Abstract
The availability of nearly complete moso bamboo genome sequences permits the detailed discovery and cross-species comparison of transposable elements (TEs) between Bambusoideae and other Poaceae species at the whole genome level. Long terminal repeat retroelements (LTR-retroelements) are the single largest components of most plant genomes and can substantially impact the genome in various ways. Through a combination of structure- and homology-based approaches, we initially investigated 982 LTR-retroelement families comprising 2,004,644 LTR-retroelement sequences, which accounted for more than 40% of the moso bamboo genome. Further analysis revealed that the ratio of solo LTRs to intact elements (S/I) in moso bamboo is significantly low (approximately 0.28:1), indicating that bamboo LTR-retroelements might have undergone relatively low frequencies of unequal recombination and illegitimate recombination. Phylogenetic analysis revealed four Ty1-copia and five Ty3-gypsy evolutionary lineages that were present before the divergence of eudicot and monocot species, but the scales and timeframes within which they proliferated significantly varied across families and lineages. Insertion time estimates showed that LTR-retroelements were amplified for approximately 0~3 million years and had longer periods of activity than those of rice and Arabidopsis. These findings suggest that the expansion of LTR-retroelements might be responsible for host large genome size during moso bamboo evolution.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Transposable elements (TEs) are DNA fragments that can move to different positions within the genome of a single cell. Two broad classes of TEs are recognized based on their mechanism of transposition. Retroelements (class I) utilize an RNA intermediate and thus require reverse transcriptase to produce a DNA copy as well as an integrase for insertion into the host genome, whereas DNA transposons (class II) move directly as DNA and require a transposase or replicase to catalyze the necessary DNA cutting and joining reactions (Feschotte et al. 2002).
Retroelements are ubiquitous in eukaryotes, both in animals and plants (Bennetzen 2000; Eickbush and Jamburuthugoda 2008), and are divided into four main groups: long terminal repeat retroelements (LTR-retroelements), tyrosine recombinase (YR) retroelements, non-LTR-retroelements, and Penelope-like retroelements (Llorens et al. 2009). LTR-retroelements mainly encode two genes (gag and pol). The gag encodes structural proteins that form the virus-like particle (VLP), whereas pol encodes enzymatic regions such as protease (PR), reverse transcriptase (RT), RNase H (RH), and integrase (INT). LTR-retroelements are further subdivided into Ty1-copia, Ty3-gypsy, and Bel/Pao superfamilies based on their structure and transposition mechanism (Llorens et al. 2009). Ty1-copia and Ty3-gypsy retroelements are particularly widely distributed in the plant kingdom (Eickbush and Jamburuthugoda 2008). The former is mainly split into four lineages, which include Sire, Oryco, Retrofit, and Tork in plants, and the latter into five lineages, which consist of Crm, Del, Reina, Athila, and Tat in plants (Llorens et al. 2011). Sire elements have an envelope (ENV) domain downstream of the RH domain and are thus considered potential retroviruses (Havecker et al. 2004, 2005). Sire elements are widespread among plant species, both in monocots (rice, maize, and sorghum) and dicots (Arabidopsis, lotus, medicago, and citrus) (Havecker et al. 2005). Oryco (Ivana) elements are found in the genome of various plant species such as Vitis vinifera, Arabidopsis thaliana, Popolus tricocarpa, and Oryza sativa (Llorens et al. 2009). No ENV domains occur in Oryco elements. Retrofit lineages, also named Ale, together with Tork lineages, belong to the Ty1-copia superfamily and have been described in the genomes of different plant species, including Zea mays, Solanum lycopersicum, V. vinifera, Nicotiana tabacum, and Vigna radiata (Llorens et al. 2009). Crm elements are plant centromere-specific Ty3-Gypsy retroelements (Gorinsek et al. 2004). The Del lineage was previously known as the Tekay lineage (Gorinsek et al. 2004) and constitutes a plant Ty3-Gypsy superfamily. All Del elements encode for the CHR domain. Similar to Del elements, Reina elements also have a CHR domain that is located proximal to the INT domain (Gorinsek et al. 2004). Athila and Tat are two ancient lineages (Wright and Voytas 2002): the former encode for an additional putative ENV domain and are considered as potential retroviruses, whereas the latter are considered as giant LTR-retroelements.
