of POPs provides an alternative method for bioremediation. One advantage of this approach is that the PBDE aerobic metabolites produced by this process show lower toxicity in ecological systems; these include hydroxylated brominated diphenyl ether, brominated phenols, and brominated catechols (Xia 2013). Complete mineralization can be brought about using aerobic PBDE biodegradation. However, little information is available on aerobic HBN PBDE biodegradation when it occurs in soils or surface water sediments. The mechanism of aerobic HBN PBDE biodegradation and its ecological physiological profile remain unclear. The objective of this study was to investigate the DBDE biodegradation using laboratory scale soil slurry microcosm systems. Removal efficiency and the kinetics of DBDE degradation were explored in a series of soil slurry microcosms. An investigation of the enzymes involved in this degradation and the identification of the functional genes involved in the process will enable the biochemical pathways used for the biodegradation of DBDEs to be identified. It should be possible to predict the pathway of DBDE biodegradation by integrating the information available on all identified metabolites in conjunction with the genes expressed as functional enzymes in the microcosms. These findings should provide suggestions that will help develop DBDE biodegradation into a practical soil slurry microcosm system.
Materials and methods
Materials
DBDE was used as the sole carbon source for the biodegradation and was obtained from Alfa Aesar (Karlsruhe, Germany, 99 % purity). In addition, a stock solution of DBDE (50 mg/L) dissolved in nonane was purchased from Cambridge Isotope Laboratories (Andover, MA, USA) and used for the GC analysis. A standard solution of a 24 PBDE congeners mixture, dissolved in isooctane-toluene (8:2, v/v), was purchased from Wellington (Guelph, Canada) and used for the GC/MS analysis; the congeners consisted of BDE-17, BDE-28, BDE-47, BDE-49, BDE-66, BDE-71, BDE-77, BDE-85, BDE-99, BDE-100, BDE-119, BDE-126, BDE-138, BDE-153, BDE-154, BDE-156, BDE-183, BDE-184, BDE-191, BDE-196, BDE-197, BDE-206, BDE-207, and BDE-209 (DBDE). The stock and experimental solutions of DBDE/PBDE congeners were prepared by serial dilution in hexane and stored in dark-brown glass containers at 4 °C to prevent photolysis of the DBDE and other PBDE congeners. The internal standards of the PBDE congeners included 13C12-BDE-28, 13C12-BDE-47, 13C12-BDE-99, 13C12-BDE-100, 13C12-BDE-153, 13C12-BDE-154, and 13C12-BDE-183 (Wellington, Guelph, Canada). 13C12-BDE-138 (Wellington, Guelph, Canada) was used as the recovery standard for the PBDE analysis. All organic solvents used in this study were of HPLC grade with a purity of >99.9 %. All other chemicals used in this study were reagent grade with a purity of >99 %. The Milli-Q water was double-distilled and deionized by a Millipore water purification system.
Bacterial-mixed cultures capable of degrading DBDE were obtained by sampling the surface river sediment under the Da-An Bridge (Da-An) and the Yi-Li bridge (Yi-Li), both in the Da-An River, Taichung City, Taiwan. In addition, activated sludge was obtained from the Nei-Hu WWTP in Taipei City, Taiwan (Nei-Hu). All three bacterial populations are able to utilize DBDE as carbon source and could degrade 93 to 95 % of the DBDE in 430 mg/kg DBDE-contaminated clay over 10 months (Chou et al. 2013). The enrichment procedure used with these bacterial populations has been described previously. Taichung natural soil (TCS), which consists of 54 % slit, 36 % clay, and 10 % sand, was selected for the DBDE biodegradation experiments involving soil slurry microcosms. TCS was collected from Taichung City, Taiwan. The background level of PBDEs in the TCS was in the nanogram per kilogram range (Supporting Information Figure S1). The BET-N2-surface area and cation exchange capacity of this soil were measured as 5.21 m2/g and 1.37 meq/100 g, respectively. The percentage of soil organic matter (SOM) in the soil was found to be 1.883 %. The soil sample was air-dried, sieved to obtain particles of less than 2.0 mm, and then stored at room temperature before being used in the experiments. The water content of the samples was measured by drying at 105 °C in an oven until all water content had disappeared based on the Taiwan EPA standard method (NIEA S280.62C). The concentrations of DBDE/PBDE congeners in each sample were converted as milligram per dry soil kilogram using the calculated water content of samples in this study.
DBDE biodegradation experiments using soil slurry microcosms
Batch biodegradation experiments using 25 mg/kg DBDE as the sole carbon source were performed in laboratory scale mixed aerobic/micro-aerobic soil slurry microcosms. Each dark-brown 125-mL serum bottle had a mixture of 4.0 g dry TCS added to it, together an appropriate amounts of DBDE and 100-mL inorganic salt growth medium (MSB) (Chang et al. 2007). The microcosms were set up by sterilizing the soil slurry bottles using an autoclave at three times. All the microorganisms within the soil slurry were killed, and this was confirmed using spread plates. The ratio of TCS to the MSB was set at 1:25 (g/mL). At the beginning of the experiments, samples from each of the three bacterial groups (OD590 = 0.020, about 1 × 109 CFU/mL growth on DBDE-MSB agar) were added separately to the soil slurry microcosms. The soil slurry microcosms, each of which had a sterilized cotton plug as their seal, were incubated on a shaker at 150 rpm (mixed aerobic under micro-aerobic conditions) at 25 °C for 6 months in a dark environment. Samples for the follow-up experimental analysis were taken from each individual microcosm. A series of samples containing the same amount of the mixed bacterial culture were collected individually from each sample at the start of the experiment and on the second, third, fourth, and sixth month of the experiment. The used samples were sterilized and discarded. A control, namely DBDE-MSB poisoned with the biocide NaN3 (ca 0.4 %, v/v) in a TCS slurry microcosm was also analyzed during this study. The SOM content of the TCS was found to be resistant to degradation by the DBDE-degrading bacteria during this experiment (data not shown).
