Abstract
Stressors of various kinds constantly affect fish both in the wild and in culture, examples being acute water temperature and quality changes, predation, handling, and confinement. Known physiological responses of fish to stress such as increases in plasma cortisol and glucose levels, are considered to be adaptive, allowing the animal to cope in the short term. Prolonged exposure to stressors however, has the potential to affect growth, immune function, and survival. Nonetheless, little is known about the mechanisms underlying the long-term stress response. We have investigated the metabolic response of juvenile Atlantic salmon (Salmo salar) to long-term handling stress by analyzing fish plasma via 1H nuclear magnetic resonance spectroscopy and ultra high performance liquid chromatography–mass spectrometry (UPLC–MS), and comparing results with controls. Analysis of NMR data indicated a difference in the metabolic profiles of control and stressed fish after 1 week of stress with a maximum difference observed after 2 weeks. These differences were associated with stress-induced increases in phosphatidyl choline, lactate, carbohydrates, alanine, valine and trimethylamine-N-oxide, and decreases in low density lipoprotein, very low density lipoprotein, and lipid. UPLC-MS data showed differences at week 2, associated with another set of compounds, tentatively identified on the basis of their mass/charge. Overall the results provided a multi-faceted view of the response of fish to long-term handling stress, indicating that the metabolic disparity between the control and stress groups increased to week 2, but declined by weeks 3 and 4, and revealed several new molecular indicators of long-term stress.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Fish in the wild or under intensive culture are constantly challenged by stressful events such as fluctuations in water quality parameters, e.g., temperature, dissolved oxygen, and pH, and exposure to contaminants, e.g., heavy metals and pesticides. In addition, fish in intensive culture can be subjected to several forms of physical insults including handling, crowding, confinement, and transport. The animals’ ability to cope with the insult will, to some extent, depend on the type, intensity and duration of the stressor (Iwama et al. 2005). Typically, fish will respond to stress by eliciting physiological responses (Barton and Iwama 1991) that are generally adaptive and help the animal to deal with the insult. Responses to stress include neuroendocrine, metabolic, and cellular changes (Bonga 1997). Plasma cortisol, catecholamine, glucose, lactate, and liver and muscle glycogen are common indicators of stress (Barton and Iwama 1991) and their relative concentrations in the plasma can be readily measured using standard techniques. Unfortunately some of the metabolites secreted into the plasma in response to stress can exceed threshold levels that are required for the normal physiological coping mechanisms and can lead to debilitating effects. For example it has been shown that high levels of metabolites such as plasma cortisol have immunosuppressive effects (Pickering and Pottinger 1989), consequently increasing the susceptibility of fish to disease.
Despite extensive studies on fish responses to acute stressors (Barton 2002; Barton and Iwama 1991), the mechanism and consequence of metabolic changes in fish exposed to chronic stress is poorly characterized. Nonetheless, it has been shown that long-term stress can have a negative impact on growth, resistance to pathogens and reproduction (Barton 2002; Hosoya et al. 2007). Eventually, fish will acclimate to long-term stress, and the classical indicators of stress such as plasma cortisol and glucose levels will return to normal (Barton et al. 1987; Pickering and Pottinger 1987). It has also been demonstrated that cortisol, glucose, and growth responses to long-term stress vary with the species. For instance, long-term term stress affects growth in haddock (Melanogrammus aeglefinus) and fish exhibit an increase in plasma cortisol (but not glucose) only during the second week of stress (Hosoya et al. 2007). In contrast, Atlantic salmon (Salmo salar) subjected to similar stress show increased plasma glucose (but not cortisol) during the first week of stress while growth remains unaffected (Fast et al. 2008). Juvenile rainbow trout (Oncorhynchus mykiss) subjected to an acute stress daily for 10 weeks did not show changes in plasma cortisol and glucose levels or growth throughout the study (Barton et al. 1987). Typically, although a stress episode that occurs repeatedly and lasts more than a few seconds is considered chronic stress (or long-term stress), there is no strict demarcation of the duration at which stress is considered long term. Various references suggest different time lengths ranging from 4 (Fast et al. 2008; Hosoya et al. 2007) to 10 (Barton et al. 1987) weeks in different fish species. Nonetheless, the classical approaches for quantifying stress remain the same.
Due to the unidimensional nature of classical methods for quantifying stress in fish, there is a risk that the measured levels of, for example, total and free plasma cortisol and glucose do not represent the whole set of organismal responses that are elicited in stressed states. In recent years however, molecular biology has witnessed a general movement towards high-throughput bio-techniques that are designed to identify and measure simultaneously the dynamics of multiple molecular components of organisms under various physical/chemical disturbances (Brown and Botstein 1999; Nicholson et al. 1999; Yates 2000). These techniques offer the potential to discover novel biomarkers of physiological perturbations to a biological system and have been successfully utilized as biomolecular markers of disease (Griffin and Kauppinen 2007; Li et al. 2002; Perou et al. 2000), toxicity (Aardema and MacGregor 2002; Coen et al. 2004; Lee et al. 2007; Viant et al. 2005; Waters et al. 2006), exposure to adverse physical and/or (bio)chemical environments (Azmi et al. 2005; Malmendal et al. 2006; Tanaka et al. 2007; Viant et al. 2003), and for bacterial characterization (Mashego et al. 2007; Soga et al. 2002; Vaidyanathan et al. 2002). In this work, we take advantage of one such technique, metabolomics, to profile fish plasma using two analytical platforms: 1H nuclear magnetic resonance (1H-NMR) spectroscopy and ultra high performance liquid chromatography–mass spectrometry (UPLC–MS).
The advantages of employing these two complementary techniques in metabolomics are well understood (Defernez et al. 2004; Lenz et al. 2004; Lindon et al. 2004). Advantages of NMR-based metabolomics include: signals that are determined by chemical structure and characteristic of particular compounds, high signal-to-noise ratio, uniform linear response of signal to analyte concentration, minimum sample preparation, and non-destruction of the samples. These advantages can be complemented by the UPLC-MS based metabolomic approach which offers rapid, sensitive and selective qualitative and quantitative analyses as well as the possibility to extend the range of compounds detected by using different ion sources (such as electrospray ionization and atmospheric chemical ionization) and/or ion modes (positive and negative) to reflect the complexity of biological samples. The unique sensitivity of MS also offers the potential to detect trace-level metabolites and provide structural information on unknown metabolites by the use of tandem MS. Both NMR and UPLC-MS are amenable to well-established multivariate statistical methods for exploratory data analysis.
1H nuclear magnetic resonance spectroscopy has been used for characterization of fluctuations in levels of small molecular weight (MW) metabolites in fish plasma resulting from exposure to pathogens (Dacanay et al. 2006; Solanky et al. 2005) and toxins (Samuelsson et al. 2006). Other NMR-based metabolomics studies on fish have mainly examined tissue homogenates for potential molecular indicators of toxicity (Viant et al. 2005) and environmental stress (Viant et al. 2003). MS has been applied for metabolomic profiling of liver extracts from fish (Stentiford et al. 2005) but has not been attempted previously with fish plasma. In this work, we profile by NMR and MS several free metabolites in plasma of juvenile Atlantic salmon (S. salar) subjected to long-term handling stress, with the goal of identifying a set of metabolites that, independently or mutually, can be used as bio-molecular indicators of the chronic stress. In line with previous work (Fast et al. 2008), 4 weeks of 15 s daily emersion of fish from water will be referred to, in this work, as chronic or long-term handling stress. Plasma was chosen due to the ease with which it can be obtained in an aquaculture setting, eliminating the need for depleting stock for purposes of routine monitoring. Sample volumes used for NMR (8.5 μl) were considerably smaller than those previously employed for such analyses.
