Abstract
Certain factors hinder the commercialization of biodesulfurization process, including low substrate-specificity of the currently reported desulfurizing bacteria and restricted mass transfer of organic-sulfur compounds in biphasic systems. These obstacles must be addressed to clean organic-sulfur rich petro-fuels that pose serious environmental and health challenges. In current study, a dibenzothiophene desulfurizing strain, Gordonia rubripertincta W3S5 (source: oil contaminated soil) was systematically evaluated for its potential to remove sulfur from individual compounds and mixture of organic-sulfur compounds. Metabolic and genetic analyses confirmed that strain W3S5 desulfurized dibenzothiophene to 2-hydroxybiphenyl, suggesting that it follows the sulfur specific 4 S pathway. Furthermore, this strain demonstrated the ability to produce trehalose biosurfactants (with an EI24 of 53%) in the presence of dibenzothiophene, as confirmed by TLC and FTIR analyses. Various genome annotation tools, such as ClassicRAST, BlastKOALA, BV-BRC, and NCBI-PGAP, predicted the presence of otsA, otsB, treY, treZ, treP, and Trehalose-monomycolate lipid synthesis genes in the genomic pool of strain W3S5, confirming the existence of the OtsAB, TreYZ, and TreP pathways. Overall, these results underscore the potential of strain W3S5 as a valuable candidate for enhancing desulfurization efficiency and addressing the mass transfer challenges essential for achieving a scaled-up scenario.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Sulfur is the third most prevalent element in petro-fuels found as organic and inorganic forms (Feng et al. 2016; Prasoulas et al. 2021; Silva et al. 2020). The combustion of sulfur-rich petro-fuels release SOX gases that can cause serious health and environmental problems (Akhtar et al. 2019; Mohebali and Ball 2016; Porto et al. 2018). To meet the set ultra-low levels of sulfur in petro-fuels, the scientists around the world are working to find effective pre-combustion ways to efficiently reduce sulfur in fuels with low operating costs (Akhtar et al. 2019; Mohebali and Ball 2008; Parveen et al. 2020; Glekas et al. 2022).
Petroleum refineries are using a chemical hydrodesulfurization (HDS) process to remove sulfur from petro-fuels, however, it is ineffective against certain organic-sulfur compounds i.e., dibenzothiophene (DBT), benzothiophene (BT), and their alkylated forms (Ferreira et al. 2017). Moreover, this method is energy-intensive, uses chemical catalysts, and releases toxic by-products such as H2S, resulting in equipment erosion and environmental pollution (Martínez et al. 2017; Rangra et al. 2018). Biodesulfurization (BDS) is a complementary alternative approach that uses particular micro-organisms to remove inorganic sulfur as well as recalcitrant organic-sulfur compounds from petro-fuels (Boniek et al. 2015; Rangra et al. 2018; Al-khazaali and Ataei 2023). In contrast to the HDS process, BDS is an eco-friendly, energy-efficient method that works under mild conditions and removes the sulfur from organic-sulfur compounds without reducing their calorific value (Aggarwal et al. 2013; Rangra et al. 2018; Silva et al. 2023). A number of bacterial genera have been reported, that can remove sulfur from organic-sulfur compounds through a well-known “4S” metabolic pathway (Akhtar et al. 2016a, b, 2018; Sadare et al. 2017). During this pathway, the DBT is sequentially converted into DBTO (DBT-Sulfoxide), DBTO2 (DBT-Sulfone), HBPSi (2-Hydroxybiphenyl-2-Sulfinite), and 2-HBP (2-Hydroxybiphenyl). The monooxygenases DszC, DszA and desulfinase DszB (Bhanjadeo et al. 2018; Kilbane and Stark 2016; Su et al. 2018; Wang et al. 2017) are involve in this pathway, which are encoded by a group of three genes dszC, dszA and dszB respectively (Bhanjadeo et al. 2018; Santos et al. 2006; Su et al. 2018).