LTR-retroelement amplification, along with polyploidization, is largely responsible for genome expansion (Bennetzen et al. 2005). For example, there are more than 26, 55, and 78% of LTR-retroelements in rice (International Rice Genome Sequencing Project 2005), maize (Schnable et al. 2009), and Sorghum (Paterson et al. 2009) genome, respectively. The aggressive proliferation of three families of LTR-retroelements resulted in the doubling of the genome size of Oryza australiensis, a wild relative of rice within the last 3 million years (Piegu et al. 2006). On the other hand,LTR-retroelements often undergo severe deletions or fragmentation via unequal homologous recombination and illegitimate recombination (Devos et al. 2002; Ma et al. 2004), which are two major mechanisms that counteract genome expansion (Bennetzen et al. 2005; Wicker and Keller 2007). Elimination through recombination frequently occurs in small genomes such as rice and Arabidopsis, but not in larger genomes such as that in maize, barley, and wheat (Bennetzen et al. 2005; Wicker and Keller 2007). In addition to their impact on genome size variations, LTR-retroelements also regulate the expression of adjacent genes in their host genomes as well as encode small RNAs that regulate specific target genes at the transcriptional and post-transcriptional levels (Kashkush et al. 2003; Kashkush and Khasdan 2007).
Moso bamboo [Phyllostachys edulis (Carrière) J. Houz.] is a large woody bamboo that has ecological, economic, and cultural value in Asia and accounts for ~70% of the total bamboo growth area (Fu 2001). The moso bamboo genome contains 24 pairs of chromosomes (2n = 48), with an approximate size of 2.075 Gb, and is diploid (Peng et al. 2013). The published high-quality draft genome sequence of moso bamboo covers 95% of the genomic region, and a total of 31,987 genes have been characterized by gene prediction modeling along with cDNA and deep RNA sequencing data. There are a few reports on TEs in moso bamboo sequences (Gui et al. 2010; Peng et al. 2013), but no detailed study on moso bamboo LTR-retroelements has been conducted to date. In the present study, LTR-retroelements in the moso bamboo genome (BambooGDB, http://www.bamboogdb.org/index.jsp, Zhao et al. 2014) were characterized. We systematically examined the structure, genomic distribution, phylogenetic diversity, and expansion pattern of moso bamboo LTR-retroelements.
Materials and methods
Identification and classification of LTR-retroelements
Genomic sequences and gene annotation information on moso bamboo were downloaded from the bamboo genome database (BambooGDB, http://www.bamboogdb.org/index.jsp, Zhao et al. 2014). A combination of structural analysis and sequence homology comparisons were performed to identify LTR-retroelements in the assembled moso bamboo genome. Initially, the LTR_STRUC program was employed for the identification of LTR-retroelements (McCarthy and McDonald 2003). All of the identified LTR-retroelements were confirmed by manual search against RepBase (version 21.02) and GenBank. The sequences that were identified as LTR-retroelements in the two databases were used in subsequent analyses. The LTR-retroelements were classified into Ty1-Copia (INT-RT-RH) and Ty3-Gypsy (RT-RH-INT) superfamilies (Llorens et al. 2011). Both superfamilies were further classified into four lineages (Sire, Oryco, Retrofit, and Tork) for the Ty1-Copia superfamily and five lineages (CRM, Del, Reina, Athila, and Tat) for the Ty3-Gypsy superfamily using BLASTX, with an e-value of <10 by submitting the identified LTR-retroelements against the cores in the Gypsy Database (GyDB) (Llorens et al. 2011).
The identified bamboo LTR-retroelements were assigned to families within the enumerated lineages on the basis of 80% identity, over at least 80 bp in at least 80% of their sequence lengths (80–80–80 rule, Wicker et al. 2007). Each LTR-retroelement family identified in the present was designated as PhXYZ#, where Ph are the two letters representing Moso bamboo (P. edulis (Carrière) J. Houz.), XYZ are the first three letters of the name of LTR-retroelement lineage, and # is the number representing the family.
To mine all the remnant LTR-retroelement sequences, the representative retroelements of families were used as the query sequence to scan the genome sequence by using RepeatMasker with Cross_Match as search engine and a cutoff of >250 (http://repeatmasker.org). The unmatched results were filtered out based on the criteria of a nucleotide identity rate of <80% and a query coverage of <80%. The workflow of identification of bamboo LTR-retroelements is summarized in Supplementary File S1.
Analysis of the structure of bamboo LTR-retroelements
The coding domains of full-length bamboo LTR- retroelements were identified using Pfam and confirmed by alignment with MUSCLE (Edgar 2004) against the domain alignment from GyDb (Llorens et al. 2011). Full-length sequences that contained the GAG, PR, RT, RH, and INT domains were designated as intact bamboo LTR-retroelements. The intact bamboo LTR-retroelements were aligned and analyzed using BioEdit (Hall 1999), and all the coding domains as well as the LTRs and target side duplications (TSDs) could been aligned. The lengths of all domains as well as the LTRs and spacer regions of each element were recorded in an Excel table, and the schematic representation of bamboo LTR-retroelement structures were drawn in Excel.
