Abstract
Biological desulfurization (biodesulfurization) of dibenzothiophene (DBT) by the 4S pathway is a model system for an enviromentally benign way to lower the sulfur content of petroleum. Despite a large amount of effort the efficiency of the 4S pathway is still too low for a commercial oil biodesulfurization process, but the 4S pathway could potentially be used now for commercial processes to produce surfactants, antibiotics, polythioesters and other chemicals and for the detoxification of some chemical warfare agents. Proteins containing disulfide bonds are resistant to temperature, pH, and solvents, but the production of disulfide-rich proteins in microbial hosts is challenging. The study of the 4S pathway can provide insights as to how to maximize the production of disulfide-rich proteins. Engineering of the operon encoding the 4S pathway to contain a greater content of methionine and cysteine may be able to link use of DBT as a sole sulfur source to increasing 4S pathway activity by increasing the nutritional demand for sulfur. This strategy could result in the development of biocatalysts suitable for use in an oil biodesulfurization process, but the study of the 4S pathway can also lead to a better understanding of microbial physiology to optimize activity of a mult-step co-factor-requiring pathway, as well as the production of highly stable industrially relevant enzymes for numerous applications.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Biodesulfurization is the enzymatic cleavage of carbon–sulfur bonds in compounds such as dibenzothiophene. The enzymes that accomplish carbon–sulfur bond cleavage, and the genes encoding these enzymes, have been well characterized in several bacterial cultures. The desulfurization operon of Rhodococcus erythropolis contains three genes, designated dszABC, that encode dibenzothiophene-5,5-dioxide monooxygenase, 2-hydroxybiphenyl-2-sulfinate sulfinolyase, and dibenzothiophene monooxygenase, respectively. The metabolic pathway consisting of these three enzymes, plus a NADH-FMN oxidoreductase, that accomplishes the progressive conversion of dibenzothiophene (DBT) to dibenzothiophene sulfoxide (DBTSO), dibenzothiophene sulfone (DBTSO2), 2-hydroxybiphenyl-2-sulfinate (HBPSi), and ultimately to 2-hydroxybiphenyl (2HBP) has been designated as the 4S pathway (Kilbane 2006). The 4S pathway is typically illustrated for the desulfurization of DBT, but it is important to keep in mind that the desulfurization enzymes can metabolize sulfides, disulfides, mercaptans, sulfoxides, sulfones, sulfinates, sulfonates, thiophenes, and benzothiophenes, so the potential applications of biodesulfurization enzymes extend to a wide range of chemicals.
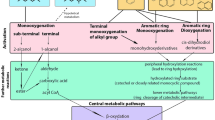
The selective removal of sulfur from petroleum can reduce air pollution resulting from the combustion of petroleum, but despite extensive research, including several genetic engineering strategies, it has not been possible to construct a biocatalyst that expresses the desulfurization trait at the levels required for a commercial oil biodesulfurization process (Debabov 2010; Alves et al. 2015; Boniek et al. 2015). However, alternative applications of biodesulfurization could support commercial processes now, and the study of biodesulfurization could lead to improved bioprocesses generally. Existing desulfurization biocatalysts can potentially be used in a variety of applications including the detoxification of some chemical warfare agents (Kilbane and Jackowski 1996), creating higher value products from organosulfur compounds in petroleum, making novel antibiotics, producing novel biodegradable plastics derived from polythioesters, and other products (Khairy et al. 2015, Kilbane 2016). However, these alternative applications for desulfurization biocatalysts have barely begun to be explored. Figure 1 illustrates a simplified representation of the biodesulfurization pathway that focuses on the sulfur atom and the carbon–sulfur bonds in organosulfur compounds and is intended to suggest the wide range of organosulfur compounds that can be metabolized by biodesulfurization enzymes and the variety of potential products that could be produced using them. Figure 1 also provides a partial listing of potential products that could be made with the assistance of desulfurization enzymes. Also shown, in some cases, are subsequent chemical (or biochemical) reactions of products produced by Dsz enzymes that introduce other functional groups such as alkyl, acyl, halogens or nitrate, particularly at locations to replace hydroxyl groups resulting from desulfurization.
Possible novel uses for desulfurization enzymes of the 4S pathway. Illustrated are the biochemical reactions in the 4S pathway listing possible substrates and the corresponding products that could be made with alternative applications. The enzymes DszA, DszB, and DszC are shown in the order in which they work in the 4S pathway with dibenzothiophene as the initial substrate. Also shown, as representative of sulfur containing compounds, is the state of the sulfur atom of dibenzothiophene at each stage of the pathway
If biotechnology is to ever to provide the widest range of products at favorable prices, it will be necessary to improve our understanding of microbial physiology and the complex interactions in biological systems. If obtaining high levels of expression of products encoded by single genes is challenging, then the efficient production of products resulting from multi-step pathways is even more challenging. The study of the 4S pathway provides a model system with unique advantages that can be used to improve our understanding of microbial physiology, how to better design microbial biocatalysts to make products resulting from multi-step pathways (Zhang et al. 2016), and how the over-expression of single genes/enzymes may be integrated into the rest of metabolism (Aggarwal et al. 2012).