One of the major bottleneck of industrial-scale application of BDS is the scarcity of bacterial strains capable of desulfurizing a range of organic-sulfur compounds i.e., DBT, BT, and their alkylated derivatives (Alves et al. 2015; Parveen et al. 2020). Another major characteristic of an ideal desulfurizing strain is its ability to produce biosurfactants (Alves et al. 2015; Parveen et al. 2020). Biosurfactants are amphiphilic compounds that could play a critical role during biodesulfurization as they increase the mass transfer of the substrate between oil and the aqueous phase (biphasic system) by decreasing the surface tension of the medium (Alves et al. 2015; Raheb et al. 2009). Trehalose-surfactants have the potential for use in cleaning oil storage tanks and microbial enhanced oil recovery because of their extraordinary capacity to reduce surface tension, environmentally benign nature and enhanced solubility of hydrophobic compounds (Sousa et al. 2020; Wang et al. 2019).
In this study, we present the research conducted on an efficient C–S bond cleaving bacterial strain W3S5 able to desulfurize various organic-sulfur compounds i.e., DBT, BT, Thiophene, and their alkyl derivatives separately as well as in mixture. Based on phylogenomic analysis, the strain has been characterized as Gordonia rubripertincta W3S5, and is found to follow the well-documented “4S” pathway of DBT desulfurization as revealed by the complete identification of dszABC genes and GC-MS analysis of the extracted metabolites. Furthermore, strain W3S5 demonstrated the ability to produce trehalose biosurfactants in the presence of dibenzothiophene, as confirmed by TLC and FTIR analyses. Moreover, the trehalose biosynthesis genes have been identified in strain W3S5 using its whole genome sequence.
Materials and methods
Chemicals
All the organic-sulfur compounds and 2-HBP used in this study were obtained from Sigma Co. (St. Louis, USA). All other media components and solvents used for the extraction of surfactants, DBT desulfurization metabolites, and GC–MS analysis were of HPLC-grade and bought from Merck Co. (Darmstadt, Germany).
Sample collection
To isolate organic-sulfur compounds metabolizing microorganisms, about 50 samples of crude-oil contaminated water, sand, soil, and sludge were taken from an oil drilling company, in sterilized bags/bottles (Supplementary Fig. 1A). The physicochemical properties, like nature of sample, total dissolved solids (TDS), dissolved oxygen (DO), temperature, electric conductivity (EC), salinity, and pH were recorded for representative samples (Supplementary Table 1), by multi-parameter (InoLab IDS Multi 9430, WTW, Germany). For soil, sand and sludge samples the parameters were calculated by making a 50% slurry (w/v) in deionized water.
Media and culture growth conditions
All the studies were carried out in a sulfur-free, chemically defined, Minimal-salt Glucose (MG) media (Akhtar et al. 2009). The composition of the MG media per Litre of distilled water was glucose: 5.0 g, KH2PO4: 2.0 g, K2HPO4: 4.0 g, MgCl2·6H2O: 0.2 g (pH 7.0), NH4Cl: 1.0 g, metal solution 10.0 mL and vitamin mixture 1.0 mL. For solid form of MG media, 0.75% of gellan gum powder was used. The vitamin mixture was composed of calcium pantothenate: 0.4 g, niacin: 0.4 g, inositol: 0.2 g, pyridoxine hydrocholride: 0.4 g, p-amino benzoic acid: 0.2 g, and cyanocobalamine: 0.0005 g per 1000 mL (distilled water) and filter sterilized through a vacuum filter assembly (LF-30, Rocker Scientific, Taiwan) containing 0.2 μm nylon membrane filter. The metal solution contained CaCl2: 2.0 g, NaCl: 1.0 g, ZnCl2: 0.5 g, FeCl2·4H2O: 0.5 g, MnCl2·4H2O: 0.5 g, Na2MoO4: 0.1 g, Na2WO4·2H2O: 0.05 g, CuCl2: 0.05 g, and 10 mL of HCl (10 M) per 1000 mL (distilled water) and filter sterilized. All the experiments were conducted in triplicates at 30 °C and 180 rpm.