The combination of NMR and MS techniques should provide diverse and comprehensive profiles of low MW metabolites present in the plasma resulting from long-term handling stress. These should reflect the interplay between metabolites resulting from enzymatic activity, and the physiological pathways that may be affected by chronic stress.
2 Materials and methods
2.1 Stress study and sampling
Protocols for animal handling were conducted according to the Canadian Council of Animal Care (CCAC) guidelines and were approved by the relevant Local Animal Care Committees. Juvenile Atlantic salmon were reared at the Marine Research Station of the National Research Council of Canada Institute for Marine Biosciences (NRC-IMB) in Ketch Harbour, NS, Canada. The fish were initially held in two 500-l flow-through (20–25 l/min) holding tanks supplied with seawater (temperature 10 ± 1°C; oxygen saturation > 90%), and fed twice daily (8:00 a.m. and 3:00 p.m.) with a commercial Atlantic salmon diet (EWOS, Vancouver, BC, Canada) at a rate of 1.5% of average body mass. Three weeks prior to experimentation, fish (weighing an average of 144.4 ± 20 g and measuring an average of 25.7 ± 2 cm long) were transferred to eight 120-l experimental tanks (20 fish/tank) and allowed to acclimate to the experimental system under the conditions described above with a daily photoperiod of 10 h light:14 h dark.
The eight tanks were randomly assigned as control or stressed groups, and there were four replicate tanks in each treatment. Following the acclimation period, fish in the stress group were subjected to a daily handling stress by netting them out of water daily for 15 s for 4 weeks. This procedure was carried out randomly either at 8:00 a.m. or 3:00 p.m., to prevent the fish from anticipating the stressor. Feedings were usually done at 8:00 a.m. and 3:00 p.m., but the timing of one of the feedings was adjusted so that it occurred 1 h after handling. During this study, fish (three per replicate tank) were sampled prior to initiation of the stress protocol (0 week) and 1, 2, 3, and 4 weeks after it began. Samples were collected just prior to the handling stress of the day, and were obtained at least 18 h after the last stress was applied. Unfortunately at week 4, two samples in the experimental group (tank 2 of 4) were found dead and therefore only 22 samples were obtained for this week.
To obtain blood samples, three fish from each tank were quickly captured, and immediately placed in a bucket containing a lethal dose (400 g/l) of tricaine methanesulfonate (Syndel Laboratories Ltd., Vancouver, BC, Canada). Blood samples were collected from the caudal vasculature immediately after the fish were anesthetized, using a 5-ml heparinized syringe (23-gauge needle). The blood was transferred to a 4-ml Vacutainer tube (Becton, Dickinson and Company, Franklin Lakes, NJ, USA) containing 60 IU of heparin, and stored on ice prior to centrifugation. Plasma was subsequently separated by centrifugation (3,000 × g for 15 min) at 4°C, and was then aliquoted into 1.5-ml microcentrifuge tubes. Samples were stored at −80°C until subsequent analysis. Weights and lengths of fish were recorded.
2.2 1H-NMR sample preparation
Aliquots (17 μl) of plasma were placed in 1.5-ml Eppendorf tubes, to which 3 μl of 10 mg/ml sodium 3-trimethylsilyl-2,2,3,3-d4-propionate (TMSP) in D2O was added and vortex-mixed. These samples were then returned to a liquid nitrogen-cooled storage Dewar flask at ca −193°C until immediately before NMR analysis, when they were removed and kept on dry ice. Individually, samples were taken from the dry ice and thawed before pipetting 10 μl into a Bruker 1-mm NMR sample tube for spectral acquisition.
2.3 NMR spectroscopy
One-dimensional 1H-NMR spectra were acquired at 4°C using a Bruker TXI 500 MHz 1-mm probe with a z-axis gradient on a Bruker Avance-DRX 500 spectrometer operating at 500.13 MHz. Whereas automatic tuning and matching was applied only to the first sample run on a particular day, each sample was shimmed individually and the receiver gain held constant for all samples. 1D 1H-NMR spectra were acquired using a 9.5-μs (90°) pulse calibrated with a 360° pulse, 7.5-kHz spectral width and a 2-s relaxation delay with water pre-saturation (PS) using a CW irradiation attenuation of 65 dB, with 128 transients and four dummy scans collected into 32-K data points. These were later zero-filled to 64 K and an exponential line-broadening of 1 Hz applied before Fourier transformation. Following manual zero- and first-order phasing, resulting spectra were calibrated (TMSP at δ = 0.0 ppm) using XWINNMR (version 2.6) before being exported to MATLAB 7.1 (MathWorks®, Natick, MA, USA) using ProMetab 1.0 software (Viant 2003) for further processing. In addition, a T2-edited Carr–Purcell–Meiboom–Gill (CPMG) sequence with PS and a 1-ms separation of 180° pulses was used to obtain another set of spectra emphasizing lower MW compounds having longer T2 relaxation times (Solanky et al. 2005).
2.4 Characterization of sample stability
The stability of fish plasma at room temperature was characterized by acquiring 15 1H-NMR spectra of plasma from each of five healthy juvenile Atlantic salmon weighing an average of 160 g (referred to as biological replicates) as described below. From each fish, ~5 ml of blood plasma was obtained and placed in a storage Dewar flask above liquid nitrogen at ~−193°C. A random sample was then taken from the liquid nitrogen and thawed for ~1 h before final preparation for NMR analysis. Subsequently, 15 sub-sample replicates (technical replicates) were created from this biological replicate and prepared as described above in the sample preparation section. A total of 75 samples were prepared; 15 technical replicates for each of the five biological replicates. These samples (hereafter referred to as NMR plasma samples) were then returned to storage over liquid nitrogen for spectral acquisition on subsequent days. Prior to spectral acquisition, several of the samples (~eight per day) were taken from the liquid nitrogen and kept in dry ice. NMR spectra were then acquired within 12 h of transferring the samples into the dry ice.
From the 15 technical replicates per fish, a random sample was chosen with which to investigate the effects of temperature-dependent temporal degradation of fish plasma as well as instrumental drift. The order of spectral acquisition for 14 of the 15 replicates was randomized, and between each, the candidate sample was run. The order was as follows: (8, 15, 2, 15, 10, 15, 7, 15, 4, 15, 3, 15, 11, 15, 14, 15, 6, 15, 12, 15, 9, 5, 13, 1) where sample 15 was the candidate sample for investigating degradation. This sample was kept refrigerated at ~4°C after every spectral acquisition. At the end of the first day the sample was left in the spectrometer overnight at 4°C and returned to the refrigerator on the morning of the following day. Three other spectra were then recorded 6 h apart and the sample returned to the refrigerator every time. On days 10 and 11 two additional spectra were acquired for each day. On day 12, two further spectra were acquired at 4°C after the sample had been left at ~21°C for 24 h and then another 12 h.