Experimental analysis
Microcosms such as those described here are consistently and generally described as aerobic but reductive debromination has been identified as a potential degradation mechanism in aerobic and micro-aerobic microenvironments within such slurries (Chou et al. 2013). The possible aerobic/micro-anaerobic products of DBDE biodegradation are measured in this study. Furthermore, the bacterial community involved in the microcosms and their physiological properties are likely to be affected by the disappearance of DBDE, the generation of biometabolites (like PBDE congers), and a range of other environmental factors, including the soil characteristics and the aerobic/micro-aerobic environment within the microcosms. Changes in the bacterial community during DBDE biodegradation were monitored by the denaturing gradient gel electrophoresis (DGGE). Physiological level characteristics were analyzed by community-level physiological profiling (CLPP) with the aim of monitoring the ecological functional potential of the microbial communities during DBDE biodegradation.
Sample pretreatment before byproducts analysis
Ultrasound-assisted extraction from the TCS slurry microcosms was selected as an appropriate approach for the analysis of DBDE and various biometabolites. For the GC analysis, both solid sample and aqueous-phase sample were separated from the soil slurry microcosm by high-speed centrifugation in a PTFE centrifuge tube (Nalgene® LabWare, USA). The DBDE and other PBDE metabolites in the TCS slurry microcosms then consisted of two phases, the separated soil sample and the aqueous-phase sample. To measure the concentrations of chemicals in of the TCS solid phase sample, precisely 1.000 g was extracted using 10 mL of solvent, which consisted of hexane and acetone (1:1, v/v); this was carried out twice for 30 min using an ultrasonic water bath at room temperature. The ultrasonic bath generated output energy of 200 W at frequency of 40 kHz. The water level in the bath was adjusted to be equal to that of the extraction solvent in the PTFE centrifuge tube. The concentration of DBDE in separated aqueous-phase TCS sample was extracted by the same method as described above. For the GC/MS analyses, purer extracts were required and an extra cleaning process was introduced involving a glass chromatography column packed with acidic silica gel. In order to prepare the acidic silica gel, 60–200 mesh neutral silica gel (Merck, Darmstadt, Germany) was added to 40 % concentrated sulfuric acid (w/w) and heated at 130 °C for 16 h. The sample extract was eluted from the acidic silica gel column using 20 mL hexane and then concentrated to near dryness by rotary evaporation. Finally, the remaining residues were dissolved in the flask to give a final volume of 1 mL using hexane and then filtered through a 0.22-μm PTFE syringe filter before being used for chromatographic analysis.
DBDE analysis
The concentration of DBDE was measured by GC (HP 5890 Series II) using a pulsed discharge electron capture detector (PDECD) (Valco Models D-2-I, Schenkon, Switzerland). The DBDE analysis used an Rxi-5HT column (Restek®, USA) for separation, with selection of the column based on the literature (Chou et al. 2013). The conditions for separation were as follows: carrier gas consisting of a mixture containing 95 % (v/v) helium (99.999 % purity) and 5 % (v/v) refined methane at high purity with the carrier gas running at a constant flow rate of 10 mL/min. Finally, 3 μL of either sample extract dissolved in hexane or standard DBDE solution dissolved in hexane was injected into the GC/PDECD in the splitless mode. A range of 95 to 105 % recovery of the amount of DBDE/PBDE congeners in the soil slurry microcosms was obtained due to the analytical error and the memory effect in the GC/PDECD after dosing with DBDE. The detection limit of this method for DBDE analysis is 0.01 mg/L.
Aerobic biometabolite analysis
Aerobic metabolites were detected by two different approaches in this study. First, the 24 PBDE congeners of DBDE biodegradation were analyzed using a GC HP 6890 (Hewlett-Packard, Avondale, MA, USA) equipped with an GC PAL autosampler (Unichrom Scientific, Shanghai, China) and coupled to an HSMS-700 high-performance double focusing magnetic sector MS (JEOL, Tokyo, Japan). A DB-5HT column was used for the separation of PBDE congeners. The sample consisted of 1 mL of the sample extract or the standard solution in hexane. Helium (purity 99.999 %) was used as the carrier gas at constant column flow (1 mL/min). The MS was operated in the selected ion monitoring mode using eight descriptors to analyze the 24 PBDE congeners with the internal standard of 13C12-PBDE congeners. The detection limit of this method for PBDE congeners analysis is 0.01 μg/L. Second, aerobic biometabolites other than PBDE congeners were detected by GC/MS (Agilent 6890/5975B, USA) using the full scan mode with the range of 50–800 amu. The column used was a capillary DB-5HT (Agilent, USA). The carrier gas was helium with the 99.999 % purity at a rate of 1 mL/min. The detector temperature and electron impact mode were 320 °C and 69.9 eV, respectively. Possible metabolites detected using the GC-MS full scan mode were identified by prediction but were not quantified.