Estimation of insertion time
Using an approach described by Ma and Bennetzen (2004), the age of the elements with two available LTR sequences were determined by comparing their 5′ and 3′ LTRs. Two LTRs were aligned using the MUSCLE program (Edgar 2004). When necessary, the alignments were manually inspected and corrected using the MEGA6 program (Tamura et al. 2013). The distance (K) between two LTRs was calculated by using the Jukes–Cantor method (Kimura and Ota 1972). An average substitution rate (r) of 1.3 × 10−8 substitutions per synonymous site per year (Ma and Jackson 2006) was used in the calculations. The time (T) since initial insertion was estimated using the formula T = k/2r.
Phylogenetic analysis
To assess the evolutionary pattern of moso bamboo LTR-retroelements, representative plant elements of four lineages of Ty1-Copia (Sire, Oryco, Retrofit, and Tork) and five lineages of Ty3-Gypsy (Crm, Del, Reina, Athila, and Tat) were downloaded from the Gypsy Database (GyDB) (http://www.gydb.org/index.php/Main_Page, Llorens et al. 2011). We also downloaded all full-length sequences of Ty1-Copia and Ty3-gypsy retroelements of A. thaliana and O. sativa from the Arabidopsis Information Resource (TAIR) (https://www.arabidopsis.org/index.jsp) and Rice Genome Annotation Project (http://rice.plantbiology.msu.edu/home_contacts.shtml), respectively. The A. thaliana and O. sativa LTR-retroelements were assigned to families by using the 80–80–80 rule (Wicker et al. 2007). The representative intact elements were selected from every family to construct the LTR-retroelement phylogenetic trees. The moso bamboo representative intact elements from each family were also selected for phylogenetic analysis.
All RT protein domain sequences were aligned using MUSCLE with default settings (Edgar 2004), and rare distinct shifts were manually adjusted. The optimal model of amino acid substitution was estimated using MEGA6 (Tamura et al. 2013) with default settings. Bootstrap neighbor-joining trees were built using the Kimura two-parameter method that was integrated in the MEGA6 program (Tamura et al. 2013).
Results
Characterization of LTR-retroelements in the moso bamboo genome
Using the LTR_STRUC program, we mined 1311 LTR-retroelement candidates in the moso bamboo genome. The 1311 LTR-retroelements were confirmed by manual search against the RepBase version 21.02 (Jurka et al. 2005) and GenBank database. The 1259 of the 1311 sequences were confirmed. Then, by using all-vs-all blastn, 982 LTR-retroelement families were classified based on the 80–80–80 rule (Wicker et al. 2007). Among the 982 LTR-retroelement families, 458 families belonged to the Ty1-copia superfamilies, of which 48 families were classified as Tork lineages, 249 families as Retrofit lineages, 60 families as Sire lineages, and 101 families as Oryco lineages. There were 501 families that belonged to Ty3-gypsy superfamilies, of which 63 families were determined to be Del lineages, 181 families belonged to Reina lineages, 71 families belonged to Crm lineages, and 186 families belonged to Tat lineages. Additionally, there were 23 families that were not homologous to the known LTR-retroelement family and were thus designated as Unknown-LTR. The general features of the representative sequence of each moso bamboo LTR-retroelement family such as element size, LTR size, and TSDs are presented in Supplementary Table S2. To retrieve partial or intact LTR-retroelements, the 982 representative sequences were used as reference queries to search the moso bamboo genome using RepeatMasker. A total of 2,004,644 LTR-retroelement sequences were detected in the moso bamboo genome, which accounted for more than 40% of the entire genome (Table 1).
Of the 2,004,644 LTR-retroelement sequences, 829,598 elements belonged to the Ty1-copia superfamily (up to 337,053,290 bp) and 1,125,018 elements were classified under the Ty3-gypsy superfamily (up to 480,215,478 bp), thereby accounting for approximately 16.43 and 23.41% of the moso bamboo genome, respectively. Among four lineages of the Ty1-copia superfamily, the content of the Sire lineage was highest (up to 6.11%) and closely followed by the Retrofit lineage (5.29%). Previous reports also showed that the Sire and Retrofit lineages were widely distributed in monocot plants such as O. sativa and Z. mays (Havecker et al. 2005; Llorens et al. 2009). Among the four lineages of the Ty3-gypsy superfamily, the content of the Tat lineage was highest (up to 9.67%) and closely followed by the Del lineage (9.53%) (Table 1). The Tat and Del lineages are plant-specific and are ubiquitous in high plant genomes (Havecker et al. 2004; Llorens et al. 2009).
The ratio of the number of Ty3-gypsy elements to the number of Ty1-copia elements in the moso bamboo was 0.8:1, which was lower than that in maize (1.6:1) (Baucom et al. 2009; Schnable et al. 2009) and much lower than that in rice (4.9:1) (International Rice Genome Sequencing Project 2005; Tian et al. 2009) and sorghum (3.7:1) (Paterson et al. 2009), but considerably higher than that in the eudicot plant, Medicago (0.3:1) (Wang and Liu 2008).