Enrichment and purification of DBT desulfurizing strains
Culture enrichment was carried out by suspending 5 g/5 mL of oil-contaminated samples such as soil, sand, sludge and water in sterilized MG media (100 mL) containing 0.5 mM of DBT (prepared in ethanol ) as the sole sulfur source and incubated at 30 °C and 180 rpm shaking (Kuhner, Switzerland). After 7 days of incubation, the culture broth (5 mL) from each flask was transferred into fresh MG medium and was sub-cultivated 3 times at the same growth conditions. At the end of 3 sub-cultivations, the cultures were analyzed for the metabolism of DBT into 2-HBP (a sulfur-free product), by conducting Gibb’s assay. For this purpose, NaHCO3: 30 µL (1 M, pH 8) and Gibb’s reagent (2,6-Dichloroquinone-4-chloroimide): 20 µL (1 mg/mL prepared in ethanol) were added into 150 µL of cell-free supernatant, mixed, and left at room temperature for 1 h. A blue color appears upon the reaction of Gibb’s reagent with aromatic hydroxyl groups, i.e., 2-HBP, which indicates the metabolism of DBT into sulfur-free compounds (Kayser et al. 1993). The development of blue color provides a qualitative assessment of a microbial culture’s capacity to desulfurize organic sulfur compounds; the darker the hue, the more effective the culture is in DBT desulfurization. To obtain the pure cultures, the DBT enriched Gibb’s positive mixed cultures (W3SN1, W3SL1, and W3S2) was serially diltued and spreaded on MG media plates. After 5 days of incubation the well seperated colonies were picked and streaked on new MG media plates for 3 times in order to confirm the culture purity. The pure cultures were cultivated in liquid MG media at 30 °C and 180 rpm for 7 days and tested for Gibbs activity. Initially 8 bacterial isolates (W3S2-P9, W3S4, W3SN5, W3SL1, W3SN1, W3S5, W3SN4, W3SN2) were purified and screened from DBT enriched Gibb’s positive mixed-cultures (see supplementary Fig. 1B, 1 C), of which the isolate W3S5 exhibited the highest level of desulfurization activity (see supplementary Fig. 1D) and was chosen for further investigations. All the cultures were stored at − 20 °C in cryo-vials containing 70% (v/v) glycerol and at 4 °C on LB/MG media slants till further use.
Morphological and molecular identification of isolated bacterium
The isolated DBT metabolizing strain was identified through morphological and molecular methods. The morphological characterization was carried out by carefully examining the colony morphology and by determining the cell shape through phase-contrast microscopy (CX41RF Olympus, Japan). Molecular characterization of the isolated strain was conducted by sequencing the 16S rRNA gene and whole genome sequencing (Parveen et al. 2021). In the present study, we have used TYGS: Type (Strain) Genome Server (https://tygs.dsmz.de/) to undertake phylogenomic analysis to identify the W3S5 strain (Meier-Kolthoff and Göker 2019). For, this purpose the fasta-formatted contig file was uploaded to TYGS genome server to obtain the genome-based phylogenetic tree using the default settings.
Genetic analysis of the dszABC operon of W3S5
The dszABC genes amplification, cloning and sequencing
DBT desulfurization genes dszA, dszB, and dszC of W3S5 strain were amplified using specific and degenerate primers through PCR amplification. The specific and the degenerate primers were designed using the 5′ and 3′ sequences of dszABC genes of Rhodococcus erythropolis IGTS8 as described in previous studies (Akhtar et al. 2014; Kilbane and Robbins 2007), and were obtained from Integrated DNA Technologies, Inc., UK (Supplementary Table 2). The detail of the PCR profile for the amplification of dszABC genes is given in Supplementary Tables 3, 4 and 5. The PCR amplicons were cloned by TA cloning using Escherichia coli TOP10 competent cells and confirmed by restriction digestion and colony PCR. The resulting clones were sequenced from Macrogen, Korea using M13 forward and reverse primers.