The DGGE method for biodiversity analysis
The genomic DNA of the microorganisms involved in DBDE biodegradation was extracted from the microcosm using a soil genomic DNA purification kit (Gene Mark, Taiwan). Bacterial 16S rDNA genes were selectively amplified from the purified DNA products by PCR. The V6-V8 region of 16S rDNA was selected using the forward primer 968 F-GC clamp (5′-CGC-CCG-GGG-CGC- GCC-CCG-GGC-GGG-GCG-GGG-GCA-CGG-GGG-GAA-CGC-GAA-GAA-CCT-TAC-3′) and the reverse primer 1392R (5′-ACG-GGC-GGT-GTG-TAC-3′) (Fang et al. 2002). The DNA product was separated by DGGE profiling using the DCodeTM Universal Mutation Detection System (Bio-Rad, USA) and a 35–55 % gradient gel at 60 °C and 110 V for 18 h. The DGGE electrophoresis gel consisted of 8 % acrylamide polymerised in the presence of denaturing agents, namely formamide and urea. The target DNA products were significant bands found to be in common across the DGGE profile; these were extracted, mixed with 10 μL of sterilized Milli-Q water, and then selectively amplified using the pair primer 968 F (5′-AAC-GCG-AAG-AAC-CTT-AC-3′) and 1392R. These PCR products were sequenced by the Mission Biotech Company, Taiwan. All sequences were compared with those of reference microorganisms from the GenBank database using BLAST. The closest 16S rDNA sequence to the 16S rRNA sequences obtained from the bacteria found within the biodegradation bacterial populations were retrieved.
Community-level physiological profiling
Phenotypic fingerprinting of the bacterial communities during DBDE biodegradation was performed by the Biolog GN2 microplate method. A soil/water sample from each microcosm consisting of 30 mL was separated by centrifugation at 5000 rpm for 30 min. The sample inocula were prepared by mixing 7.5 mL of centrifuged aqueous solution and 12.5 mL PBS. Each mixture of 150 μL was then added to each well of the microplate. This was followed by incubation at 25 °C. The average absorbance value of the microtiter plates after 72 h was measured at 590 and 700 nm (reference wavelength) in the MicroStation™ ID system (Biolog, USA). The CLPP was processed by principal component analysis (PCA) based on the average well color. The CLPP method consisted of the following steps. First, the average well color development (AWCD) was calculated using the absorbance values at wavelengths of 590 and 700 nm. Second, PCA was performed from the AWCD results using Ward’s method (Chang et al. 2007). The minimum eigenvalue in the PCA was set to be 0.000 in order to examine all principal component variances. Each point in the CLPP represents a BioLog pattern, and the distances between points approximate to the pattern similarities.
Detection of genes encoding hydrocarbon aromatic dioxygenases
Genomic DNA samples were assessed to detect the presence of genes encoding functional enzymes related to DBDE biodegradation. Since the chemical structure of DBDE is composed of two benzene rings, it is possible to have serial benzene ring-cleavage reactions by dioxygenases under aerobic conditions. Since the enzymes needed for DBDE biodegradation are not well understood, the functional genes involved in the aerobic biodegradation of compounds with a similar aromatic structure to POPs, such as polychlorinated biphenyl and PAH, were selected for analysis in this study. Table S1 lists primer pairs able to detect the following functional genes that are likely to be involved in DBDE biodegradation in this study. These are catechol 1, 2-dioxygenase (EC 1.13.11.1, C12O), catechol 2, 3-dioxygenase (EC 1.3.11.2, C23O), protocatechuate 3, 4-dioxygenase (EC 1.3.11.3, C34O), aromatic ring-hydroxylating dioxygenase (Nid A), and Rieske iron-sulfur dioxygenase (Rf).
Results
Biodegradation of DBDE in soil slurry microcosms
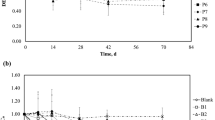
Figure 1 demonstrates the mixing aerobic/micro-aerobic biodegradation of DBDE as sole carbon source in soil slurry microcosms. The average sum concentration of DBDE consists of the amounts present in both the aqueous-phase sample and the solid-phase sample of each microcosm. Different bacterial sources resulted in different rate constants for the DBDE biodegradation. The best removal rate of DBDE in this study was by the Da-An bacterial group, which was able to degrade 80 % of the DBDE in 6 months (data not show). The rate constants for a pseudo first-order DBDE degradation reaction were found to be as follows: Da-An (0.0093/day, r 2 = 0.9977) > Nei-Hu (0.0066/day, r 2 = 0.9835) > Yi-Li (0.0054/day, r 2 = 0.9444). Aerobic biodegradation of HBN PBDE over a short period of time is usually difficult. In this study, the rate constants for the degradation of DBDE in our soil slurry microcosms are higher by 1.22- to 3.35-folds compared to previous study in similar circumstance. The aerobic degradation of DBDE followed the first-order kinetics with constant only detected between 2.77 × 10−3/day and 3.79 × 10−3/day and the half-lives of DBDE degradation ranged between 6.0 and 8.2 months (Stiborova et al. 2015).
DBDE biodegradation in a TCS/water microcosm using various mixed cultures. The initial concentration of DBDE was 25 mg kg−1. The Y-axis is the ratio (%) of the remaining concentration of DBDE divided by the initial concentration. The net concentration of DBDE is summed by adding together the separated soil sample and the aqueous sample
GC-MS biometabolites during DBDE biodegradation
Table 1 summarizes the major m/z fragments detected after DBDE biodegradation using full scan mode GC-MS in the selected microcosm samples. Major ion fragments (m/z) at the 57.0, 72.9 (or 73.0), 146.9 (or 147.0), 206.9, 280.9, 354.9 (or 355.0), and 428.9 (or 429.1) were frequently observed (see Supporting Information Fig. S2). According to the most recent mechanism of mixing aerobic/micro-aerobic biodegradation of DBDE/PBDE congers, two major reactions play an important role. These are firstly, the cleavage of benzene structure by hydroxylation, and secondly, reductive debromination (Xia 2013). It was difficult to identify unknown biometabolites in addition to the PBDE congeners due to the wide range of possibilities. For example, any bromine located on the structure of BDE-209 could be substituted by OH group due to hydroxylation under aerobic biodegradation. Some biometabolites with OH group did not have commercial standards for the GC-MS analysis. The possible metabolites associated with many of these ion fragments are predicted in this study. These include various hydroxylated-substituted brominated diphenyl ethers, namely C12H7OBr3·H2O (MW = 425), C12O3Br2H6 (MW = 358), C12H8O3Br2 (MW = 280), and C6H5O3Br (MW = 205), based on the major m/z fragments (see Supporting Information Fig. S2). However, these possible byproducts need to be identified precisely by GC-MS using appropriate standard at a future time.