Because sulfur is an essential nutrient for microbial growth, the desulfurization pathway can also be used in natural selection experiments to obtain derivative cultures with increased levels of expression of the desulfurization trait and in the process, reveal insights into sulfur metabolism. A synthetic gene encoding a high-sulfur-content-polypeptide has been inserted into the desulfurization operon and used in directed evolution experiments to simultaneously increase the nutritional demand for sulfur while selecting for cultures that utilize organically bound sulfur more efficiently such that a 20-fold increase of desulfurization activity was achieved (Pan et al. 2013).
Proteins that contain increased numbers of disulfide bonds show increased resistance to temperature, pH, and solvents, so this is relevant to industrial biotechnology generally; in addition, many venoms (Klint et al. 2013) and some therapeutic peptides contain high concentrations of sulfur (Kaspar and Reichert 2013; Yacoub et al. 2016). The expression of proteins that contain high concentrations of cysteine/methionine in microbial hosts is challenging, but the use of desulfurization-competent hosts to study the expression of high-sulfur-content-polypeptides allows for a novel approach to learn more about gene expression, protein folding, and protein secretion. It is these applications of biodesulfurization that hold the greatest promise for increasing our fundamental knowledge in microbiology, and eventually developing a commercially viable biodesulfurization process for petroleum.
Use of desulfurization enzymes for production of high value products from petroleum
Any process for the development of petroleum faces extreme challenges because huge volumes of oil must be treated inexpensively. The biotechnology industry generally is more focused on the production of low amounts of high value products such as pharmaceuticals (Biotechnology Market 2014). Despite several decades of research no currently known biocatalyst is capable of desulfurizing petroleum in an economically viable process (Kilbane 2006; Debabov 2010; Alves et al. 2015; Boniek et al. 2015). However, if the biodesulfurization pathway could be used for different applications, to make lower volumes of higher value products, then it may be possible to develop an economically viable process today that will produce a profit and support additional research to improve the biodesulfurization pathway/process. This, in turn, may eventually enable the development of a practical process for the biodesulfurization of petroleum.
Possible alternative applications that could be considered using biodesulfurization biocatalysts or enzymes include production of surfactants, and various specialty chemicals derived from organosulfur compounds present in petroleum. Potential processes for the desulfurization of petroleum include oxidative desulfurization and reactive adsorption (Stanislaus et al. 2010; Jiang et al. 2016). While these processes result in the production of low sulfur fuels, they decrease fuel yields because the organosulfur compounds are physically removed by solvent extraction or adsorption. Thus, the resulting waste streams contain enriched concentrations of organosulfur compounds and could subsequently be treated using biodesulfurization to produce sulfoxides, sulfones, sulfonates, phenols, and phenyl styrenes.
Sulfoxides and sulfones can be created in chemical processes that oxidize organosulfur compounds using ultrasound, peroxides, and other oxidizing techniques (Stanislaus et al. 2010; Bhasarkar et al. 2015; Jiang et al. 2016). Petroleum desulfurization is required to comply with environmental regulations for gasoline and diesel fuels, but hydrodesulfurization, the current method used at the industrial scale, is too expensive for high sulfur oils (Kilbane 2006; Alves et al. 2015; Boniek et al. 2015). A hybrid process that uses new chemical processes for the desulfurization of oil combined with biodesulfurization to make specialty chemicals from organosulfur waste products could simultaneously produce low sulfur fuels as well as an assortment of higher value byproducts. Sulfoxide and sulfone compounds have increased solubility in polar solvents as compared with hydrocarbons, allowing them to be removed by solvent extraction or adsorption (Stanislaus et al. 2010). If adsorption is used, the sulfoxides and sulfones could be eluted using solvents.
The beneficial use of this organosulfur material has hardly been examined, but especially with the low price of petroleum the production of higher value products derived from oil is worthy of investigation. The sulfoxide and sulfone compounds could potentially be used directly as surfactants, but subsequent treatment using biodesulfurization creates possible additional product options. Mixtures of organosulfur compounds will contain compounds that are more reactive and compounds that are somewhat recalcitrant to biodesulfurization (Zhang et al. 2013). If a biodesulfurization catalyst containing DszC and DszA, but lacking DszB, is used, then the most reactive compounds will be converted to sulfonates, yielding a mixture of sulfoxide, sulfone, and sulfonate compounds that could be fractionated by distillation or solvent extraction to yield different product streams.