2.5 UPLC-MS sample preparation
Plasma samples were protein-precipitated using acetonitrile (ACN; HPLC grade, Caledon Laboratories, Georgetown, ON, Canada) in a 4:1 v/v ratio (ACN:sample). The mixture was vortexed for 30 s, allowed to stand for 5 min and centrifuged at 10,000 rpm for 2 min. The supernatant was reconstituted in deionized water from a MilliQ system in a 1:2 v/v ratio and filtered through Microcon YM-3 centrifugal filter devices, MW cut-off 3,000 Da (Amicon Bioseparations, Millipore, Billerica, MA, USA) previously rinsed with deionized water to eliminate traces of glycerol. The filtrate was dried and reconstituted in the mobile phase prior to UPLC-MS analysis.
2.6 UPLC-MS analysis
Chromatographic separation was performed on a Waters Acquity UPLC™ system (Waters, Milford, MA, USA) comprising a compartment with two flow pumps that deliver a parallel binary gradient (binary solvent manager), a high capacity system for preparation and injection of samples (sample manager), and a column compartment that allows automated switching among several columns (column manager) together with the 2996 PDA detector. Waters Acquity UPLC BEH C18 (1.7 μm 2.1 × 100 mm) and HSS T3 (1.8 μm 2.1 × 100 mm) columns were used. The UPLC system was coupled to a 4000 QTRAP mass spectrometer (AB/Sciex, Concord, ON, Canada) with a Turbo IonSpray™ source operating in the positive ion mode. The source temperature was set to 600°C, curtain gas to 10 psi and the ion spray voltage to 5,000 V. Pressures of 50 and 60 psi were employed for the nebulizer gas the turbo gas, respectively. The declustering potential was set to 75 V and collision energy of 10 eV was employed. Mass spectra were acquired in the enhanced mass scan mode from 90 to 1,000 mass/charge (m/z).
Plasma samples were reconstituted in the mobile phase (A = 0.1% formic acid in deionized water and B = 0.1% formic acid in ACN) and kept at 18°C in the sample manager prior to injection onto the column. A sample volume of 5 μl was injected. The BEH C18 column was kept at 30°C and a linear gradient comprising an equilibration at 5% B for 0.65 min followed by a gradient of 5–60% B between 0.65–3.77 min was employed. The composition was held at 60% B for 1.04 min and then returned to 5% B at 4.84 min at a flow rate of 0.4 ml/min. For the HSS T3 column, the column temperature was maintained at 40°C and a linear gradient with an equilibration at 100% A for 1 min, a gradient of 0–60% B between 1–4 min followed by 60–100% B between 4–5 min and, finally, 100–0% B in 1.5 min at a flow rate of 0.6 ml/min was used.
6α-Methylprednisolone (≥97%, Sigma-Aldrich, St Louis, MO, USA) was injected prior of analysis and after every six samples to monitor retention time consistency and fluctuations in instrumental response. This compound was also used as internal standard, added to plasma samples in a concentration of 3 μg/ml.
2.7 NMR data analysis
The large number of data points per spectrum (65,536), was reduced to 860 points by restricting the analysis of the data to the 0.2–4.5 ppm frequency range and binning by integrating all the data points within a 0.005-ppm chemical shift window into one. This eliminated peak registration shifts that might be induced by pH and other variations. Outside of this range, the spectra were generally devoid of signals apart from the water peak.
The data were normalized to unit sum and mean centered before analysis by principal components analysis (PCA) via singular value decomposition in MATLAB 7.1 (MathWorks, Natick, MA, USA). In addition, due to the unsupervised nature of PCA, partial least squares for linear discriminant analysis (PLS-DA; Ståhle and Wold 1987) was employed. This method was only applied in certain instances to eliminate separation ambiguity in cases where the information about group differences appeared in the smaller eigenvalues, i.e., when other factors influenced the separation of samples into more than the two known treatment classes.
Prior to PCA, the data were inspected for outliers using Hotelling’s T 2 values and Q residuals (Macho et al. 2001) as well as plots of Studentized residuals vs. residual leverage (Beebe et al. 1989). Any spectrum which was flagged as an outlier was inspected to determine the cause for its outlying tendency. In cases where the variability of particular “outlier” samples could not be shown to originate from errors in acquiring the spectra, they were included in the analysis although a replicate sample was prepared afresh and its spectrum acquired. The data were then normalized to unit sum, to eliminate variability due to the uncertainty in sample concentration introduced at the preparation.
2.8 MS data analysis
Ultra high performance liquid chromatography-mass spectrometry data were first analyzed in the time dimension followed by the m/z dimension. For the time dimension analysis, the total ion chromatograms (TICs) were extracted from Analyst™ software (Version 1.4.2; AB/Sciex) as text files and exported to MATLAB. Each chromatogram was subsequently segmented into 200 time dimension bins between 0.6 and 5.6 min for the BEH C18 column, and between 0.4 and 5.4 min for the HSS T3 column, which correspond to bin widths of 1.5 s. The area under each bin was integrated to yield a 1 × 200 vector containing intensity descriptors of the original TIC for each sample. The data were compiled into a 118 × 200 matrix and each row was normalized to unit sum before mean-centering. For the m/z dimension, mass spectra from Analyst were exported to MATLAB where the spectrum for each sample for a determined range of time was extracted as follows: intensities at each m/z (90–1,000 amu) were integrated over a given range of time (1.5 s) for each sample thus obtaining the spectrum in that period of time as intensities vs. m/z for each sample. These data were then normalized to unit sum and mean centered prior to multivariate analysis.
3 Results and discussion
3.1 Characterization of sample stability by 1H-NMR
As literature studies involving fish plasma are scarce (Samuelsson et al. 2006; Solanky et al. 2005), the degradation rate of fish plasma was investigated by 1H-NMR to establish its stability as a biofluid for metabolomics experiments. A PCA model was developed for the binned, normalized PS spectra collected for plasma stability characterization. The scores plot of the first two principal components (inset in Fig. 1) indicated differences between samples whose spectra were collected immediately after thawing from liquid nitrogen and those obtained after the sample was refrigerated for several days. Spectra that were acquired on days 1 and 2 after the sample was removed from liquid nitrogen were clustered at a low value of PC1, whereas spectra acquired following storage of sample in the refrigerator for over 10 days, and those left at room temperature ~21°C, clustered at successively higher values.
Scores plot of plasma spectra of five replicate fish. Technical replicates for individual fish samples are clustered together and are labeled F1–F5. Replicate F5 was used for the investigation of plasma degradation. Spectra of the sample used for this study cluster in closer proximity to the other F5 samples. Inset: scores plot of the plasma sample spectra acquired on different days. On day 1 all the samples were kept refrigerated at 4°C after every spectral acquisition. On day 2 the samples were also refrigerated although sample 11 had stayed in the spectrometer overnight. Samples were then stored in the refrigerator and spectra acquired 10 days later. On days 10 and 11, the spectra were kept in the refrigerator between spectral acquisitions (6 h apart) while on day 12 the samples were kept on the counter top at room temperature and pressure (RTP, i.e., ~21°C and ~101.3 kPa respectively) all day prior to spectral acquisition
Combination and re-analysis by PCA of these spectra with those acquired for all five fish plasma samples (Fig. 1) indicated (a) that the variability between technical replicates was not significant compared to the differences between individual biological sample replicates, and (b) that refrigeration of plasma at ~4°C for less than 12 days is unlikely to affect its quality and stability for metabolomics studies.