The 2,004,644 LTR-retroelement sequences could be classified into three types, including full-length elements, solo LTRs, and truncated elements. The ratio of the number of solo LTRs to the number of full-length elements (S/I) in moso bamboo is approximately 0.28:1 (Table 2). Among the eight lineages, the highest S/I ratio was observed in the Retrofit lineage (0.55), whereas the lowest S/I ratio was detected in the Reina lineage (0.034) (Table 2).
Structural features of LTR-retroelements in moso bamboo
We identified all the internal coding domains (GAG, PR, RT, INT, RH, ENV, and CHR) of the representative full-length sequences of LTR-retroelements. The ENV domain was identified in all sire elements and the CHR domain in both the Del and Reina elements (Table 1). There were 124 sequences of 458 representative Ty1-copia retroelement sequences and 286 sequences of 501 representative Ty3-Gypsy retroelement sequences that contain all domains (GAG, PR, RT, RH, INT, /ENV, /CHR). The 410 intact retroelement sequences were further analyzed in terms of structural features.
There was a general pattern in the overall size of the elements from various families. In both superfamilies, two families were determined to be very long (Sire and Del, 7.2–17.5 kb) and two families were relatively short (Oryco and Reina, 4.7–8.9 kb) (Fig. 1a, b). Differences in the total length were mainly due to variations in LTR size and the presence and size of spacer regions between the internal coding domains, rather than the gag/pol coding regions (Fig. 1a, b).

The proliferation spectrum of LTR-retroelements in moso bamboo
Although the eight evolutionary lineages were identified in moso bamboo, the proliferation spectrum of LTR-retroelements was highly variable among lineages. The number of families within lineages and the copy numbers of intact and solo elements within lineages identified in the species are listed in Table 2 (truncated elements were not considered here). Among the four Ty1-copia lineages in moso bamboo, Retrofit has the largest number of LTR-retroelement families (249), accounting for 54.37% of the Ty1-copia families (458) analyzed. However, the 249 families only contained 18.86% (621) of the 3292 Ty1-copia elements. In contrast, Sire consisted of the highest number of Ty1-copia elements (1295, 39.34%); however, these elements belonged to only 60 families. More dramatic differences were observed among the four Ty3-gypsy lineages in moso bamboo. For example, the Tat lineage contained 186 (37.13%) of the 501 Ty3-gypsy families analyzed, but the lineage comprised only 864 elements (19.89% of the 4343 Ty3-gypsy elements). The Crm lineage showed the highest number of elements, accounting for 34.72% of the Ty3-gypsy elements that were identified in the moso bamboo genome. The number of elements of LTR-retroelements within individual lineages reflects their recent amplification, whereas the number of families within individual lineages represents ancient divergent activities. Hence, the above observations suggest that different lineages and families of LTR-retroelements have distinct activities for division and expansion over evolutionary time.
Estimation of insertion date
We estimated the insertion time of 7845 intact and solo LTR-retroelements in moso bamboo. Figure 2 shows that most of the elements (91%) were amplified around 0~3 Mya. The distribution of insertion times of LTR-retroelements in every lineage was represented by a single-peak parabolic curve. These results indicated that the eight lineages were active and amplified within the same evolutionary timeframes.
Evolutionary dynamics of moso bamboo LTR-retroelements
To understand the evolutionary dynamics, history, and fates of bamboo LTR-retroelements in moso bamboo over evolutionary time, we used the relatively conserved RT domains from individual elements for phylogenetic analysis. Of the 982 families identified in the moso bamboo genome, 124 Ty1-copia and 286 Ty3-gypsy family representative sequences that consisted of a complete RT domain were used to construct phylogenetic trees. The other family representative sequences contained deletions of the RT domains.
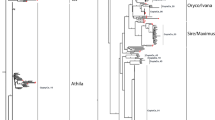
For the Ty1-copia phylogenetic tree, we grouped 94 Ty1-copia retroelements in A. thaliana, 85 retroelements in O. sativa, 124 retroelements in moso bamboo, and 23 other plant typical Ty1-copia elements into four distinct lineages, namely, Sire, Oryco, Retrofit, and Tork (Fig. 3a). All families were shared by these three species, which showed that these four lineages all existed before the divergence of eudicots and monocots (Fig. 3a).