Similarly, if a biocatalyst containing the intact desulfurization pathway is used, the most reactive organosulfur compounds will be converted to phenol and phenyl styrene compounds derived from dibenzothiophene-like and benzothiophene-like precursors, respectively (Wang et al. 2013). Furthermore, the chemical products resulting from the desulfurization of asymmetrically alkylated dibenzothiophenes can yield products that have hydroxyl groups added in specific locations with high yields (Onaka et al. 2001). The phenyl styrene compounds could be polymerized to yield plastic material, while the phenolic compounds, besides being polymerized, may be useful as fungicides, disinfectants, preservatives, dyes and antioxidants, and can be reacted to add nitrate, halogen, alkyl or acyl groups to create a wide range of chemical derivatives (Li and Chan 2007).
Possible uses of biodesulfurization enzymes in neutralizing toxins, and in production of novel antibiotics and polymers
Biodesulfurization can be used for the detoxification of some chemical warfare agents and other toxic chemicals (Kilbane and Jackowski 1996). The desulfurization-competent culture Rhodococcus erythropolis IGTS8 has been demonstrated to cleave carbon–sulfur bonds in the chemical warfare agent known as sulfur mustard or mustard gas (2,2′-dichlorodiethyl sulfide) resulting in detoxification, and may also be capable of detoxifying the chemical warfare nerve agent VX (o-ethyl S-[2-9diisopropylamino) ethyl] methylphosphonothioate) (Kilbane and Jackowski 1996). Strong oxidizing agents or incineration are commonly used to decontaminate and destroy chemical warfare agents, but harsh chemicals used in this way can be toxic themselves, may create toxic byproducts, and are too chemically reactive to safely decontaminate people, animals, and sensitive equipment (Prokop et al. 2006). However, enzymes and biocatalysts are non-toxic, non-corrosive, and non-flammable so they can be safely used for decontamination of skin and could be included in advanced textiles to make self-decontaminating materials and clothing to protect against chemical warfare agents and other toxic chemicals (Prokop et al. 2006). Stockpiles of VX and other chemical warfare agents are known to exist in Russia and elsewhere, and chemical warfare agents were allegedly used in Syria as recently as 2015 (Chivers 2015).
Biodesulfurization could also be used for the development of new antibiotics. Excessive and irregular use of antibiotics causes emergence of multiple drug resistance in bacteria. Antibiotic resistant bacteria are increasingly common and are becoming a global health crisis (Blair et al. 2015). Therefore, there is an urgent need to exploit alternative approaches for controlling such superbugs. Antibiotics in clinical use today are commonly chemical derivatives of parent antibiotic compounds. For example, there are dozens of chemical derivatives of penicillin (Blair et al. 2015). Chemical modification of antibiotics is often employed to develop derivatives that overcome the resistance of bacteria to the antibiotic compound originally used. However, creating chemical modifications of antibiotics can be challenging, because the antibiotic activity of the compound must be preserved. Introducing some new functional groups at precise locations in molecules while avoiding modifications to other regions of molecules may not be possible or cost effective with traditional chemical synthesis methods (Li and Chan 2007). Biochemical reactions, however, can provide highly selective modifications.
Penicillin, bacitracin, cephalothin, cephalexin and sulfanilamide are antibiotics that contain carbon–sulfur bonds, so biodesulfurization enzymes could be used to create novel chemical variants that may have the ability to treat microbial infections that are resistant to current antibiotics. The selective oxidation of sulfur and/or the selective cleavage of carbon–sulfur bonds in antibiotic molecules could provide a convenient means of creating novel chemical derivatives of antibiotics. The hydroxyl groups added to molecules as a consequence of desulfurization can subsequently be reacted to introduce nitrate, halogen, alkyl or acyl groups at specific locations (Li and Chan 2007).
The microbial production of biodegradable plastics includes biochemical production of triacylglycerols (Kurosawa et al. 2010), poly-lactic acid (Liaud et al. 2015), and sulfur-containing polythioesters (Khairy et al. 2015). Polythioesters can be used as thermoplastics and to make biodegradable polymer films. Thioesters are produced in microbial fatty acid synthesis pathways. Microbial enzymes are used now to metabolize organosulfur compounds to make novel monomers that can be polymerized to make polythioesters (Khairy et al. 2015), and desulfurization enzymes could potentially also be used to modify organosulfur compounds to produce novel compounds suitable for polymerization, as well as to modify polythioesters after polymerization to selectively oxidize sulfur atoms or cleave carbon–sulfur bonds. This could aid in the subsequent addition of various chemical functional groups and the production of polythioester derivatives with a range of novel properties.