3.2 1H-NMR spectra of plasma from control and stressed groups of fish
Standard 1H-NMR PS spectra of plasma typically include broad peaks from high MW compounds such as proteins and lipids, which may partially obscure sharper signals from the low MW compounds emphasized by CPMG. Thus, it was anticipated that combining data acquired with both pulse sequences (Supplementary Figure 1) would provide a more comprehensive 1D 1H-NMR “picture” of the molecular status of fish plasma (Solanky et al. 2005).
Prior to the application of stress (week 0), plasma samples were obtained from a subset of the control and stress fish groups. PCA scores based on the 1H-NMR PS spectra confirmed that there was no initial difference between these two groups (Supplementary Figure 2). Differences were observed in the scores plot (PC1 vs. PC2) for weeks 1 and 2 shown in Fig. 2a, b, respectively; the corresponding loadings plots on the principal components along which the separation is observed are shown in Fig. 2c, d.
Principal components analysis of chronic handling stress for 1H-NMR data: dark filled triangle represents control group, dark filled square represents stress group. (a) Scores plot for week 1 plasma spectra; (b) scores plot for week 2 plasma spectra; (c) and (d) respective loadings. There is a clear distinction between the plasma spectra of two classes, i.e., control and stress groups at these 2 weeks. Identified metabolites are labeled numerically: (1) O-phosphocholine (O-PC), (2) lactate, (3) low density lipoprotein, (4) very low density lipoprotein, (5) α, β carbohydrates and amino acid residues, (6) alanine, (7) lipid, (8) valine, and (9) Trimethylamine-N-oxide (TMAO)
The scores plot for week 1 plasma spectra (Fig. 2a) revealed two major clusters of spectra for the plasma drawn from stressed (squares) and control (triangles) groups of fish. The two groups of fish were separated along PC2. The appearance of two sub-clusters of the control group may reflect unattributed physiological differences between individual fish. To restrict ourselves to modeling only the effects of stress on the metabolic profiles of plasma, a supervised method of classification, PLS-DA, was employed for analyzing the spectra acquired for week 1 samples. Technical replicates had been acquired for this particular week, thus it was possible to develop a classification model using one set of spectra for training (with internal cross-validation), and the other set for prediction (Fig. 3).
Partial least squares for linear discriminant analysis scores and loadings plot for week 1 sample. (a) PLS-DA scores, PC2 vs. PC1: dark filled circle represents training set (control group), dark filled square represents training set (stress group), dark filled diamond represents prediction set (control group), and dark filled triangle represents prediction set (stress group). (b) PLS-DA loadings plot along the first PC. This analysis was done in order to supervise the model in order to ensure that the separation is only due to the effects of stress
Analysis of spectra for week 2 plasma (Fig. 2b) revealed two distinct clusters for control and stress groups separated along PC1. The separation between the two fish groups at week 2 was more marked than at week 1, perhaps indicating a larger difference between the metabolite profiles of stress and control groups at week 2.
In contrast to weeks 1 and 2, it was not possible to identify clear differences between metabolite profiles in the plasma of control and experimental groups at week 3 by the methods employed here (Supplementary Figure 3). For week 4 (Supplementary Figure 4), a pattern of differential metabolite profiles was observed with control samples to the negative side of PC1. The general separation in week 4 was not as clear as those observed in weeks 1 and 2 however.
The results suggest that the metabolic disparity between the control and stress groups is observable at week 1 and becomes more prominent at week 2, but diminishes by weeks 3 and 4, conceivably due to some acclimation to the stress. At week 4 however, it was still possible to observe differences between the two groups. When the stress samples from all the weeks were combined and analyzed together, there was no observable pattern to depict a temporal fluctuation in the metabolite profiles. A similar lack of temporal organization was observed when control samples from all the weeks were combined and analyzed together. Nonetheless, when stressed samples from weeks 1 and 2 were analyzed together, it was possible to discern some classification among the samples (Supplementary Figure 5). However, when the stress samples from all the weeks were combined, no observable classification was found (Supplementary Figure 6). At individual experimental weeks, we were able to determine which metabolites changed in concentration in response to handling stress with the aid of the loadings plots (Fig. 2c, d), shown for the PC along which a separation occurred. Thus, Fig. 2c is a plot of the loadings on PC2 for week 1 spectra, and its interpretation is only meaningful in conjunction with the corresponding scores plot (Fig. 2a). Under this representation, one can see which metabolite peaks within the NMR spectra exhibit intensity changes that influence the separation between the plasma of control and experimental groups, providing a metabolite profile associated with a treatment. This is depicted in Fig. 2c, d for 2 weeks of chronic handling stress. In particular, Fig. 2c depicts chemical shifts with positive loadings which correspond to metabolites that have elevated concentrations in the stressed group relative to the control group while negative loadings correspond to metabolites whose concentration is decreased in the stressed group relative to the control. These metabolites were identified by use of Chenomx® software (Chenomx, Inc., Edmonton, AB, Canada), literature (Nicholson and Foxall 1995) and by spiking suspected metabolites into the plasma to evaluate chemical shift similarities. Moreover, given that the separation along PC2 encompasses three sub-clusters, i.e., one cluster of stress samples and another cluster which is further split into two sub-classes of control samples, we eliminated the third cluster by restricting our model to focus on only differences due to treatment. This was done using a supervised cluster analysis approach, PLS-DA (Fig. 3). The PLS-DA loadings are shown along the PC1 (Fig. 3b), where the metabolite profile created is associated primarily with stress. It is important to note that the results shown in this figure were obtained with a PLS-DA model that was developed after internal cross-validation (leave-one-out cross-validation) with an “independent” test set (technical replicates) used for prediction. Diagnostic plots for this analysis, i.e., prediction probability curves, classification and prediction of samples into two classes by the model and receiver operator characteristic (ROC) curves as well as the classification of sham data (using the same model) are shown, respectively, (Supplementary Figure 7a–c).
For week 2 (Fig. 2d), the loadings presented are along PC1 and their direction is a lot clearer than that depicted in Fig. 2c for week 1. More metabolite peaks can be discerned from this loadings plot compared to week 1, further indicating increased metabolic disparity between control and stress groups during the second week of handling stress. In this figure, the chemical shifts with negative loadings correspond to metabolites whose concentration is elevated in the stress group relative to control while those chemical shifts with positive loadings correspond to metabolites whose concentration is decreased.
The metabolite profile illustrated in Fig. 2c, d was derived from spectra acquired using a single pulse experiment with water PS. PCA was also performed on CPMG data in a similar approach as described for the PS spectra presented above. This technique emphasizes spectra of low MW in comparison to high MW compounds, reducing the prevalence of broad resonances. However, with CPMG spectra we could only observe a difference between stress and control groups by PCA in week 2, further affirming our earlier observation that the most prominent class differences occurred at this time. This observation implies that the low MW metabolites have a more important role in differentiating stressed samples in week 2 as opposed to week 1. The scores and corresponding loadings plots for the CPMG data are shown in Fig. 4 in which specific metabolite resonances are labeled in accordance with Figs. 2c, d and 4. Concentration trends relative to the control condition are presented in Table 1. The significance of these metabolite changes, in relation to the physiological status of the fish given the applied stress, remains to be established.