PBDE congeners during DBDE biodegradation
Figure 2 shows the presence of increasing concentrations of PBDE congeners containing three to seven bromines (tri-BDE to hepta-BDE) after DBDE biodegradation for 6 months. A limited number of the PBDE congeners (ng/L level) can be found in the TCS control, but their presence does not seem to have had an effect on the biodegradation of 25 mg kg−1 DBDE in this study (see Supporting Information Fig. S1). A number of PBDE congeners, especially hepta-BDE and hexa-BDE, were found to accumulate during DBDE biodegradation. The highest concentrations found in the Yi-Li, Da-An, and Nei-Hu groups were measured during the fourth month of biodegradation and these reached maxima of 79.28, 91.07, and 77.83 ng/g, respectively. The ability to carry out DBDE bio-debromination by these bacterial groups seemed to vary during the DBDE biodegradation. For example, the Da-An group gave highest concentrations of 17.65 ng/g penta-BDE, 15.63 ng/g hexa-BDE, and 21.53 ng/g hepta-BDE during the second month; while the Nei-Hu group gave highest concentrations of 11.50 ng/g tetra-BDE and 17.30 ng/g penta-BDE during the fourth month.
Bacterial communities present during DBDE biodegradation
Figure 3 shows the DGGE profiles obtained after DBDE biodegradation during the 6 months of incubation. Table 2 shows the number of the DGGE bands detected across the three soil slurry microcosms, which acts as a measure of richness with respect to biodiversity. The bacterial populations obtained originally from the WWTP and from the river sediments showed varying levels of similarity. Furthermore, the patterns of bands during the DBDE biodegradation differed across the three microcosms over time. The DGGE bands of the Yi-Li bacterial group remained relatively constant (13-11-12 bands) over the whole microcosm biodegradation. The DGGE bands of the Da-An bacterial population were found to have increased in number from 10 bands in the third month to 12 bands in the fourth month. On the other hand, there was a decrease in the number of DGGE bands present in the Nei-Hu bacterial population over 4 months of incubation, from 12 bands in the third month to 8 bands in the fourth month. One major band was found on all of the various DGGE profiles (see Fig. 3 (d)), and thus, this bacterium seems to be present in all soil slurry microcosms during DBDE biodegradation. This organism was identified as being a Pseudomonas sp. (99 % similarity, NCBI Accession No. KC013941).
The 16S rRNA DGGE profiles obtained using DNA extracted from the mixed cultures present during DBDE biodegradation in a TCS/water microcosm. a Bands from the Yi-Li and Da-An microcosms at fourth months that are in the same position and thus probably very similar in sequence based on the DGGE analysis. b Bands from the Da-An and Nei-Hu microcosms at fourth months that are in the same position and thus probably very similar in sequence based on the DGGE analysis. c Bands from the Nei-Hu and Yi-Li microcosms at fourth months that are in the same position and thus probably very similar in sequence based on the DGGE analysis. d The brightest DGGE band present in all samples was identified as a Pseudomonas sp.
CLPP and hydrocarbon aromatic oxygenase gene analysis
The maximum utilization of the 95 Biolog carbon sources during POPs biodegradation was determined in a manner similar to that of a previous study (Chang et al. 2007). In this study, maximum utilization of Biolog carbon sources was found to be 80 for the Yi-Li bacterial population. There was significant variation in the Biolog utilization of carbon sources across the bacterial populations, and this involved both polymers and carboxylic acid. The Da-An bacterial population and the Nei-Hu bacterial population gave lower maximum utilization of carbon source values of 74 and 76, respectively. Figure 4 demonstrates the CLPP findings using two-dimensional PCA with a good explanatory value (88.97 %). The three bacterial populations can be seen to be initially part of the same group (group 1) within the profile. The Da-An and the Nei-Hu continue to demonstrate similar patterns (group 2) for their profiles during DBDE biodegradation over time. However, the Yi-Li bacteria population diverges over time in terms of utilization of the Biolog GN2 carbon sources and becomes an independent group (group 3) within the profile.
Genes encoding C12O, C23O, C34O, Nid A, and Rf are well known to be involved in the biodegradation of aromatic hydrogen compounds and have been shown to bring about the oxidative cleavage of catechol and LBN PBDE congers in a number of different species. In this study, positive PCR reactions for C23O during the third and fourth months suggest that this enzyme may be involved in the aerobic biodegradation of DBDE/PBDE congeners in all microcosms (Supporting Information Fig. S3). On the other hand, the genes encoding C12O, C34O, NidA, and Rf were not detected by PCR as being present in the three different bacterial populations at any time during the aerobic DBDE biodegradation process.