Biodesulfurization competent hosts as model systems for efficient production of proteins with high sulfur content
General considerations
The introduction of cysteine residues and disulfide bonds between two cysteine residues within a protein can greatly stabilize a protein, making it better able to tolerate temperature, pH, and exposure to solvents and other chemicals (Northfield et al. 2014). The introduction of disulfide bonds is not the only way to produce more stable proteins, but it is a possible way to create more stable proteins. The isolation of thermophiles capable of biodesulfurization has been reported but this has not resulted in biocatalysts with high biodesulfurization activity, and there is currently no understanding of what structural features in desulfurization enzymes from thermophiles contribute to improved thermal stability (Kayser et al. 2002; Li et al. 2003; Kirimura et al. 2004; Ohshiro et al. 2005; Wang et al. 2015; Yu et al. 2015). More robust/stable enzymes are better suited to industrial processes as they can potentially support higher catalytic rates and can operate at conditions where substrates have higher solubility (Northfield et al. 2014). Durable enzymes that allow bioprocesses to run for longer times without enzyme replacement, and with the ability to tolerate product recovery approaches all contribute to more favorable economics (Klint et al. 2013). However, the introduction of cysteine residues at many locations within a protein can lead to decreased levels of enzymatic activity, and the microbial production of enzymes containing high concentrations of cysteine residues can be challenging (Klint et al. 2013).
The improved microbial production of sulfur-containing compounds would benefit from a better understanding of microbial physiology generally, and sulfur metabolism specifically, and studies of desulfurization-competent microorganisms can provide that information. Figure 2 illustrates how biodesulfurization can be used as a model system to better understand microbial physiology, particularly as it relates to the optimum functioning of multi-step cofactor requiring pathways, sulfur metabolism, production of sulfur-rich proteins, disulfide bond formation, and the secretion of sulfur containing proteins.
Flow chart illustrating the use of desulfurization competent bacterial cultures combined with directed evolution, transcriptomics, proteomics, and metabolic flux modeling to create improved biodesulfurization cultures, improved bioprocesses for multiple applications, and the improved microbial production of stable enzymes, sulfur rich therapeutic proteins, defensins, antibiotics, venoms, and other products
Biotechnology can produce high concentrations of many proteins encoded by single genes, as effective gene expression systems have been developed for several microbial hosts such as Escherichia coli (Gopal and Kumar 2013). However, the cost effective production of proteins still poses significant challenges in many cases, and yields of some recombinant proteins in E. coli are just milligrams per liter. Gene stability, translational stability, codon usage patterns, posttranslational processing, endogenous proteases, transport and localization, growth rate control, and co-expression of other genes are some of the factors impacting gene expression (Liu et al. 2015; Zhou et al. 2016). For example, even when the same gene expression system is used in three different E. coli strains, a range of yields of recombinant proteins is obtained due to variations in growth rate, plasmid stability, metabolic stress level, protein degradation rates, and other factors (Marisch et al. 2013).
E. coli is the most widely used host for the production of recombinant proteins and prokaryotic systems are less expensive than mammalian cells or fungi, but the production of sulfur-rich proteins in E. coli is problematic (Klint et al. 2013). If prokaryotic hosts could be used for the production and the correct folding and disulfide bond formation of sulfur-rich proteins, it would greatly benefit biotechnology. In this regard the study of the expression of sulfur-rich proteins in a host in which sulfur is supplied by the enzymes of the desulfurization pathway provides a valuable tool to increase our understanding of microbial cell physiology and thus lead to more efficient production of such proteins.
Cysteine scanning mutagenesis
To identify amino acid residues that are important for enzyme activity and to identify locations within a protein where cysteine can be introduced without sacrificing enzymatic activity, the process of cysteine scanning mutagenesis can be employed (Frillingos et al. 1998; Vecchiarelli and Funnell 2013; Ohnishi et al. 2014). Cysteine scanning mutagenesis is the systematic replacement of amino acids in a protein with cysteine to subsequently observe the effect on enzymatic activity and protein stability. Because of the difficulty of producing high concentrations of proteins containing high cysteine concentrations in microbial hosts (Liu et al. 2015), cysteine scanning mutagenesis is most commonly used to establish structure–function relationships in proteins and to create stable protein crystals that can be used to determine the 3-dimensional structure of proteins (Frillingos et al. 1998; Ohnishi et al. 2014). However, if high yields of cysteine-containing active, stable, enzymes could be produced in microbial hosts, they could benefit multiple bioprocesses; a combination of cysteine scanning mutagenesis and next-generation desulfurizing hosts could accomplish this.