Scores (PC2 vs. PC1) and loadings plot on the first PC for week 2 CPMG spectra. Spectra acquired using this pulse sequence showed a classification only visible among week 2 samples. (a) The PC2 vs. PC1 scores showing a clear distinction between stressed and control samples along the first PC: dark filled square represents stress group, dark filled circle represents control group. (b) Loadings along the first PC that highlight different spectral regions emphasized by the pulse program. The numerical labeling of metabolites on this figure is consistent with Fig. 2
3.3 UPLC-MS analysis of plasma from control and stressed fish
UV-detected profiles for plasma samples of control and stressed fish obtained using the Waters Acquity UPLC BEH C18 and the Waters Acquity UPLC HSS T3 columns showed no marked differences for weeks 0–4. TICs generated by UPLC-MS using either the BEH C18 or HSS T3 columns showed some small changes in the intensities when comparing control and stressed samples at different retention times. PCA was applied to the TICs (PCA-TIC) and where this analysis revealed differences, PCA over the mass spectra (PCA-Spec) was applied to find the metabolites responsible for those differences.
Principal components analysis-total ion chromatograms of plasma samples for weeks 0–4, analyzed on the BEH C18 and HSS T3 columns, showed classification in week 2 only (Fig. 5a, b). Loadings plots for week 2 for samples analyzed on the BEH C18 and HSS T3 columns are shown in Fig. 5c, d, respectively. The loadings plot for samples analyzed on the BEH C18 column (Fig. 5c) revealed the time points (1.1375, 1.1625, 1.1875, 2.4125, 2.5375, 2.6875, 2.7625, 2.7875, 2.8625, 2.8875 min) responsible for the classification between groups. PCA-Spec was performed considering the following time ranges (which include the time points indicated in Fig. 5c): 1.1125–1.2375, 2.3875–2.4375, 2.4875–2.6125, 2.6625–2.7375, 2.7375–2.8125, and 2.8375–2.9125 min. The time points responsible for the classification in week 2 for samples analyzed on the HSS T3 column indicated by the loadings plot (Fig. 5d) were: 1.9625, 1.9875, 2.1625, 2.2375, 2.2625, 3.3875, 3.4125, 3.8625, and 3.8875 min, and the following time ranges were considered for PCA of the spectra: 1.8750–2.0375, 2.1125–2.2875, 3.3625–3.4375, and 3.8375–3.9125 min.
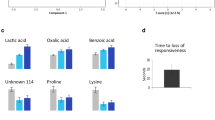
Principal components analysis of the mass spectra for the time ranges above indicated the m/z values for metabolites associated with the stress response and their relative changes in levels between plasma samples from control and stressed fish (Table 1). Figures 6 and 7 illustrate control/stress differences in average mass spectrum levels analyzed on the BEH C18 and HSS T3 columns during week 2 of this study. These metabolites were indicated by the loadings plots for the analysis of all plasma mass spectra in the time range indicated for the loadings of the PCA-TIC.
Average mass spectrum of plasma samples from control (solid lines) and stressed (dashed lines) fish analyzed on the BEH C18 column showing differences in levels for the following metabolites: (a) m/z = 877.4, (b) m/z = 430.3, (c) m/z = 651.4. Intensities shown on the y-axis correspond to the scale factor corrected and normalized raw intensities and errors bars represent 1 SD
Average mass spectrum of plasma samples from control (solid lines) and stressed (dashed lines) fish analyzed on the HSS T3 column showing differences in levels for the following metabolites: (a) m/z = 310.1, (b) m/z = 870.5, (c) m/z = 589.3. Intensities shown on the y-axis correspond to the scale factor corrected and normalized raw intensities and errors bars represent 1 SD
3.4 1H-NMR and UPLC-MS results
Several variables that contribute significantly to the discrimination of the control group of fish from the chronic handling stress group were identified in these non-targeted studies (Table 1). Through 1H-NMR profiling with a single-pulse sequence, it was possible to discriminate between stress and control groups in weeks 1 and 2. UPLC-MS and 1H CPMG NMR showed differences in the metabolic profiles of the two groups only at week 2, possibly because these two approaches favor lower MW compounds (the plasma was physically filtered prior to UPLC-MS to remove molecular components greater than 3,000 Da and the mass scan range was limited to 1,000 Da). The different metabolites involved in the separation between control and stressed groups show the complementary nature of 1H NMR and UPLC-MS techniques.
During the 2 weeks in which the fish demonstrated a measurable perturbation to their (plasma) metabolic balance (weeks 1 and 2), PCA of the NMR results showed an increase in concentration of O-phosphocholine (O-PC) in the stressed fish relative to controls, the identity of the characteristic singlet –N+(CH3)3 peak at 3.23 ppm being confirmed by spiking. In mammalian cells, phosphorylation of choline by choline kinase forms O-PC, which is converted to cytidyldiphosphate choline (Wong et al. 2000), then to phosphatidylcholine (PtdCho), a major phospholipid component of cell membranes (Eliyahu et al. 2007). A long-term stress study in rats has shown increased plasma choline levels at 44 days after initiation of the stressor (Teague et al. 2007). The authors suggested this change could be the result of an overall increase in plasma membrane turnover and/or PtdCho breakdown. Therefore it is possible that the increase in O-PC that we observed was due to the breakdown of PtdCho. In fact, it has been shown in mammalian cells that PtdCho can be hydrolyzed to form PC (Ramoni et al. 2001). Although the functional significance of the increased plasma O-PC levels remains unknown, our data suggest that this metabolite could potentially be used as a biomarker of long-term stress in fish.
Another obvious change observed in the metabolic profiles of the stressed fish was an increase in multiplet peaks in the chemical shift range 3.5–3.9 ppm, consistent with the presence of carbohydrates. The relative change in concentration (for week 1 samples) is assessed more readily from the PLS-DA loadings shown in Fig. 3b, clearly showing an elevation in plasma glucose and other carbohydrates, consistent with the observations at week 2 as well as with established knowledge about the relationship between glucose concentrations and the physiological response of fish to stress. Elevated plasma glucose level is a classical indicator of stressed states in fish (Barton 2002). Hyperglycemia in fish is mediated by hormones such as cortisol (Mommsen et al. 1999), and it is important to provide alertness and energy for the animals to cope with the stressor. Although sometimes there is no change in plasma glucose levels during long-term stress studies in fish (Hosoya et al. 2007), the results we obtained are not surprising. Interestingly, this study showed that the metabolic profiling approach seems to give a more thorough view of the glycemic response in fish. For instance, the determination of plasma glucose levels (as the only measure of fish response to stress) of the samples collected in this study using a modified Trinder enzymatic assay (Fast et al. 2008) showed that glucose concentrations were significantly increased in stressed fish only at week 1, and remained constant between the stressed and control groups at all other times. In contrast, using only 8.5 μl of plasma we were able to detect by PCA of 1H-NMR spectra, a general elevation in the levels of several carbohydrates including glucose in stressed fish at both weeks 1 and 2. This is in spite of the fact that glucose was not the main metabolite assumed, a priori, to be an indicator of stress in fish.