Discussion
Biometabolite analysis of the soil slurry microcosms
Different reductive metabolized products were detected during the DBDE biodegradation in the microcosm. Specifically, biological reductive debromination seems to occur in the micro-aerobic environment of soil slurry microcosms. Similar to the situation with polychlorinated biphenyl, HBN-PBDE congeners (including DBDE) are usually biodegraded anaerobically (Ding et al. 2013). Some facultative bacteria are able to debrominate DBDE/PBDE congeners in micro-environments such as the soil particles in our microcosms. For example, previous studies have demonstrated that octa-BDE and hepta-BDE congeners are present when the aerobic debromination of DBDE is taking place (Deng et al. 2011). A pure bacterial species Pseudomonas aeruginosa was identified as being involved in the main aerobic transformation mechanism of BDE-209, which is debromination (Shi et al. 2013). Aerobic biotransformation of LBN-PBDE congeners has been found to be proportional to the degree of debromination (Robrock et al. 2009). Furthermore, based on our knowledge of the aerobic biodegradation of halogenated aromatic compounds, the bromines within PBDE congeners usually may be replaced by a number of different functional groups, for example, −OH, −CH3, and −OCH3. For example, a pure culture of Sphingomonas sp. PH-07 strain was found to be able to aerobically degrade LBN-PBDE congeners to a number of predicted metabolites such as bromophenol, monohydroxy-bromodiphenyl ether, dihydroxy-bromodiphenyl ether, and dibromophenol (Kim et al. 2007). Similarly, Burkholderia xenovorans LB400 has been shown to have the ability to biodegrade a mono-BDE to a hydroxylated mono-BDE metabolite (Robrock et al. 2009). In the present study, soil slurry microcosms were found to increase significantly the degradation rate of DBDE and to generate a complex mixture of biometabolites, including bromobenzene, brominated phenol, bromocatechol, and dihydroxy-diphenyl ether.
Serial chemical reactions, such as those associated with aerobic bio-oxidation reactions and biological reductive debromination, are expected to occur in parallel using the system explored in this study. It can be seen from our results that the concentration of PBDE congeners varied across all samples during DBDE biodegradation. For example, the concentration of hexa-BDE in 0 month was 5–8 ng/g while it was only 0–2.5 ng/g in second month. The concentration of hepta-BDE after second month was also lower than the result for the control at 0 months. The hexa-BDE and hepta-BDE could be debrominated to other LBN-BDE or converted to various hydroxylated metabolites in parallel. The amounts of the hexa-BDE and hepta-BDE present at the second month are lower than that at 0 months in this study. Unfortunately, the full-scan GC-MS byproducts found in this study cannot be completely identified by theoretical prediction based on their m/z profiles obtained. This is in agreement with previous findings whereby unbalance amounts of bromide were also detected during the aerobic biodegradation of mono-BDE and di-BDE by specific bacteria (Robrock et al. 2009). Carrying out a mass balance of the transformation of DBDE to PBDE as part of this study is not possible; this is because only a small amount of PBDE congeners was found compared to the theoretical yield if all the added DBDE is considered. The total concentration of all PBDE congeners was found to be highest at the end of experiments (sixth month) and to be in the range 15.42 to 24.09 ng/g. Thus, it is also possible that DBDE is converted to a number of unknown brominated metabolites, which are then utilized by a range of different catabolic pathways. Similar results, whereby bromine-containing unknown byproducts were identified, occurred during the biodegradation of 100 μg/L DE-71, which is a penta-BDE mixture consisting of BDE-47, BDE-99, BDE-100, BDE-85, BDE-154, and BDE-155; identification in this study involved GC-TOF-MS and ICP-MS (Vonderheide et al. 2006). In other systems, the increase in bromide that occurs during DBDE biodegradation has been found not to parallel the decrease in DBDE that occurs (Gerecke et al. 2006; Chou et al. 2013).
Biodiversity and the bacterial physiological characteristics involved in DBDE biodegradation
The different soil slurry microcosms have different abilities to remove DBDE, and these differences in bacterial populations are reflected in the PCR-DGGE analysis. Interestingly, the DGGE patterns of the three bacterial populations changed significantly during DBDE biodegradation and the new patterns remained distinct. Some bands disappeared and some bands remained constant in the DGGE profile. For example, at the fourth month of biodegradation, the Yi-Li bacterial population and the Da-An bacterial population can be seen to have seven bands in common during biodegradation (see Fig. 3 (a)), while, in contrast, the Da-An bacterial population and the Nei-Hu bacterial population seemed to have only three bands in common (see Fig. 3 (b)), while the Yi-Li bacterial population and Nei-Hu bacterial population seemed to have only two bands in common (see Fig. 3 (c)). Towards the beginning of DBDE biodegradation, the Yi-Li, Da-An, and Nei-Hu populations were found to have eight, four, and four bands in common, respectively. This implies that the microbial populations were different when initially sampled and, despite this, all three bacterial populations were able to biodegrade DBDE/PBDE congers.
The Biolog system has been reported to be suitable for the monitoring of microbial community in terms of functional potential during environmental change (Frac et al. 2012). When compared, the biodiversity richness index does not seem to be significantly related to performance during DBDE biodegradation as measured by CLPP, which may be because three distinct bacterial populations were present at the beginning of DBDE biodegradation and various distinct and different strains seem to have been selected during biodegradation. A previous study has shown that the presence of 1 ng/g of DBDE does affect the proteobacterial population of contaminated soils (Zhu et al. 2010). PBDE is known to change the composition of bacterial communities, and this is also true for the various PBDE congeners that are present (Yen et al. 2009). When the three microcosms are classified using CLPP, the changes in bacterial community seem to be irrelevant. For example, the biodiversity of the Yi-Li bacterial population is greater than that of the other two bacterial populations, and this might imply that the bacterial species within the Yi-Li population should be able to utilize more of the 95 carbon sources provided on the Biolog GN2 microplates, even though the DBDE biodegradation rate of this population is not the best; however, this was found not to be true. It is important to note that, in general, the biomass and respiration of bacterial populations are lower when their physiological activities are inhibited by pollutants that they cannot utilize (Bǔnemann et al. 2004). On the other hand, there is physiological stimulation when pollutants are able to contribute to bacterial growth. Specific bacteria within the Yi-Li population were able to perform well in the presence of DBDE as measured by CLPP. Furthermore, while the Da-An and Nei-Hu bacterial populations utilized different resources, they had few species in common; nevertheless, their CLPP findings are similar. It would seem that, because of the relatively high utilization of Biolog 95 carbon sources by the three bacterial populations in this study, ecological toxicity is not a significant factor during DBDE biodegradation.