Cysteine scanning mutagenesis can be used to find the best locations for the introduction of cysteine residues while retaining enzymatic activity, and this is a well-developed available technology, but what is lacking are microbial production hosts that can produce high yields of disulfide-rich proteins (Klint et al. 2013; Northfield et al. 2014). The study of the desulfurization operon provides a way to develop such hosts. For example, the production of a small, disulfide-rich protein from a synthetic gene inserted within the desulfurization operon of a Rhodococcus species growing with dibenzothiophene as the sole source of sulfur has been investigated; when paired with directed evolution, it resulted in 20-fold improvement in the desulfurization activity of the host (Pan et al. 2013).
Improved, next generation biodesulfurization hosts
While conventional methods of genetic manipulation of the 4S pathway such as increased copy number, use of alternative promoters, and the use of alternative microbial hosts all failed to yield biocatalysts with superior desulfurization activity (Kilbane 2006; Debabov 2010; Alves et al. 2015; Boniek et al. 2015), the novel use of a sulfur-rich protein combined with directed evolution (as described above and below) provides an example of a promising approach for developing improved desulfurization biocatalysts; still, even more improvements are needed. Biodesulfurization research would benefit from a systems biology approach. While a metabolic flux model of biodesulfurization has been developed, genome-scale transcription, proteomics, or metabolomics studies have not been reported. Some microbial strains are reported to contain an extended 4S pathway such that 2HBP is converted to 2-methyl biphenyl (Yu et al. 2015) or biphenyl (Akhtar et al. 2009); however, the genes/enzymes responsible for extended 4S pathways have not been identified.
The sulfur-rich polypeptide (“S1”) inserted into the dsz operon in prior directed evolution experiments had no enzymatic activity, but contained a signal sequence to promote the secretion of the protein, decreasing the ability of the culture to break down the protein and recycle the sulfur-containing amino acids (Pan et al. 2013). When the microbial culture was grown with DBT as the sole source of sulfur, it was dependent upon the production of the desulfurization enzymes of the 4S pathway. Because the sulfur-rich polypeptide gene was inserted within the desulfurization operon, the increased production of desulfurization enzymes also resulted in the increased production of the sulfur-rich polypeptide, thereby increasing the nutritional demand for sulfur. This created selective pressure for the culture to create more enzymatically efficient desulfurization enzymes and/or utilize sulfur more efficiently (Pan et al. 2013).
The next generation improvement of this experimental approach could be to introduce more cysteine (and methionine) residues into the desulfurization proteins themselves, at locations that preserve enzymatic activity (Fig. 2). The desulfurization genes can’t be modified, like the sulfur-rich polypeptide S1, to include signal sequences for the extracellular secretion of these enzymes, because the enzymes require cofactors, so there is still value in utilizing sulfur-rich polypeptides like S1 that can be engineered to be secreted from the cell. This would provide a useful model system to study how to produce high concentrations of sulfur-rich enzymatically active, stable enzymes, and the secretion of sulfur-rich polypeptides (Klint et al. 2013) (see below). The use of desulfurization-competent hosts to study the expression of high-sulfur-content-polypeptides allows for a novel approach to learn more about gene expression, protein folding, disulfide bond formation, and protein secretion. The lessons learned from such a system could be applied to enable the production more generally of industrially useful enzymes that have been modified using cysteine scanning mutagenesis to produce more stable derivatives.
Biodesulfurization competent hosts as model systems for efficient production of bioactive polypeptides with high sulfur content
Venoms
Another relevant application of biodesulfurization is that polypeptides that contain a high concentration of cysteine/methionine include some of the most potent venoms (Klint et al. 2013) and some therapeutic compounds such as bacteriocins, defensins, host defense peptides, and various synthetic peptides with anti-microbial, anti-fungal, anti-viral, and anti-cancer properties (Yacoub et al. 2016; Kaspar and Reichert 2013; Northfield et al. 2014). Venom proteins contain from 17 to 76 amino acids and from 2 to 6 disulfide bonds, such that the percentage of cysteine in venom proteins ranges from 10.5 to 23.5 % with most at 17–21 % (Klint et al. 2013). They are commonly secreted by animal cells and are inherently soluble and stable to temperature, pH, solvents, ionic strength, and detergents (Klint et al. 2013).
Besides being used for the production of anti-venoms, venom proteins can have therapeutic uses and some can be used as bioinsecticides (Klint et al. 2013). It would be much easier to obtain large quantities of venom proteins if they could be produced by microbial hosts rather than breeding and milking snakes, spiders, lizards, Komodo dragons, snails and other venomous animals; however, producing disulfide-rich proteins in microbial hosts can be challenging (Klint et al. 2013). The formation of disulfide bonds in E. coli occurs in the periplasm, so getting the protein to the correct cellular location is required, plus the challenge of formation of the correct disulfide bond combinations. A protein with 4 disulfide bonds could form up to 105 different isomers (Klint et al. 2013). A better understanding of the production, translocation, and disulfide bond formation of sulfur rich proteins that could be derived from the study of next generation biodesulfurization cultures could enable better and cheaper production of sulfur-rich venom proteins. Furthermore, desulfurization enzymes can be used to create derivatives of venom proteins that may have useful properties.