Phylogenetic relationship of Ty1-copia (a) and Ty3-gypsy elements (b) identified in moso bamboo, rice, and Arabidopsis. The RT amino acid sequences of individual elements were used to construct the phylogenetic trees. Robustness of the nodes was estimated by 500 bootstrap replications. Individual LTR-retroelements in moso bamboo, rice, and Arabidopsis are indicated by solid black dots, blue solid diamonds, and pink solid squares, respectively. a The plant typical Ty1-copia elements derived from four lineages are represented by green hollow triangles. Four lineages, including Oryco, Sire, Tork, and Retrofit, were marked by brown, green, red, and purple branches, respectively. b The plant typical Ty3-gypsy elements derived from five lineages are represented by green hollow triangles. Five lineages, including Reina, Crm, Del, Athila, and Tat, are marked by brown, dark green, red, light green, and blue branches, respectively
For the Ty3-gypsy phylogenetic tree, there were 59 Ty3-gypsy retroelements in A. thaliana, 108 retroelements in O. sativa, 286 retroelements in moso bamboo, and 25 other plant typical Ty3-gypsy elements, which grouped into five lineages, including Crm, Del, Reina, Athila, and Tat (Fig. 3b). These five lineages all existed before the divergence of eudicots and monocots, but the Athila family was not identified in the moso bamboo (Fig. 3b).
There were two distinctive phenomena in two phylogenetic trees. One is that the bamboo LTR-retroelements and rice LTR-retroelements frequently clustered together, whereas Arabidopsis LTR-retroelements clustered together themselves. The other is that there was an abundance of very identical LTR-retroelement sequences among the three species (e.g., Oscopia44, PhOry93, and Atcopia205 with the similarity of more than 95% each other) and the presence of relative diverse LTR-retroelement sequences in individual species (e.g., PhRet191, PhSir30, and PhTor40 with the similarity of less than 75% each other).
Discussion
LTR-retroelements predominate in the moso bamboo genome
Most genome projects identify and annotate TEs using homology and de novo methods (Hoen et al. 2015), but the most accurate assessment of TE landscapes is currently only possible through a combination of de novo and homology-based repeat identification in conjunction with an additional manual curation step (Platt et al. 2016). One reason for this is that longer elements are often recovered as multiple smaller fragments by both types of searches. Peng et al. (2013) annotated moso bamboo LTR-retroelements with LTRharvest and LTR_FINDER soft. In the study, a combination of structure-based and homology-based approaches was also employed to identify LTR-retroelements in the moso bamboo genome sequence. Initially, the representative sequences of LTR-retroelements with intact LTR sequences were identified by the structure-based approach, LTR_STRUC program (McCarthy and McDonald 2003). After filtering out the false-positive sequences, the rest of the sequences were characterized for LTR, TSD sequences, and GAG, PR, RT, RH, and INT domains. Then, the complete RT domains of individual elements from moso bamboo, A. thaliana, and O. sativa were used to construct phylogenetic trees to display the evolutionary dynamics of LTR-retroelements. After that, the identified moso bamboo LTR-retroelements were classed into individual superfamilies, lineages, and families of LTR-retroelements. To display the proliferation spectrum of bamboo LTR-retroelements, the full-length elements, solo LTRs, and truncated elements of each superfamily, lineage, and family of LTR-retroelements in the moso bamboo genome were identified using RepeatMasker. This is the first study to perform a systematical survey of the structure, genomic distribution, phylogenetic diversity, and expansion pattern of moso bamboo LTR-retroelements Supplementary File 1).
By using a combination of structure-based and homology-based approaches, a total of 2,004,644 LTR-retroelement sequences were identifiNed in the moso bamboo genome. The content of LTR-retroelement sequences in moso bamboo was up to 40.35%, which is slightly higher than that reported by Peng et al. (37.3%, 2013). The content of all LTR-retroelements in the moso bamboo genome is far lower than that in Z. mays (79%) (Schnable et al. 2009) and S. bicolor (55%, Paterson et al. 2009) and significantly higher than that in O. sativa (26%, International Rice Genome Sequencing Project 2005).
Some LTR-retroelements might be missed due to the incompleteness of the moso bamboo genome. The moso bamboo genome assembly is highly fragmented (>9000 scaffolds). Although we combined the structure-based and homology-based approaches in the identification of LTR-retroelements, the fragmentation of the moso bamboo genome could still cause an underestimation of LTR-retroelement contents. In addition to full-length elements and solo LTRs, massive truncated elements were detected. These truncated elements might be due to the difficulty in assembling full-length TEs, which might have resulted in the inclusion of a high number of truncated elements in the original assembly. Therefore, the 40.35% content of LTR-retroelement sequences identified in this study may be a conservative figure and more LTR-retroelements may be detected in the future with the availability of a higher-quality moso bamboo genome.