Other metabolites in plasma (Table 1) showing concentration changes in response to stress as identified by NMR/PCA were: lactate, lipoproteins, lipids, alanine, valine, and trimethylamine-N-oxide (TMAO). Plasma lactate and alanine levels decreased, relative to control levels, by week 1 of stress, and increased at week 2. Lactate and several amino acids, including alanine and valine, are important precursors of glucose formation through gluconeogenesis (Walter et al. 2006). Thus, the decreased levels of these metabolites suggest that gluconeogenesis was being stimulated during week 1. Rats also show decreased levels of these metabolites following acute stress (Teague et al. 2007). The increased relative levels of these metabolites in the fish plasma during week 2 of stress may indicate a decrease in gluconeogenesis, as the animals were likely acclimating to the daily stressor. After the week 2 the levels of these metabolites were no longer different from the control group.
Relative levels of plasma low density lipoprotein (LDL) and very low density lipoprotein (VLDL) increased during week 1 of stress, and decreased by week 2. It has been suggested that variations in lipoprotein and lipid levels during stress are associated with their increased or decreased metabolism (Teague et al. 2007), which is likely a reflection of the animals trying to deal with, and adapt to, the stressor. Changes in lipoprotein profiles during stressful events have been reported not only in mammals (Hershock and Vogel 1989) but also in fish (Atlantic salmon) challenged with a bacterial pathogen (Solanky et al. 2005).
Relative levels of plasma TMAO also increased during weeks 1 and 2 of stress. TMAO is found in fish and mammals and it is believed to have an important role in osmoregulation. The origin and synthetic pathways of TMAO are still controversial topics (Seibel and Walsh 2002; Samerotte et al. 2007). Seibel and Walsh (2002) hypothesized that accumulation of TMAO is a by-product of the production and storage of acylglycerol lipids, and is derived at least in part from the hydrolysis of PtdCho, which releases choline. Choline is converted to trimethylamine (TMA, a toxic product), which is then oxidized by trimethylamine oxygenase to TMAO (Seibel and Walsh 2002). It is possible that the accumulation of TMAO seen in this study was in part due to this route (lipid metabolism). One of the metabolites we identified, that had its relative levels increased, was O-PC, which is one of the breakdown products of PtdCho. This metabolite, can release choline, which can be converted either to O-PC (Eliyahu et al. 2007) or TMAO.
To assess the extent to which some of these metabolites varied over the course of the experiment, 1H-NMR spectra of four random samples, one from each stress group in the four experimental time points, were acquired under quantitative conditions (Burton et al. 2005). Owing to the difficulty associated with integrating peaks in a complex mixture such as plasma, we chose resonances of the known markers of stress (lactate ~4.12 ppm and glucose ~4.65 ppm) which are clear of other signals, as well as O-PC (~3.23 ppm), identified by NMR as a major contributor to the observed difference between the two fish groups at weeks 1 and 2. The O-PC peak is somewhat overlapped with other signals from TMAO and choline. Relative quantitation of this compound was therefore conducted using two approaches. First, its signal was integrated assuming that the overlapping “impurity” peaks were of constant relative area. The resulting integrals were then normalized to the total spectral intensity for each sample. In the second approach, the main peak was fitted to a Gauss–Lorentz function (Seah and Brown 1998) before integrating the area under the fitted curve. Using both approaches, the trends of the integrals were similar and only those obtained by the first are shown in Supplementary Figure 8. Note that these integrals were scaled by the integral of the TMSP peak in each sample. It is important also to note that these are not absolute concentrations and the plots are only intended to capture the general characteristics displayed by the temporal fluctuations in metabolite levels in a relative sense.
Supplementary Figure 8 indicates that the concentrations of these three metabolites in stressed groups increase in week 1, are maximized in week 2 coinciding with the greatest discrimination obtained via PCA and decrease in weeks 3 and 4. O-PC showed a greater relative concentration increase than lactate or glucose, which were comparable. In contrast, similar measurements on control samples indicated that the relative changes in the concentrations of this metabolite over time were negligible.
Ultra high performance liquid chromatography-mass spectrometry combined with PCA indicated that a considerable number of metabolites (Table 1, Supplementary Figure 9a) changed during long-term handling stress. Differences between levels in samples from stressed compared to control animals were only significant in week 2. Provisional identification of these metabolites on the basis of precise m/z was not possible, owing to the limited mass accuracy of the instrument used. The most prominent metabolites that increased in response to stress had m/z 651.4 and m/z 825.6 (non-polar, separated on the BEH C18 column), and m/z 870.5 and m/z 431.2 (polar, separated on the HSS T3 column). A few metabolites had decreased levels (m/z 430.3, m/z 310.1 and m/z 120.1). Examples of temporal behavior of levels are given (Supplementary Figure 9a–d) for some metabolites that showed less pronounced changes between control and stressed fish.
4 Concluding remarks
Metabolic profiling as a means of studying stress in fish and other organisms offers a complementary approach to the targeted measurement of known biomarkers. The expectation is that use of techniques that respond in one measurement to a large proportion of the metabolites present in a biofluid, combined with statistical analysis, will show patterns of concentration change correlated to the stress that can be interpreted in terms of physiological changes and also act as biomarkers of the stressed condition.
Our study of long-term handling stress in Atlantic salmon has applied both 1H-NMR and UPLC-MS to the same plasma samples and used exploratory data analysis to highlight differences between classes. The fact that each technique has yielded a different set of compounds as discriminants between stressed fish and controls is expected, considering that the NMR profile was obtained with intact samples whereas mass spectrometry was performed on filtered and separated components from UPLC. The total ion current vs. time was used as a first level of analysis. 1H-NMR profiles are close to uniform in response (signal area per 1H nucleus) vs. concentration, highly reproducible for a given sample, and are obtained within a few minutes with minimal sample preparation. However, there is difficulty in detecting metabolites at low concentrations particularly where their resonances overlap those of higher-level components. UPLC-MS is well-suited for detecting compounds at very low concentrations, but the large number of stages involved before detection requires great care to ensure proportionality of response to concentration for each individual metabolite, and the proportionality constant will vary from one compound to another, and from one instrument to another, owing to differences in ionization conditions.
1H-NMR profiles of salmon plasma samples were stable over a period of at least 2 days when stored at 4°C, and even after 10–12 days, the cluster of PCA scores for replicates from a particular fish were well separated from clusters for other individuals. Consequently, differences between groups of stressed and control fish were not influenced by sample handling. Unsupervised PCA scores and loadings comparisons between NMR spectra from stressed and control fish groups revealed classifiable differences in week 1 and more pronounced differences in week 2. The latter were associated with increased stress-induced levels of O-PC, lactate, carbohydrates including glucose, alanine, valine and TMAO, and decreased LDL, VLDL, and lipid. Such differences were not detectable at weeks 3 and 4.
A similar pattern of time variation of concentration of a different set of metabolites was detected by UPLC-MS, again showing maximum differences between stressed and control groups at week 2. Data to assign these metabolites, e.g., exact mass and/or fragmentation patterns, were not obtainable with the instrumentation used.
References
Aardema, M. J., & MacGregor, J. T. (2002). Toxicology and genetic toxicology in the new era of “toxicogenomics”: Impact of “-omics” technologies. Mutation Research, 499, 13–25. doi:10.1016/S0027-5107(01)00292-5.
Azmi, J., Connelly, J., Holmes, E., Nicholson, J. K., Shore, R. F., & Griffin, J. L. (2005). Characterization of the biochemical effects of 1-nitronaphthalene in rats using global metabolic profiling by NMR spectroscopy and pattern recognition. Biomarkers, 10, 401–416. doi:10.1080/13547500500309259.