Bacteriocins
Bacteriocins are a diverse family of proteins produced by various microorganisms that have anti-microbial activity (Cascales et al. 2007). Moreover, bacteriocins are sulfur-containing polypeptides, and a highly conserved N-terminal sequence in bacteriocins contains a cysteine residue (Cascales et al. 2007). While disulfide bonds are generally not found in bacteriocins, some, such as nisin and subtilin, include the uncommon sulfur-containing amino acid lanthionine, which is produced by post-translational modification (Paul and van der Donk 2005). The study of the production of bacteriocins in next generation biodesulfurization cultures could provide insight into microbial physiology, specifically an improved understanding of sulfur metabolism in microbial cultures. Moreover, cysteine scanning mutagenesis can be helpful in discerning the structure–function relationships of the regions of bacteriocins responsible for antibiotic activity; this knowledge could be helpful not only in the design of bacteriocin molecules that may have clinical relevance, but may also be useful in the development of novel sulfur-containing therapeutics generally (Northfield et al. 2014).
Many therapeutic proteins are cysteine-rich/disulfide-rich, but they don’t generally contain lanthionine. The knowledge gained in the study of bacteriocins can assist in the development of novel sulfur-rich therapeutic peptides that also contain lanthionine, and similarly, the introduction of disulfide bonds in bacteriocins using insights gained through the study of venoms and disulfide-rich therapeutic proteins may allow the development of novel bacteriocins. Furthermore, the use of desulfurization enzymes can selectively modify bacteriocins and disulfide-rich therapeutic proteins, and these chemical derivatives can be tested to identify proteins with novel properties.
Defensins
Defensins include a wide variety of small cysteine-rich cationic proteins that protect both vertebrates and invertebrates from infections of bacteria, fungi and some viruses, and some defensins are reported to have anti-cancer properties (Yacoub et al. 2016). The interest in defensins by the pharmaceutical industry is increasing, along with increasing interest in host defense peptides (Kaspar and Reichert 2013). The latter are synthetic peptides built to mimic defensins and generally contain only 12–50 amino acids and a high percentage of cysteine residues (Yacoub et al. 2016). Many peptide therapeutics have received Federal Drug Administration approval for use in the USA (Kaspar and Reichert 2013), so learning from nature which protein structures yield antibiotic or therapeutic properties, as well as stability, and then using that knowledge to make useful medicines has already begun. Desulfurization enzymes can be used to make novel derivatives of defensins and host defense peptides, and the study of desulfurization competent cultures can lead to increased yields of sulfur-rich proteins in bioprocesses using microbial cultures.
Conclusion
Desulfurization biocatalysts have been trying to address one of the most difficult challenges for any process, namely to treat extremely large volumes at low cost. Desulfurization biocatalysts can be used for applications besides the desulfurization of petroleum, and while these alternative applications hold great promise, they have been largely ignored. Existing desulfurization biocatalysts could potentially be used now for commercial processes to produce surfactants, antibiotics, polythioesters and other chemicals, and for the detoxification of some chemical warfare agents. These and other alternative uses for desulfurization biocatalysts warrant further investigation.
The study of the 4S pathway provides a model system with unique advantages that can be used to improve our understanding of microbial physiology generally, sulfur metabolism specifically, and how to better design microbial biocatalysts to make products resulting from multi-step pathways. There are numerous types of sulfur-rich and/or disulfide-rich proteins with known medical or industrial applications, but using microbial hosts to produce these compounds needs improvement. The study of microbial cultures containing the 4S pathway and growing with organosulfur compounds as their sole source of sulfur provides a useful approach to improving the production of such proteins.
There is still much to learn from venoms, bacteriocins, defensins, and host defense peptides as we seek to develop improved antibiotics, anti-fungal, anti-virus, and anti-cancer compounds. The low cost production of these compounds, many of which will be cysteine-rich and disulfide-rich, is an additional challenge that can be met, at least in part, with the use of microbial processes. However, producing large quantities of peptides that are potentially toxic to the host, and possess a variety of disulfide isomers in a microbial production system will not be easy. The same can be said for the production of highly stable sulfur rich enzymes. But the use of desulfurization-competent hosts to study the expression of high-sulfur-content-polypeptides allows for a novel approach to learn more about gene expression, protein folding, disulfide bond formation and protein secretion. It is these applications of biodesulfurization that hold the greatest promise for increasing our fundamental knowledge in microbiology, and perhaps eventually developing a commercially viable biodesulfurization process for petroleum.