Identifying the complete dszABC gene sequences by whole-genome sequence analysis
The draft genome sequencing (WGS) of the W3S5 strain was achieved through Illumina (NovaSeq) platform (2 × 100 bp paired-end) by Macrogen Inc. Korea. To identify the complete dszABC gene sequences in W3S5 the PCR amplified partial dszABC gene sequences were used to conduct a gene similarity search in the draft genome sequence of W3S5 using RAST-alignment tool. The identified complete sequences of dszA, dszB and dszC genes were submitted in GenBank database at NCBI (www.ncbi.nlm.nih.gov).
Phylogenetic analysis of dszABC gene
The deduced complete dszABC gene sequences were used for phylogenetic analysis to find out the evolutionary relationship between W3S5 and other bacterial desulfurization genes. The homologous dsz gene sequences were retrieved from BLASTn analysis (available at www.ncbi.nlm.nih.gov) and applied to build the phylogenetic tree through MEGA-X program (Tamura and Nei 1993; Tamura et al. 2013). A multiple-sequence alignment was generated through ClustalW using default parameters and the resulting alignment was employed to build the phylogenetic tree using Maximum Likelihood method, Tamura-Nei-model, and bootstrap approach (100 replicates). Initial tree(s) for the heuristic search were obtained by applying the Neighbor-Joining method to a matrix of pairwise distances estimated using the Maximum Composite Likelihood (MCL) approach.
Desulfurization of various organic-sulfur compounds
To investigate the broad substrate specificity, the W3S5 culture was initially examined to desulfurize various organic-sulfur compounds individually. The strain was cultivated in MG-medium with 0.2 mM concentrations of DBT (Dibenzothiophene), DBTS (Dibenzothiophene Sulfone), 4-MDBT (4-Methyldibenzothiophene), 2,8-DMDBT (2,8-Dimethyldibenzothiophene), BT (Benzothiophene), 3-MBT (3-Methylbenzothiophene), TH (Thiophene), 2-MTH (2-Methylthiophene), and 3-MTH (3-Methylthiophene). To determine whether these compounds were metabolized, the bacterial cell growth (OD660 nm) and the production of the phenolic end-product, 2-HBP/2-HBPeq (OD595 nm), were recorded every 24 h. Furthermore, the isolate was also investigated to desulfurize organic-sulfur compounds in combination as well. For this purpose, the strain W3S5 was cultivated in MG media containing 0.1 mM concentrations of each DBT, 3-MDBT, DBTS, BT and 3-MBT, making up a total concentration of 0.5 mM in the medium. The growth (OD660 nm) and the 2-HBP production (OD595 nm) was recorded every 24 h.
Detection of biosurfactants produced by W3S5 during DBT desulfurization
Qualitative tests for detecting the biosurfactants
Oil displacement and Emulsification index (% EI24) tests were performed for the detection of biosurfactants produced by W3S5 strain, as described by Tabatabaee et al. (2005). For oil displacement test, poured 50 mL of distilled water into a petri dish (100 mm × 15 mm) and then added 20 µL of crude-oil to the surface of water and allowed it to settle at room temperature for 2 min. Dropped 10 µL of culture extract (grown in 0.2 mM DBT supplemented MG media for 5 days) onto the oil surface, that immediately created a clear halo on the surface of oil. The diameter of the clear halo was noted under visible light and used to calculate the oil displacement area (ODA) by using Eq. 1.
The Emulsification index (% EI24) was estimated by mixing culture supernatant and the kerosene oil in equal volumes (1:1) in a glass tube (125 mm × 15 mm). After being vortexed for 3 min, the glass tubes were left at room temperature for 24 h. The EI24 index was then calculated following Eq. 2, by measuring the total height of the liquid column and the height of the emulsified layer using a ruler scale.