Facultative Pseudomonas spp., which were common to all three microcosms, are able to be active under micro-aerobic environment in the microcosm system. Such facultative species are able to easily degrade DBDE under different oxygen-reduction profiles (ORPs) with different biochemical reactions, including biological debromination and hydroxylation. It can be inferred that there was no significant influence on the efficiency of the aerobic degradation by these facultative species. Many Pseudomonas species have been identified as present and widely distributed in DBDE/PBDE congers-contaminated soil. For example, an isolated stain of Pseudomonas stutzeri has been shown to be able to utilize BDE-47 as a sole carbon source and bring about 97.94 % biodegradation after 2 weeks (Zhang et al. 2013). Pseudomonas spp. were found to be the most dominant species in the soil after spiking with BDE-209 and BDE-15 at 1, 10, and 100 mg/kg for 180 days (Liu et al. 2011). Aerobic degradation of the mixture of 50 mg/kg PBDE congeners including BDE-15 and BDE-28, BDE-47, BDE-99, and BDE-100 was occurred by Pseudomonas sp., isolated from river sediment (Yang et al. 2015). In addition, the pure C23O enzyme obtained from Pseudomonas cepacia Et 4 is able to aerobically biodegrade the possible end metabolic byproducts of DBDE/PBDE congeners biodegradation, including 2,3-dihydroxydiphenyl ether, catechol, and 3-substituted catechols (Pfeifer et al. 1993).
The mechanism by which DBDE biodegradation takes place in soil slurry microcosms
Figure 5 outlines the possible mechanism of DBDE biodegradation in our soil slurry microcosms and is based on the formation of biometabolites and signals obtained for encoded hydrocarbon aromatic genes related to functional enzymes, namely specific monooxygenases or dioxygenases in this study. The pathway of DBDE biodegradation is likely to be complex in soil slurry microcosms. Initially, biological reductive debromination seems to be the major mechanism involved in the aerobic biotransformation of DBDE. Various amounts of PBDE congeners, including HBN to LBN PBDE congeners, are gradually generated in the oxygen-free or micro-oxygen microenvironment of the soil slurry. These LBN PBDE metabolites might also have been debrominated to diphenyl ether by reductive dehalogenation of the PBDE congeners. Second, the aromatic cleavage of LBN PBDE seems then to occur via a series of reactions involving hydroxylation. Various aerobic bromine-contained metabolites, including hydrolated brominated diphenyl ether, brominated phenols, and brominated catechol, are then detected as being present during the present study. For example, aerobic microorganisms in microcosms seem to degrade LBN PBDE congeners using the 2,3-dioxygenases present in the genomes of the various organism present in the microcosms based on the exact locations of 2, 3-unbrominated carbons and a meta-cleavage of DBDE/PBDE congeners during aerobic hydroxylation in this study. A wide variety of 2,3-dioxygenase genes have been reported from different bacteria involved in the biodegradation of PBDE congeners. For example, an increase in expression of 30-fold to 3000-fold was found for biphenyl and ethylbenzene dioxygenases during the transformation of mono- through penta-BDE congeners (Robrock et al. 2011). Third, debromination seems to continually occur via attacks on the various bromine-contained metabolites present, which are then metabolized into aromatic hydrocarbon products with a simpler structure (e.g., benzoate, catechol, etc.). Finally, eventual mineralization of less toxic aromatic hydrocarbon products by C23O is expectedly to occur via the TCA cycle or the Entner-Doudoroff pathway.
Proposed mechanisms for DBDE biodegradation in the soil slurry microcosms. Asterisks indicate identified biometabolites in this study. Solid lines indicate identified pathway in this study. Dotted lines indicate proposed pathway from other references (Xia 2013). C23O = catechol 2,3-oxygenase
Conclusions
This research demonstrates that soil slurry microcosms can be an effective approach to DBDE biodegradation. A possible mechanism for DBDE biodegradation in soil slurry microcosms is proposed, and this includes biological debromination and hydroxylation. Genes encoding specific enzymes that may be involved in DBDE biodegradation were identified by PCR. C23O were found to be present in the genomes of the bacteria making up all three bacterial populations. These enzymes have been shown previously to be involved in the benzene cleavage of DBDE/PBDE congeners. Many byproducts, including brominated products, dihydroxydiphenyl ether, and others were identified as PBDE congeners in the soil slurry microcosms, suggesting that a mixture of aerobic and micro-aerobic conditions was present. Ecological toxicity does not seem to be a significant factor in these systems based on the fact that a wide range of carbon sources was found to be utilized during the DBDE biodegradation. Laboratory scale soil slurry microcosms are thus useful for demonstrating processes that could occur when this process is carried out on a larger scale. The findings of this research will help to evaluate the possibility of using a practical soil slurry microcosm for DBDE remediation. Nevertheless, more in-depth studies are needed in the near future. These should include, for example, experiments where DBDE bioremediation is executed using in situ or ex situ soil slurry microcosms. Such systems would also be improved by the enrichment and isolation of the specific species of bacteria that are involved in the degradation of DBDE and application of these isolates to bioremediation in natural environments contaminated by DBDE. Furthermore, the microbial community diversity changes that occur in these systems need to be monitored in depth using a molecular approach.