Eight evolutionary lineages were identified in moso bamboo (Sire, Oryco, Retrofit, Tork of Ty1-copia and Crm, Del, Reina, Tat of Ty3-gypsy). Although the Athila family is widespread in the plant kingdom and also occurs in monocot plants such as sorghum and rice (Steinbauerova et al. 2011), it was not identified in the moso bamboo genome. One possible reason is that Athila elements are not suitable for identification using the LTR_STRUC program. The Athila elements are usually 8.5–12 kb in size and have relatively long LTR sequences (1.5–2.5 kb, Wright and Voytas 2002), which might be difficult to be recognized by the LTR_STRUC program. Other software with more adjustable parameters may be employed for the identification of Athila elements. On the other hand, Athila elements might be missed due to the incompleteness and fragmentation of the moso bamboo genome. Additionally, there are 23 families and more than 50,000 sequences (0.52%) which were homologous to unknown LTR-retroelements of other species and were named Unknown-LTR. Unknown-LTR sequences in moso bamboo are less than those in maize (181 families, 31.31%) (Schnable et al. 2009).
Among the eight evolutionary lineages, the scales and timeframes of activity for proliferation of LTR-retroelements vary tremendously among lineages. A similar scenario was seen in rice and soybean (Du et al. 2010). There are three lineages (i.e., Ivana/Oryco, Ale/Retrofit, and Reina) that contain the highest number of families of LTR-retroelements in rice and soybean (Du et al. 2010). In contrast, there are similar three lineages (i.e., Retrofit, Reina, and Tat) that contain the highest number of families of LTR-retroelements in moso bamboo (Table 2). Arabidopsis has significantly fewer families in the nine lineages than rice, soybean, and moso bamboo due to overall low activities for LTR-retroelement expansion (Du et al. 2010). The results showed that the moso bamboo genome has the consistently high expansion activities of LTR-retroelements over evolutionary time, which are likely responsible for the current large size of the moso bamboo genome (Du et al. 2010).
Moso bamboo LTR-retroelements might undergo a relative low frequency of unequal recombination and illegitimate recombination
The unequal recombination and illegitimate recombination is thought to be a major process for formation of solo LTRs in plants (Devos et al. 2002; Ma et al. 2004). Our data showed that the ratio of the number of solo LTRs to the number of intact elements (S/I) in moso bamboo is approximately 0.28:1. This estimate is significantly lower than reported in rice (1.62:1) (Tian et al. 2009). These findings thus indicate that the moso bamboo genome might undergo unequal recombination and illegitimate recombination at a relatively low frequency, which is one of the main mechanisms for confrontation of LTR-retroelement expansion activities (Devos et al. 2002; Ma et al. 2004).
LTR-retroelements remain relative long existence in moso bamboo genome
We estimated the insertion times of bamboo LTR-retroelements. The largest number of bamboo LTR-retroelements was estimated to be inserted within 0~3 Mya (Fig. 2). The results showed that there might be several bamboo LTR-retroelements that recently transposed.
Additionally, the distribution of insertion times of bamboo LTR-retroelements was represented by a single-peak parabolic curve with a relatively long time span, which was similar to that of barley and wheat (Wicker and Keller 2007). The pattern of insertion times of rice and soybean LTR-retroelements was exponentially distributed (Wicker and Keller 2007; Du et al. 2010). Although the available data set was extremely small to draw definite conclusions, the observed distribution curve of LTR-retroelement insertions may reflect its very long existence in the moso genome (Wicker and Keller 2007), which might be due to the low frequency of unequal recombination and illegitimate recombination for bamboo LTR-retroelements (above discussion).
Bamboo LTR-retroelements are structurally diverse and have evolutionary lineages that are shared between monocots and eudicots
Previous surveys on LTR-retroelements in plants have defined four major common evolutionary Ty1-copia lineages and five major common evolutionary Ty3-gypsy lineages (Llorens et al. 2011). Four lineages (i.e., Sire, Oryco, Retrofit, and Tork) of Ty1-copia and four lineages of Ty3-gypsy (i.e., Crm, Del, Reina, and Tat) were identified in the moso bamboo genome, all of which originated prior to the divergence of monocots and eudicots (Llorens et al. 2009).
Among most families, the bamboo LTR-retroelements and rice LTR-retroelements were frequently clustered together, whereas the Arabidopsis LTR-retroelements clustered together with themselves, which indicated that most lineages have diverged after the division between the monocots and eudicots (Fig. 3). However, the Tork lineage is an exception. LTR-retroelements of the Tork lineage from three species clustered together without discrimination, which indicated that the evolution of the Tork lineage was relatively more conserved compared with other lineages (Llorens et al. 2009).
Comparative genome analysis suggested that the moso bamboo diverged from Oryzoideae more than 48 Mya (Peng et al. 2013). Interestingly, there was an abundance of near identical LTR-retroelement sequences between rice and moso bamboo and the presence of relatively diverse LTR-retroelement sequences in individual rice and moso bamboo species (Fig. 3). This might be due to divergent evolutionary rate, stochastic loss, vertical extinction, or horizontal transfer of LTR-retroelements (Du et al. 2010).