Barton, B. A. (2002). Stress in fishes: A diversity of responses with particular reference to changes in circulating corticosteroids. Integrative and Comparative Biology, 42, 517–525. doi:10.1093/icb/42.3.517.
Barton, B. A., & Iwama, G. K. (1991). Physiological changes in fish from stress in aquaculture with emphasis on the response and effects of corticosteroids. Annual Review of Fish Diseases, 10, 3–26. doi:10.1016/0959-8030(91)90019-G.
Barton, B. A., Schreck, C. B., & Barton, L. D. (1987). Effects of chronic cortisol administration and daily acute stress on growth, physiological conditions, and stress responses in juvenile rainbow trout. Diseases of Aquatic Organisms, 2, 173–185. doi:10.3354/dao002173.
Beebe, R. K., Pell, J. R., & Seasholtz, M. B. (1989). Chemometrics: A practical guide. New York: Wiley.
Bonga, S. E. W. (1997). The stress response in fish. Physiological Reviews, 77, 591–625.
Brown, P. O., & Botstein, D. (1999). Exploring the new world of genome with DNA microarrays. Nature Genetics, 21, 33–37. doi:10.1038/4462.
Burton, I. W., Quilliam, M. A., & Walter, J. A. (2005). Quantitative 1H NMR with external standards: Use in preparation of calibration solutions for algal toxins and other natural products. Analytical Chemistry, 77, 3123–3131. doi:10.1021/ac048385h.
Coen, M., Ruepp, S. U., Lindon, J. C., Nicholson, J. K., Pognan, F., Lenz, E. M., et al. (2004). Integrated application of transcriptomics and metabonomics yields new insight into the toxicity due to paracetamol in the mouse. Journal of Pharmaceutical and Biomedical Analysis, 35, 93–105. doi:10.1016/j.jpba.2003.12.019.
Dacanay, A., Knickle, L., Solanky, K. S., Boyd, J. M., Walter, J. A., Brown, L. L., et al. (2006). Contribution of the type III secretion system (TTSS) to virulence of Aeromonas salmonicida subsp. salmonicida. Microbiology, 152, 1847–1856. doi:10.1099/mic.0.28768-0.
Defernez, M., Gunning, Y. M., Parr, A. J., Shepherd, L. V. T., Davies, H. V., & Colquhoun, I. J. (2004). NMR and HPLC-UV profiling of potatoes with genetic modifications to metabolomic pathways. Journal of Agricultural and Food Chemistry, 52, 6075–6085. doi:10.1021/jf049522e.
Eliyahu, G., Kreizman, T., & Degani, H. (2007). Phosphocholine as a biomarker of breast cancer: Molecular and biochemical studies. International Journal of Cancer, 120, 1721–1730. doi:10.1002/ijc.22293.
Fast, M. D., Hosoya, S., Johnson, S. C., & Afonso, L. O. B. (2008). Cortisol response and immune-related effects of Atlantic salmon (Salmo salar Linnaeus) subjected to short- and long-term stress. Fish & Shellfish Immunology, 24, 194–204. doi:10.1016/j.fsi.2007.10.009.
Griffin, J. L., & Kauppinen, R. A. (2007). Tumour metabolomics in animal models of human cancer. Journal of Proteome Research, 6, 498–505. doi:10.1021/pr060464h.
Hershock, D., & Vogel, W. H. (1989). The effects of immobilization stress on serum triglycerides, NEFAs, and total cholesterol in male rats after dietary modifications. Life Sciences, 45, 157–165. doi:10.1016/0024-3205(89)90290-7.
Hosoya, S., Johnson, S. C., Iwama, G. K., Gamperl, A. K., & Afonso, L. O. B. (2007). Changes in free and total plasma cortisol levels in juvenile haddock (Melanogrammus aeglefinus) exposed to long-term handling stress. Comparative Biochemistry and Physiology, 146, 78–86. doi:10.1016/j.cbpa.2006.09.003.
Iwama, G. K., Afonso, L. O. B., & Vijayan, M. M. (2005). Stress in fish. In D. H. Evans & J. B. Claiborne (Eds.), The physiology of fishes (3rd ed., pp. 319–342). Florida: CRC Press.
Lee, S. H., Woo, H. M., Jung, B. H., Lee, J., Kwon, O. S., Pyo, H. S., et al. (2007). Metabolomic approach to evaluate the toxicological effects of nonylphenol with rat urine. Analytical Chemistry, 79, 6102–6110. doi:10.1021/ac070237e.
Lenz, E. M., Bright, J., Knight, R., Wilson, I. D., & Major, H. (2004). A metabonomic investigation of the biochemical effects of mercuric chloride in the rat urine using 1H NMR and HPLC-TOF/MS: Time dependant changes in the urinary profile of endogenous metabolites as a result of nephrotoxicity. Analyst (London), 129, 535–541. doi:10.1039/b400159c.
Li, J., Zhang, Z., Rosenzweig, J., Wang, Y. Y., & Chan, D. W. (2002). Proteomics and bioinformatics approaches for identification of serum biomarkers to detect breast cancer. Clinical Chemistry, 48, 1296–1304.
Lindon, J. C., Holmes, E., Bollard, M. E., Stanley, E. G., & Nicholson, J. K. (2004). Metabonomics technologies and their applications in physiological monitoring, drug safety assessment and disease diagnosis. Biomarkers, 9, 1–31. doi:10.1080/13547500410001668379.
Macho, S., Sales, F., Callao, M. P., Larrechi, M. S., & Rius, F. X. (2001). Outlier detection in the ethylene content determination in propylene copolymer by near-infrared spectroscopy and multivariate calibration. Applied Spectroscopy, 55, 1532–1536. doi:10.1366/0003702011953766.
Malmendal, A., Overgaard, J., Bundy, J. G., Sorensen, J. G., Nielsen, N. C., Loeschcke, V., et al. (2006). Metabolomic profiling of heat stress: Hardening and recovery of homeostasis in Drosophila. American Journal of Physiology: Regulatory, Integrative and Comparative Physiology, 291, R205–R212. doi:10.1152/ajpregu.00867.2005.
Mashego, M. R., Rumbold, K., De Mey, M., Vandamme, E., Soetaert, W., & Heijnen, J. J. (2007). Microbial metabolomics: Past, present and future methodologies. Biotechnology Letters, 29, 1–16. doi:10.1007/s10529-006-9218-0.
Mommsen, T. P., Vijayan, M. M., & Moon, T. W. (1999). Cortisol in teleost: Dynamics, mechanism of action, and metabolic regulation. Reviews in Fish Biology and Fisheries, 9, 211–268. doi:10.1023/A:1008924418720.
Nicholson, J. K., & Foxall, P. J. D. (1995). 750 MHz 1H and 1H–13C NMR spectroscopy of human blood plasma. Analytical Chemistry, 67, 798–811. doi:10.1021/ac00101a004.
Nicholson, J. K., Lindon, J. C., & Holmes, E. (1999). Metabonomics: Understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data. Xenobiotica, 29, 1181–1189. doi:10.1080/004982599238047.