References
Aggarwal S, Karimi IA, Kilbane JJ II, Lee DY (2012) Roles of sulfite oxidoreductase and sulfite reductase in improving desulfurization by Rhodococcus erythopolis. Mol BioSyst 8:2724–2732
Akhtar N, Ghauri MA, Anwar MA, Akhtar K (2009) Analysis of the dibenzothiophene metabolic pathway in a newly isolated Rhodococcus spp. FEMS Microbiol Lett 301:95–102
Alves L, Paixao SM, Pacheco R, Ferreira AF, Silva CM (2015) Biodesulfurization of fossil fuels: energy, emissions and cost analysis. RSC Adv 5:34047–34057
Bhasarkar JB, Dikshit PK, Moholkar VS (2015) Ultrasound assisted biodesulfurization of liquid fuel using free and immobilized cells of Rhodococcus rhodochrous MTCC 3552: a mechanistic investigation. Bioresour Technol 187:369–378
Biotechnology Market (2014) Biopharmacy, bioservices bioagriculture, bioindustrial, fermentation, DNA sequencing, tissue engineering, regeneration, analysis and segment forecasts to 2020. Grand View Research Inc, San Francisco
Blair JMA, Weber MA, Baylay AJ, Ogbolu DO, Piddock LJV (2015) Molecular mechanisms of antibiotic resistance. Nat Rev Microbiol 13:42–51
Boniek D, Figueredo D, Batista dos Santos AF, de Resende Stoianoff MA (2015) Biodesulfurization: a mini review about the immediate search for the future technology. Clean Technol Environ Policy 17:29–37
Cascales E, Buchanan SK, Duché D, Kleanthous C, Lloubes R, Postle K, Riley M, Slatin S, Cavard D (2007) Colicin biology. Microbiol Mol Biol Rev 71:158–229
Chivers CJ (2015) Large stockpiles of VX and other chemical warfare agents are known to exist in Russia. New York Times, New York
Debabov VG (2010) Microbial desulfurization of motor fuel. Appl Biochem Microbiol 46:733–738
Frillingos S, Sahin-Toth M, Wu J, Kaback HR (1998) Cys-scanning mutagenesis: a novel approach to structure-function relationships in polytopic membrane proteins. FASEB J 12:1281–1299
Gopal GJ, Kumar A (2013) Strategies for the production of recombinant protein in Escherichia coli. Protein J 32:419–425
Jiang B, Yang H, Zhang L, Zhang R, Sun Y, Huang Y (2016) Efficient oxidative desulfurization of diesel fuel using amide-based ionic liquids. Chem Eng J 283:89–96
Kaspar AA, Reichert JM (2013) Future directions for peptide therapeutics development. Drug Discov Today 18:807–817
Kayser KJ, Cleveland L, Park H-S, Kwak J-H, Kolhatkar A, Kilbane JJ II (2002) Isolation and characterization of a moderate thermophile, Mycobacterium phlei GTIS10, capable of dibenzothiophene desulfurization. Appl Microbiol Biotechnol 59:737–745
Khairy H, Wubbeler JH, Steinbuchel A (2015) Biodegradation of the organic disulfide 4,4′-dithiodibutyric acid by Rhodococcus spp. Appl Environ Microbiol 81:8294–8306
Kilbane JJ II (2006) Microbial biocatalyst developments to upgrade fossil fuels. Curr Opin Biotechnol 17:305–314
Kilbane JJ II (2016) Future applications of biotechnology to the energy industry. Front Microbiol 7(Article 86):1–4
Kilbane JJ II, Jackowski K (1996) Biocatalytic detoxification of 2-chloroethyl ethyl sulfide. J Chem Technol Biotechnol 65:370–374
Kirimura K, Harada K, Iwasawa H, Tanaka T, Iwasaki Y, Furuya T, Ishii Y, Kino K (2004) Identification and functional analysis of the genes encoding dibenzothiophene-desulfurizing enzymes from thermophilic bacteria. Appl Microbiol Biotechnol 65:703–713
Klint JK, Senff S, Saez NJ, Seshadri R, Lau HY, Bende NS, Undheim EAB, Rash LD, Mobli M, King GF (2013) Production of recombinant disulfide-rich venom peptides for structural and functional analysis via expression in the periplasm of E. coli. PloS One 8:e63865
Kurosawa K, Boccazzi P, deAlmeida NM, Sinskey AJ (2010) High-cell-density batch fermentation of Rhodococcus opacus PO630 using a high glucose concentration for triacylglycerol production. J Biotechnol 147:212–218
Li C-H, Chan T-H (2007) Comprehensive organic reactions in aqueous media. Wiley-Interscience, Wiley, Hoboken, NJ
Li FL, Xu P, Ma CQ, Luo LL, Wang XS (2003) Deep desulfurization of a hydrodesulfurization-treated diesel oil by a facultative thermophilic Mycobacterium sp. X7B. FEMS Microbiol Lett 223:301–307