Conclusions
Considering the high proportion of LTR-retroelements in the moso bamboo genome, our work contributes to a greater understanding of their impact on genome organization and evolution. We initially investigated 982 LTR-retroelement families comprising 2,004,644 LTR-retroelement sequences, which accounted for more than 40% of the moso bamboo genome. Further analysis revealed that bamboo LTR-retroelements had longer period of activity than those of rice and Arabidopsis and might undergo a relatively low frequency of unequal recombination and illegitimate recombination. The knowledge gained from this study constitutes a valuable resource for both the improvement in genome annotation and investigations of the contributions of LTR-retroelements to moso bamboo genome evolution.
References
Baucom RS, Estill JC, Chaparro C, Upshaw N, Jogi A, Deragon JM, Westerman RP, Sanmiguel PJ, Bennetzen JL (2009) Exceptional diversity, non-random distribution, and rapid evolution of retroelements in the B73 maize genome. PLoS Genet 5:e1000732
Bennetzen JF (2000) Transposable element contributions to plant gene and genome evolution. Plant Mol Biol 42:251–269
Bennetzen JL, Ma J, Devos KM (2005) Mechanisms of recent genome size variation in flowering plants. Ann Bot 95:127–132
Devos KM, Brown JK, Bennetzen JL (2002) Genome size reduction through illegitimate recombination counteracts genome expansion in Arabidopsis. Genome Res 12:1075–1079
Du J, Tian Z, Hans CS, Laten HM, Cannon SB, Jackson SA, Shoemaker RC, Ma J (2010) Evolutionary conservation, diversity and specificity of LTR-retrotransposons in flowering plants: insights from genome-wide analysis and multi-specific comparison. Plant J 63:584–598
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797
Eickbush TH, Jamburuthugoda VK (2008) The diversity of retroelements and the properties of their reverse transcriptases. Virus Res 134:221–234
Feschotte C, Jiang N, Wessler SR (2002) Plant transposable elements: where genetics meets genomics. Nat Rev Genet 3:329–341
Fu J (2001) Chinese moso bamboo: its importance. Bamboo 22:5–7
Gorinsek B, Gubensek F, Kordis D (2004) Evolutionary genomics of chromoviruses in eukaryotes. Mol Biol Evol 21:781–798
Gui YJ, Zhou Y, Wang Y et al (2010) Insights into the bamboo genome: syntenic relationships to rice and sorghum. J Integr Plant Biol 52:1008–1015
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Havecker ER, Gao X, Voytas DF (2004) The diversity of LTR retrotransposons. Genome Biol 5:225
Havecker ER, Gao X, Voytas DF (2005) The Sireviruses, a plant-specific lineage of the Ty1/copia retrotransposons, interact with a family of proteins related to dynein light chain. Plant Physiol 139:857–868
Hoen DR, Hickey G, Bourque G, Casacuberta J, Cordaux R, Feschotte C, Fiston-Lavier AS, Hua-Van A, Hubley R, Kapusta A, Lerat E, Maumus F, Pollock DD, Quesneville H, Smit A, Wheeler TJ, Bureau TE, Blanchette M (2015) A call for benchmarking transposable element annotation methods. Mob DNA 6:1–9
International Rice Genome Sequencing Project (2005) The map-based sequence of the rice genome. Nature 436:793–800
Jurka J, Kapitonov VV, Pavlicek A, Klonowski P, Kohany O, Walichiewicz J (2005) Repbase update, a database of eukaryotic repetitive elements. Cytogenet Genome Res 110(1-4):462–467
Kashkush K, Khasdan V (2007) Large-scale survey of cytosine methylation of retrotransposons and the impact of readout transcription from long terminal repeats on expression of adjacent rice genes. Genetics 177:1975–1985
Kashkush K, Feldman M, Levy AA (2003) Transcriptional activation of retrotransposons alters the expression of adjacent genes in wheat. Nat Genet 33:102–106
Kimura M, Ota T (1972) On the stochastic model for estimation of mutational distance between homologous proteins. J Mol Evol 2:87–90
Llorens C, Muñoz-Pomer A, Bernad L, Botella H, Moya A (2009) Network dynamics of eukaryotic LTR retroelements beyond phylogenetic trees. Biol Direct 4:41
Llorens C, Futami R, Covelli L et al (2011) The gypsy database (GyDB) of mobile genetic elements: release 2.0. Nucleic Acids Res 39:70–74
Ma J, Bennetzen JL (2004) Rapid recent growth and divergence of rice nuclear genomes. Proc Natl Acad Sci U S A 101:12404–12410
Ma J, Jackson SA (2006) Retrotransposon accumulation and satellite amplification mediated by segmental duplication facilitate centromere expansion in rice. Genome Res 16:251–259
Ma J, Devos KM, Bennetzen JL (2004) Analyses of LTR-retrotransposon structures reveal recent and rapid genomic DNA loss in rice. Genome Res 14:860–869