To compare the results, a positive control (1% Sodium dodecyl sulfate) and a negative control (fresh culture medium) were also used. These experiments were repeated for three times to take an average of the recorded values.
Solvent extraction of biosurfactants
To characterize the biosurfactants produced by W3S5 strain, the culture was cultivated in 100 mL of MG medium in triplicates, added with DBT to a final concentration of 0.2 mM, and grown for 5 days until it reached the log-phase. The whole culture broth from the log-phase was used to extract the biosurfactants using an equal volume of chloroform: methanol solution (2:1) thrice by shaking the culture broth vigorously every time to allow the separation of two layers until no further color appears in the culture broth. The extracted biosurfactants concentrated to dryness in a rotary vacuum evaporator (G3 Heidolph, Germany) and were re-suspended in chloroform: methanol solution (2:1) for further analysis (Wang et al. 2019).
Purification of biosurfactants by thin-layer chromatography (TLC)
The extracted biosurfactants were purified by Thin-Layer Chromatography (TLC) using pre-coated aluminum sheets (Silica gel 60-F254 plates, Merck, Germany). To separate the crude biosurfactants, 10 µL of the sample was carefully loaded on the TLC plate, run for 90 min using butanol:pyridine:water (15:30:20, v/v) solution as a mobile phase system, and left at room temperature to dry for 10 min. The dried TLC plate was colored developed by spraying with 0.5% of KMnO4 solution prepared in 1 N NaOH, carefully scratched the well separated bands from the TLC plate and dissolved in chrolorom:methanol (2:1) solvent till further use to perform Fourier Transform Infrared Spectroscopy (FT-IR) analysis.
Identifying the trehalolipid-related genes in W3S5 by analyzing the genome sequence
To identify the trehalose-surfactant related genes in W3S5, its whole-genome sequence was analyzed through different online genome annotation programs such as ClassicRAST (version 2.0): Rapid Annotation using Subsystem Technology (https://rast.nmpdr.org/), BV-BRC: Bacterial and Virus Bioinformatics Resource Centre (https://www.bv-brc.org/), BlastKOALA: BLAST- KEGG Orthology And Links Annotation (https://www.kegg.jp/blastkoala/) and NCBI-PGAP: NCBI Prokaryotic Genome Annotation Pipeline (https://www.ncbi.nlm.nih.gov/genome/) using default settings. The RAST and BV-BRC annotation servers accepts FASTA formatted contig file while BlastKOALA uses FASTA formatted amino acid sequence file as input query. In case of NCBI-PGAP, the annotation for a genome sequence becomes available at NCBI when the whole genome sequence is submitted to NCBI genome server as FASTA formatted contig file.
Analytical methods
To measure the bacterial cell growth, the optical-density (OD660 nm) of the culture broth was recorded through Double-Beam UV–Visible Spectrophotometer (Camspec M350, UK), and the pH was noted using a multi-parameter (InoLab IDS Multi 9430, WTW, Germany). The concentration of 2-HBP (end product) was measured by performing Gibb’s assay and taking the optical density of the resulting blue-colored solution at 595 nm followed by comparing the OD values against 2-HBP standard-curve. The qualitative estimation of DBT conversion into 2-HBP and the characterization of desulfurization metabolites was conducted through High-Performance Liquid Chromatography (HPLC) followed by Gas Chromatography-Mass Spectrometry (GC–MS) analysis. For this purpose, the W3S5 culture was inoculated in MG media (100 mL) containing 0.2 mM of DBT (sole sulfur source) and