Perou, C. M., Sørlie, T., Eisen, M. B., van den Rijn, M., Jeffery, S. S., Rees, C. A., et al. (2000). Molecular portraits of human breast tumors. Nature, 406, 747–752. doi:10.1038/35021093.
Pickering, A. D., & Pottinger, T. G. (1987). Crowding causes prolonged leucopenia in salmonid fish, despite interrenal acclimation. Journal of Fish Biology, 30, 701–712. doi:10.1111/j.1095-8649.1987.tb05799.x.
Pickering, A. D., & Pottinger, T. G. (1989). Stress responses and disease resistance in salmonid fish: Effects of chronic elevation of plasma cortisol. Fish Physiology and Biochemistry, 7, 253–258. doi:10.1007/BF00004714.
Ramoni, C., Spadaro, F., Menegon, M., & Podo, F. (2001). Cellular localization and functional role of phosphatidylcholine-specific phospholipase C in NK Cells. Journal of Immunology (Baltimore, MD.: 1950), 167, 2642–2650.
Samerotte, A. L., Drazen, J. C., Brand, G. L., Seibel, B. A., & Yancey, P. H. (2007). Correlation of trimethylamine oxide and habitat depth within and among species of teleost fish: An analysis of causation. Physiological and Biochemical Zoology, 80, 197–208. doi:10.1086/510566.
Samuelsson, L. M., Förlin, L., Karlsson, G., Adolfsson-Erici, M., & Larsson, D. G. J. (2006). Using NMR metabolomics to identify responses of an environmental estrogen in blood plasma of fish. Aquatic Toxicology (Amsterdam, Netherlands), 78, 341–349. doi:10.1016/j.aquatox.2006.04.008.
Seah, M. P., & Brown, M. T. (1998). Validation and accuracy of software for peak synthesis in XPS. Journal of Electron Spectroscopy and Related Phenomena, 95, 71–93. doi:10.1016/S0368-2048(98)00204-7.
Seibel, B. A., & Walsh, P. J. (2002). Trimethylamine oxide accumulation in marine animals: Relationship to acylglycerol storage. The Journal of Experimental Biology, 205, 297–306.
Soga, T., Ueno, Y., Naraoka, H., Ohashi, Y., Tomita, M., & Nishioka, T. (2002). Simultaneous determination of anionic intermediates for Bacillus subtilis metabolomic pathways by capillary electrophoresis electrospray ionization mass spectrometry. Analytical Chemistry, 74, 2233–2239. doi:10.1021/ac020064n.
Solanky, K. S., Burton, I. W., MacKinnon, S. L., Walter, J. A., & Dacanay, A. (2005). Metabolic changes in Atlantic salmon exposed to Aeromonas salmonicida detected by 1H-nuclear magnetic resonance spectroscopy of plasma. Diseases of Aquatic Organisms, 65, 107–114. doi:10.3354/dao065107.
Ståhle, L., & Wold, S. (1987). Partial least squares analysis with cross-validation for the two-class problem: A Monte Carlo study. Journal of Chemometrics, 1, 185–196. doi:10.1002/cem.1180010306.
Stentiford, G. D., Viant, M. R., Ward, D. G., Johnson, P. J., Martin, A., Wenbin, W., et al. (2005). Liver tumors in wild flatfish: A histopathological, proteomic, and metabolomic study. OMICS: A Journal of Integrative Biology, 9, 281–299. doi:10.1089/omi.2005.9.281.
Tanaka, Y., Higashi, T., Rakwal, R., Wakida, S.-I., & Iwahashi, H. (2007). Quantitative analysis of sulfur-related metabolites during cadmium stress response in yeast by capillary electrophoresis-mass spectrometry. Journal of Pharmaceutical and Biomedical Analysis, 44, 608. doi:10.1016/j.jpba.2007.01.049.
Teague, C. R., Dhabhar, F. S., Beckwith-Hall, B., Powell, J., Cobain, M., Singer, B., et al. (2007). Metabonomic Studies on the physiological effects of acute and chronic psychological stress in Sprague-Dawley rats. Journal of Proteome Research, 6, 2080–2093. doi:10.1021/pr060412s.
Vaidyanathan, S., Kell, D. B., & Goodacre, R. (2002). Flow-injection electrospray ionization mass spectrometry of crude cell extracts for high-throughput bacterial identification. Journal of the American Society for Mass Spectrometry, 13, 118–128. doi:10.1016/S1044-0305(01)00339-7.
Viant, M. R. (2003). Improved methods for the acquisition and interpretation of NMR metabolomic data. Biochemical and Biophysical Research Communications, 310, 943–948. doi:10.1016/j.bbrc.2003.09.092.
Viant, M. R., Bundy, J. G., Pincetich, C. A., de Ropp, J. S., & Tjeerdema, R. S. (2005). NMR-derived developmental metabolic trajectories: An approach for visualizing the toxic actions of trichloroethylene during embryogenesis. Metabolomics, 1, 149–157. doi:10.1007/s11306-005-4429-2.
Viant, M. R., Rosenblum, E. S., & Tjeerdema, R. S. (2003). NMR-based metabolomics: A powerful approach for characterizing the effects of environmental stressors on organism health. Environmental Science and Technology, 37, 4982–4989. doi:10.1021/es034281x.
Walter, J. A., Ewart, K. V., Short, C. E., Burton, I. W., & Driedzic, W. R. (2006). Accelerated hepatic glycerol synthesis in rainbow smelt (Osmerus mordax) is fuelled directly by glucose and alanine: A 1H and 13C nuclear magnetic resonance study. The Journal of Experimental Zoology, 305, 480–488.
Waters, N. J., Waterfield, C. J., Farrant, R. D., Holmes, E., & Nicholson, J. K. (2006). Integrated metabonomic analysis of bromobenzene-induced hepatotoxicity: Novel induction of 5-oxoprolinosis. Journal of Proteome Research, 5, 1448–1459. doi:10.1021/pr060024q.
Wong, J. T., Chan, M., Lee, D., Jiang, J. Y., Skrzypczak, M., & Choy, P. C. (2000). Phosphatidylcholine metabolism in human endothelial cells: Modulation by phosphocholine. Molecular and Cellular Biochemistry, 207, 95–100. doi:10.1023/A:1007054601256.
Yates, J. R., III. (2000). Mass spectrometry: From genomics to proteomics. Trends in Genetics, 16, 5–8. doi:10.1016/S0168-9525(99)01879-X.
Acknowledgments
The authors wish to thank Corey Coldwell, Laura Garrison and Ron Melanson for fish husbandry and maintenance of aquatic facilities, Joseph Hui for mass spectrometry, Dr Susan Douglas for helpful comments and Dr Kirty Solanky for initial sample collection. Funding for this project was provided by the Genomics and Health Initiative of the National Research Council of Canada.
Author information
Authors and Affiliations
Corresponding author
Additional information
Major contributions were made by Tobias K. Karakach (NMR, data analysis and manuscript preparation), Elizabeth C. Huenupi (UPLC-MS and data analysis), and Luis O.B. Afonso (stress experiments and fish husbandry).
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Karakach, T.K., Huenupi, E.C., Soo, E.C. et al. 1H-NMR and mass spectrometric characterization of the metabolic response of juvenile Atlantic salmon (Salmo salar) to long-term handling stress. Metabolomics 5, 123–137 (2009). https://doi.org/10.1007/s11306-008-0144-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11306-008-0144-0