McCarthy EM, McDonald JF (2003) LTR_STRUC: a novel search and identification program for LTR retrotransposons. Bioinformatics 19:362–367
Paterson AH, Bowers JE, Bruggmann R et al (2009) The Sorghum bicolor genome and the diversification of grasses. Nature 457:551–556
Peng Z, Lu Y, Li L et al (2013) The draft genome of the fast-growing non-timber forest species moso bamboo (Phyllostachys heterocycla). Nat Genet 45:456–461
Piegu B, Guyot R, Picault N et al (2006) Doubling genome size without polyploidization: dynamics of retrotransposition-driven genomic expansions in Oryza australiensis, a wild relative of rice. Genome Res 16:1262–1269
Platt RN, Blanco-Berdugo L, Ray DA (2016) Accurate transposable element annotation is vital when analyzing new genome assemblies. Genome Biology and Evolution 8(2):403–410
Schnable PS, Ware D, Fulton RS et al (2009) The B73 maize genome: complexity, diversity, and dynamics. Science 326:1112–1115
Steinbauerova V, Neumann P, Novak P, Macas J (2011) A widespread occurrence of extra open reading frames in plant Ty3/gypsy retrotransposons. Genetica 139(11–12):1543–1555
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol Bio Evol 30:2725–2729
Tian Z, Rizzon C, Du J, Zhu L, Bennetzen JL, Jackson SA, Gaut BS, Ma J (2009) Do genetic recombination and gene density shape the pattern of DNA elimination in rice long terminal repeat retrotransposons? Genome Res 19:2221–2230
Wang H, Liu JS (2008) LTR retrotransposon landscape in Medicago truncatula: more rapid removal than in rice. BMC Genomics 9:382
Wicker T, Keller B (2007) Genome-wide comparative analysis of copia retrotransposons in Triticeae, rice, and Arabidopsis reveals conserved ancient evolutionary lineages and distinct dynamics of individual copia families. Genome Res 17:1072–1081
Wicker T, Sabot F, Hua-Van A et al (2007) A unified classification system for eukaryotic transposable elements. Nat Rev Genet 8:973–982
Wright DA, Voytas DF (2002) Athila4 of Arabidopsis and Calypso of soybean define a lineage of endogenous plant retroviruses. Genome Res 12:122–131
Zhao H, Peng Z, Fei B, Li L, Hu T, Gao Z, Jiang Z (2014) BambooGDB: a bamboo genome database with functional annotation and an analysis platform. Database Vol article ID:bau006
Acknowledgements
We thank LetPub (www.letpub.com) for its linguistic assistance during the preparation of this manuscript.
Authors’ contribution
M. B. Zhou—designation of the experiments, identification and classification of LTR-retroelement, and writing of paper; B. J. Hu, Y. H. Zhu—estimation of LTR-retroelement insertion date; all authors read and approved the paper.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Funding
Funding the Talents Program of the Natural Science Foundation of Zhejiang Province (Grant No. LR12C16001), and The National Natural Science Foundation of China (Grant Nos. 31470615 and 31270645) supported this study.
Data Archiving Statement
Genomic sequences and gene annotation information of moso bamboo presented in this report are available in the bamboo genome database (BambooGDB, http://www.bamboogdb.org/index.jsp). The plant representative elements of four lineages of Ty1-Copia (Sire, Oryco, Retrofit, and Tork) and five lineages of Ty3-Gypsy (Crm, Del, Reina, Athila, and Tat) are available in the Gypsy Database (GyDB) (http://www.gydb.org/index.php/Main_Page). All full-length sequences of Ty1-Copia and Ty3-gypsy retroelements of A. thaliana and O. sativa are available in the Arabidopsis Information Resource (TAIR) (https://www.arabidopsis.org/index.jsp) and Rice Genome Annotation Project (http://rice.plantbiology.msu.edu/home_contacts.shtml), respectively.
Additional information
Communicated by C. Chen
Electronic supplementary material
Supplementary file S1
An overview of the workflow for the identification of moso bamboo LTR-retroelements. (DOC 37 kb)
Supplementary table S2
Representative sequence of each moso bamboo LTR-retroelement family, including element sizes, LTR sizes, TSD sizes, element sequences, LTR sequences, TSD sequences, primer binding site (PBS) sequences, and polypurine tract (PPT) sequences. (XLS 377 kb)
Rights and permissions
About this article
Cite this article
Zhou, M., Hu, B. & Zhu, Y. Genome-wide characterization and evolution analysis of long terminal repeat retroelements in moso bamboo (Phyllostachys edulis). Tree Genetics & Genomes 13, 43 (2017). https://doi.org/10.1007/s11295-017-1114-3
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11295-017-1114-3