incubated at 30 °C and 180 rpm. After 3 days of incubation, 50 mL of culture broth was removed for metabolite extraction. To obtain the cell-free supernatant the culture-broth was centrifuged at 13,000×g, 4 °C, and 15 min followed by acidification to pH 2.0 using HCl (6 M) solution. The DBT desulfurization metabolites were extracted from acidified supernatant with 30 mL of ethyl-acetate and concentrated to dryness by evaporating the sample through Rotary vacuum evaporator (G3 Heidolph, Germany). The extracted metabolites were dissolved in acetonitrile solvent and filtered using nylon filters (0.2 µm pore size). The HPLC (Perkin-Elmer Series 200, USA) analysis was performed at 245 nm using the Hypersil-C18-column (Thermo Hypersil-Keystone, UK). The acetonitrile:water (60:40, v/v) solution was used as the mobile-phase system for HLPC analysis, and the samples were run at the rate of 1 mL per min to obtain the bands of DBT metabolites. The DBT desulfurization metabolites were further characterized using a Triple Quad 7000D GC–MS equipment from Agilent-Technologies (Little Falls, California, USA) equipped with an Agilent-Technologies 7683B series auto-injector and an Agilent-Technologies 5975 inert XL Mass selective detector. The samples (1.0 µL) were injected into HP-5MS column (30 m × 0.25 mm, film thickness 0.25 mm; Little Falls, CA, USA) in the split-mode keeping a split ratio of 1:10. The GC/MS analysis was performed using an electron-ionization system (70 eV) and keeping the column-oven temperature at 60 ºC for 1 min, followed by ramping at 10 °C per min to 325 °C, for 10 min. Helium was employed as a carrier-gas at a flow rate of 0.5 mL per min. The mass-scanning range was adjusted as 50 to 550 m/z, whereas the injector and Mass Spectrum transfer-line temperatures were adjusted to 220 and 290 °C, respectively. The extracted metabolites were characterized by comparing their mass-spectra with standards as well as NIST02.L and WILEY7n.L mass-spectral libraries. Fourier-transform infrared spectroscopy (FT-IR) was used to characterize the extracted/ TLC-purified biosurfactants produced by the W3S5 strain. The FT-IR analysis was performed in a dry atmosphere using Perkin-Elmer grating 1430 IR (Norwalk, CT). Data were collected in the range of 400–4000 wavenumbers (cm−1) having a resolution of 4 cm−1 and processed with IR analytical software. Samples were developed by diffusing the solid extract homogeneously in a potassium bromide matrix.
Results
Isolation and identification
To obtain efficient DBT desulfurizing bacteria, a total of 50 crude-oil contaminated samples were screened using MG media containing 0.5 mM of DBT as a sole sulfur source. Out of eight bacterial cultures, W3S5 exhibited maximum desulfurization activity (see Supplementary Fig. 1D). The 2-HBP bar graph in Fig. 1B represents that isolate W3S5 successfully desulfurized nearly all the DBT into 2-HBP within 72 h of incubation to obtain sulfur for its growth. This phenomenon was further evaluated by growing the culture in MG-medium containing either an organic-sulfur source (DBT) or a carbon source (glucose) or both. Figure 1C demonstrate that the isolate W3S5 growth was highest when both glucose and DBT were supplied in the medium, and reduced significantly when only DBT or glucose were added in the medium. These findings confirm that W3S5 requires both a carbon source and an organic-sulfur source for its optimum growth. The isolate shows a distinctive colony morphology when grown on LB agar plates and appeared as coccoid shape under phase-contrast microscope (Fig. 1A).