Abbreviations
- AWCD:
-
Average well color development
- CLPP:
-
Community-level physiological profiling
- DBDE:
-
Decabromodiphenyl ether
- DGGE:
-
Denaturing gradient gel electrophoresis
- HBN:
-
High bromine number
- HRMS:
-
High resolution mass spectrometer
- LBN:
-
Low bromine number
- MSB:
-
Inorganic salt growth medium
- PBDE:
-
Polybrominated diphenyl ethers
- PCA:
-
Principal component analysis
- PDECD:
-
Pulsed discharge electron capture detector
- POPs:
-
Persistent organic pollutants
- SOM:
-
Soil organic matte
- TCS:
-
Taichung natural soil
References
Albina ML, Alonso V, Linares V, Bellés M, Sirvent JJ, Domingo JL, Sánchez DL (2010) Effects of exposure to BDE-99 on oxidative status of liver and kidney in adult rats. Toxicology 271:51–56
Bǔnemann EK, Bossio DA, Smithson PC, Frossard E, Oberson A (2004) Microbial community composition and substrate use in a highly weathered soil as affected by crop rotation and P fertilization. Soil Biol Biochem 36:889–901
Chang Y-T, Lee J-F, Hung C-H (2007) PAH biodegradation in surfactant-water systems based on the theory of cohesive energy density (CED). J Chem Technol Biotechnol 82:442–452
Chen C, Zhao H, Chan J, Qiao X, Xie Q, Zhang Y (2012a) Polybrominated diphenyl ethers in soils of the modern Yellow River delta, China: occurrence, distribution and inventory. Chemosphere 88:791–797
Chen Q, Yu L, Yang L, Zhou B (2012b) Bioconcentration and metabolism of decabromodiphenyl ether (BDE-209) result in thyroid endocrine disruption in Zebrafish Larvae. Aquat Toxicol 110–111:141–148
Chou H-L, Chang Y-T, Liao Y-F, Lin C-H (2013) Biodegradation of decabromodiphenyl ether (BDE-209) by bacterial mixed cultures in a soil/water system. Int Biodeterior Biodegrad 85:671–682
Cincinelli A, Martellini T, Misuri L, Sweetman E, Laschi S, Palchetti I (2012) PBDEs in Italian sewage sludge and environmental risk of using sewage sludge for land application. Environ Pollut 161:229–234
Deng D, Guo J, Sun G, Chen X, Qiu M, Xu M (2011) Aerobic debromination of Deca-BDE: isolation and characterization of an indigenous isolate from a PBDE contaminated sediment. Int Biodeterior Biodegrad 65:465–469
Ding C, Chow WL, He J (2013) Isolation of Acetobacterium sp. strain AG, which reductively debrominates octa- and pentabrominated diphenyl ether technical mixtures. Appl Environ Microbiol 79:1110–1117
Fang HHP, Zhang T, Liu H (2002) Microbial diversity of a mesophilic hydrogen-producing sludge. Appl Microbiol Biotechnol 58:112–118
Fangstrom B, Hovander L, Bignert A, Athanassiadis I, Linderholm L, Grandjean P, Weihe P, Bergman A (2005) Concentrations of polybrominated diphenyl ethers, polychlorinated biphenyls, and polychlorobiphenylols in serum from pregnant faroese women and their children 7 years later. Environ Sci Technol 39:9457–9463
Frac M, Karolina O, Lipiec J (2012) Community level physiological profiles (CLPP), characterization and microbial activity of soil amended with diary sewage sludge. Sensors 12:3253–3268
Frederiksen M, Vorkamp K, Mathiesen L, Mose T, Knudsen LE (2010) Research placental transfer of the polybrominated diphenyl ethers BDE-47, BDE-99 and BDE-209 in a human placenta perfusion system: an experimental study. Environ Heal 9:32
Gerecke AC, Giger W, Hartmann PC, Heeb NV, Kohler H-PE, Schmid P, Zennegg M, Kohler M (2006) Anaerobic degradation of brominated flame retardants in sewage sludge. Chemosphere 64:311–317
Gevao B, Ghadban AN, Uddin S, Jaward FM, Bahloul M, Zafar J (2011) Polybrominated diphenyl ethers (PBDEs) in soils along a rural–urban-rural transect: sources, concentration gradients, and profiles. Environ Pollut 159:3666–3672
Kim Y-M, Nam I-H, Murugesan K, Schmidt S, Crowley DE, Chang Y-S (2007) Biodegradation of diphenyl ether and transformation of selected brominated congeners by Sphingomonas sp. PH-07. Appl Microbiol Biotechnol 77:187–194
Li Y, Lin T, Chen Y, Hu L, Guo Z, Zhang G (2012) Polybrominated diphenyl ethers (PBDEs) in sediments of the costal East China Sea: occurrence, distribution and mass inventory. Environ Pollut 171:156–161
Liang SX, Gao HX, Zhao YY, Ma XM, Sun HW (2010) Effects of repeated exposure to decabrominated diphenyl Ether (BDE-209) on mice nervous system and its self-repair. Environ Toxicol Pharmacol 29:297–301
Liu L, Zhu W, Xiao L, Yang L (2011) Effect of decabromodiphenyl ether (BDE 209) and dibromodiphenyl ether (BDE 15) on soil microbial activity and bacterial community composition. J Hazard Mater 186:883–890
Mackay D, Shiu WY, Ma K-C, Lee SC (2006) Handbook of physical-chemical properties and environmental fate for organic chemicals. Volume II-Halogenated Hydrocarbons, 2nd edn. CRC Press, Taylor & Francis Group, FL
Moon HB, Choi M, Yu J, Jung RH, Choi HG (2012) Contamination and potential sources of polybrominated diphenyl ethers (PBDEs) in water and sediment from the artificial lake Shihwa, Korea. Chemosphere 88:837–843
Nyholm JR, Lundberg C, Andersson PL (2010) Biodegradation kinetics of selected brominated flame retardants in aerobic and anaerobic soil. Environ Pollut 158:2235–2240