Liaud N, Rosso M-N, Fabre N, Crapart S, Herpoel-Gimbert I, Sigoillot J-C, Raouche S, Levasseur A (2015) L-lactic acid production by Aspergillus braziliensis overexpressing the heterologous ldha gene from Rhizopus oryzae. Microb Cell Fact 14:66
Liu JL, Goldman ER, Zabetakis D, Walper SA, Turner KR, Shriver-Lake LC, Anderson GP (2015) Enhanced production of a single domain antibody with an engineered stabilizing extra disulfide bond. Microb Cell Fact 14:158
Marisch K, Bayer K, Cserjan-Puschmann M, Luchner M, Striedner G (2013) Evaluation of three industrial Escherichia coli strains in fed-batch cultivations during high-level SOD protein production. Microb Cell Fact 12:58
Northfield SE, Wang CK, Schroeder CI, Durek T, Kan MW, Swedberg JE, Craik DJ (2014) Disulfide-rich macrocyclic peptides as templates in drug design. Eur J Med Chem 77:248–257
Ohnishi S, Hays A, Hagenbuch B (2014) Cysteine scanning mutagenesis of transmembrane domain 10 in organic anion transporting polypeptide 1B1. Biochemistry 53:2261–2270
Ohshiro T, Ishii Y, Matsubara K, Ueda K, Izumi Y, Kino K, Kirimura K (2005) Dibenzothiophene desulfurizing enzymes from moderately thermophilic bacterium Bacillus subtilis WU-S2B: purification, characterization and over-expression. J Biosci Bioeng 100:266–273
Onaka T, Kobayashi M, Ishii Y, Konishi J, Matuhashi K (2001) Selective cleavage of the two C-S bonds in asymmetrically alkylated dibenzothiophenes by Rhodococcus erythropolis KA2-5-1. J Biosci Bioeng 92:80–82
Pan J, Wu F, Wang J, Xu L, Khayyat NH, Stark BC, Kilbane JJ II (2013) Enhancement of desulfurization activity by enzymes of the Rhodococcus dsz operon through coexpression of a high sulfur peptide and directed evolution. Fuel 112:385–390
Paul M, van der Donk WA (2005) Chemical and enzymatic synthesis of lanthionines. Mini Rev Org Chem 2:23–37
Prokop Z, Oplustil F, DeFrank J, Damborsky J (2006) Enzymes fight chemical weapons. Biotechnol J 1:1370–1380
Stanislaus A, Marafi A, Rana MS (2010) Recent advances in the science and technology of ultra low sulfur diesel (ULSD) production. Catal Today 15:1–68
Vecchiarelli AG, Funnell BE (2013) Probing the N-terminus of ParB using cysteine-scanning mutagenesis and thiol modification. Plasmid 70:86–93
Wang W, Ma T, Lian K, Zhang Y, Tian H, Ji K, Li G (2013) Genetic analysis of benzothiophene biodesulfurization pathway of Gordonia terrae strain C-6. PloS One 8:e84386
Wang J, Davaadelger B, Salazar JK, Butler RR, Pombert JF, Kilbane JJ II, Stark BC (2015) Isolation and characterization of an interactive culture of two Paenibacillus species with moderately thermophilic desulfurization ability. Biotechnol Lett 37:2201–2211
Yacoub HA, El-Hamidy SM, Mahmoud MM, Baeshen MN, Almehdar HA, Uversky VN, Redwan EM, Al-Maghrabi OA, Elazzaz AM (2016) Biocidal activity of chicken defensin-9 against microbial pathogens. Biochem Cell Biol 94:176–187
Yu B, Tao F, Li F, Hou J, Tang H, Ma C, Xu P (2015) Complete genome sequence of Mycobacterium goodie X7B, a facultative thermophilic biodesulfurization bacterium with industrial potential. J Biotechnol 212:56–57
Zhang SH, Chen H, Li W (2013) Kinetic analysis of biodesulfurization of model oil containing multiple alkyl dibenzothiophenes. Appl Microbiol Biotechnol 97:2193–2200
Zhang J, Quan C, Wang C, Wu H, Li Z, Ye Q (2016) Systematic manipulation of glutathione metabolism in Escherichia coli for improved glutathione production. Microb Cell Fact 15:38
Zhou Y, Liu P, Gan Y, Sandoval W, Katakam AK, Reichelt M, Rangell L, Reilly D (2016) Enhancing full-length antibody production by signal peptide engineering. Microb Cell Fact 15:47
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kilbane, J.J., Stark, B. Biodesulfurization: a model system for microbial physiology research. World J Microbiol Biotechnol 32, 137 (2016). https://doi.org/10.1007/s11274-016-2084-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11274-016-2084-6