Pfeifer F, Trüper HG, Klein J, Schacht S (1993) Degradation of diphenyl ether by Pseudomonas cepacia Et4: enzymatic release of phenol from 2, 3-dihydroxydiphenyl ether. Arch Microbiol 159:323–329
Qiu M, Chen X, Deng D, Guo J, Sun G, Mai B, Xu M (2012) Effects of electron donors on anaerobic microbial debromination of polybrominated diphenyl ethers (PBDEs). Biodegradation 23:351–361
Robrock KR, Coelhan M, Sedlak DL, Alvarez-Cohen L (2009) Aerobic biotransformation of polybrominated diphenyl ethers (PBDEs) by bacterial isolates. Environ Sci Technol 43:5705–5711
Robrock KR, Mohn WW, Eltis LD, Alvarez-Cohen L (2011) Biophenyl and ethylbenzene dioxygenases of Rhodococcus jostii RHA1 transform PBDEs. Biotechnol Bioeng 108:313–314
Saegusa Y, Fujimoto H, Woo GH, Ohishi T, Wang L, Mitsumori K, Nishikawa A, Shibutani M (2012) Transient aberration of neuronal development in the hippocampal dentate gyrus after developmental exposure to brominated flame retardants in rats. Arch Toxicol 86:1431–1442
Shao J, White CC, Dabrowski MJ, Kavanagh TJ, Eckert ML, Gallagher EP (2008) The role of mitochondrial and oxidative injury in BDE 47 toxicity to human fetal liver hematopoietic stem cells. Toxicol Sci 101:81–90
Shi G, Yin H, Ye J, Peng H, Li J, Luo C (2013) Aerobic biotransformation of decabromodiphenyl ether (PBDE-209) by Pseudomonas aeruginosa. Chemosphere 93:1487–1493
Stiborova H, Vrkoslavova J, Lovecka P, Pulkrabova J, Hradkova P, Hajslova J, Demnerova K, Stiborova (2015) Aerobic biodegradation of selected polybrominated diphenyl ethers (PBDEs) in wastewater sewage sludge. Chemosphere 118:315–321
Tokarz JA, Ahn MY, Leng J, Filley TR, Nies L (2008) Reductive debromination of polybrominated diphenyl ethers in anaerobic sediment and a biomimetic system. Environ Sci Technol 42:1157–1164
Toms L-ML, Mortimer M, Symons RK, Paepke O, Mueller JF (2008) Poly-brominated diphenyl ethers (PBDEs) in sediments by salinity and land-use type from Australia. Environ Int 34:58–66
U.S. EPA (2008) (Environmental Protection Agency), Toxicological review of decabromodipheny ether (BDE-209), EPA/635/R-07/008F, Washington DC
Viberg H, Fredriksson A, Jakobsson E, Orn U, Eriksson P (2003) Neurobehavioral derangements in adult mice receiving decabrominated diphenyl ether (PBDE 209) during a defined period of neonatal brain development. Toxicol Sci 76:112–120
Vonderheide AP, Mueller-Spitz SR, Meija J, Welsh GL, Mueller KE, Kinkle BK, Shanna JR, Caruso JA (2006) Rapid breakdown of brominated flame retardants by soil microorganisms. J Anal At Spectrom 21:1232–1239
Wang P, Zhang Q-H, Wang T, Chen W-H, Ren D-W, Li Y-M, Jiang G-B (2012) PCBs and PBDEs in environmental samples from King George Island and Ardley Island Antarctica. RSC Adv 2:1350–1355
Wei HA, Aziz-Schwanbeck C, Zou Y, Corcoran MB, Poghosyan A, Li A, Rockne KJ, Christensen ER, Sturchio NC (2012) Polybromodiphenyl ethers and decabromodiphenyl ethane in aquatic sediments from southern and eastern Arkansas. U.S.A. Environ Sci Technol 46:8017–8024
Xia X (2013) Microbial degradation of polybrominated diphenyl ethers: current and future. J Bioremed Biodeg 4:e130
Yang C-W, Huang H-W, Chao W-L, Chang B-V (2015) Bacterial communities associated with aerobic degradation of polybrominated diphenyl ethers from river sediments. Environ Sci Pollut Res 22:3810–3819
Yen JH, Liao WC, Chen WC, Wang YS (2009) Interaction of polybrominated diphenyl ethers (PBDEs) with anaerobic mixed bacterial cultures isolated from river sediment. J Hazard Mater 165:518–524
Yogui GT, Sericano JL (2009) Polybrominated diphenyl ether flame retardants in the U.S. marine environment: a review. Environ Int 35:655–666
Zhang S, Xia X, Xia N, Wu S, Gao F, Zhou W (2013) Identification and biodegradation efficiency of a newly isolated 2, 2’, 4, 4’-tetrabromodiphenyl ether (BDE-47) aerobic degrading bacterial strain. Int Biodeterior Biodegrad 76:24–31
Zhu W, Liu L, Zou P, Xiao L (2010) Effect of decabromodiphenyl ether (BDE 209) on soil microbial activity and bacterial community composition. World J Microbiol Biotechnol 26:1891–1899
Acknowledgments
The financial support for this study was provided by the Ministry of Science and Technology (previously National Science Council), Taiwan, and this is gratefully acknowledged. The project numbers are NSC99-2627-B- 031–002 and MOST102-2221-E-031-001-MY2. Part of the results/discussions was presented at the 1st International Conference on Emerging Contaminants, Kaohsiung, in Taiwan on October, 2013. The authors thank Mrs. Liao, Yi-Fen for her great efforts during the experiments.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interests
The authors have declared no conflict of interest.
Additional information
Responsible editor: Philippe Garrigues
Rights and permissions
About this article
Cite this article
Chou, HL., Hwa, MY., Lee, YC. et al. Microbial degradation of decabromodiphenyl ether (DBDE) in soil slurry microcosms. Environ Sci Pollut Res 23, 5255–5267 (2016). https://doi.org/10.1007/s11356-015-5767-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11356-015-5767-x