Phenotypic traits and DBT desulfurization estimation of strain W3S5. A The pure culture of W3S5 showing distinctive colony morphology with characteristics such as pink colored, elevated, viscous texture, and entire at the edges, when grown on LB agar plates (i), the cells appeared as circular, non-motile, non-spore-forming, and coccoid shape, when observed under phase-contrast microscope, magnification 100 X, Ph:3, (ii). B Estimation of cell growth and 2-HBP production by W3S5 strain when cultivated in liquid MG medium supplemented with 0.2 mM of DBT and 5% of glucose. (C) A comparison of cell growth and 2-HBP production when W3S5 was cultivated in MG medium with DBT only (organic-sulfur source), glucose only (carbon source), or DBT plus glucose (organic-sulfur source plus carbon source). The SD (n = 3) is shown as error bars
For genome-based identification of strain W3S5, the phylogenomic analysis was carried out through TYGS: Type (Strain) Genome Server (https://tygs.dsmz.de). The phylogenomic tree in Fig. 2 demonstrates that the strain W3S5 clusters with the Gordonia rubripertincta NBRC 101,908 and Gordonia rubripertincta ATCC 14,352 type strains, with a bootstrap value of 97, a δ value of 0.087, and sharing approximately the same G + C content of 67% with genome sizes of 4.8, 5.6, and 5.2 Mb (see Fig. 2, label 1–5) respectively. The number of coded proteins for Gordonia rubripertincta NBRC 101,908, Gordonia rubripertincta ATCC 14,352 and W3S5 are also nearly the same i.e., 5023, 4802 and 4326 (see Fig. 2, label-6) respectively. From these results it can be concluded with high confidence that strain W3S5 is taxonomically more related to Gordonia rubripertincta.
The TYGS genome-based phylogenomic analysis of W3S5 strain. The phylogenomic tree was constructed using GBDP distances estimated from genome sequences and was deduced using FastME 2.1.456. The branch support was calculated using GBDP pseudo-bootstrap values from 100 replications and are shown above each branch, whereas the branch lengths are scaled with reference to GBDP distance formula d5. The clustering produced 10 species and subspecies clusters. The colored leaf-labels in the figure represent various aspects of the genome annotations and the same color codes have been used to identify the species and subspecies within the same taxon, such as label 1 and 2 in the figure shows clusters of same dDDH (digital DNA-DNA Hybridization) species and subspecies, respectively. The percent G + C content of each species is represented on label 3 by light to dark blue color codes, with the lowest to greatest values. Similarly, labels 4, 5, and 6 with the brown, black, and mustard color codes, respectively, denote the delta value, genome size, and number of proteins for each species. The length of the colored boxes varies to show the range of values. Whereas label 7 designates the type of strain at each branch, such as Type-Strain, Type-Specie, or User-Strain, with each denoted by a white box, a red circle and a plus sign respectively
Characterization of DBT desulfurization pathway in W3S5 by GC–MS analysis of the extracted metabolites. A Gas-Chromatogram of DBT desulfurization extracted metabolites. B, C, D Mass-spectrum of main ion peaks 184.1, 216.1, and 170.2, identified as DBT, DBTO2 and 2-HBP respectively. E Proposed “4S” pathway of DBT desulfurization in W3S5 strain, interpreted from GC–MS analysis. The DBT is first converted into DBT-sulfoxide (DBTO) by DBT monooxygenase DszC through addition of an oxygen molecule. The DBTO is transformed into DBT sulfone (DBTO2) by the same enzyme through addition of another oxygen molecule. The DBT monooxygenase DszA then converts DBTO2 into sulfinate HBPSi (2-Hydroxybiphenyl-2-Sulfinite. Finally, the HBPSi is metabolized into 2-Hydroxybiphenyl (2-HBP)) and sulfate by desulfinase DszB enzyme by cleaving the C–S bond
Elucidation of the DBT desulfurization pathway through metabolic and genetic analysis
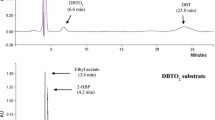
The desulfurizing metabolites produced by W3S5 strain during the desulfurization of DBT were identified using HPLC and GC–MS analysis of extracted metabolites. Initially, the HPLC analysis was performed, which revealed the existence of DBT, DBTO2, and 2-HBP in the extracted metabolites, when compared to the standards. The further characterization of DBT desulfurization metabolites was conducted through GC–MS analysis of the extracted metabolites. In GC chromatogram, a strong peak was detected at a retention time (Rt) of 6.461 min along with some minor peaks (Fig. 3A). When the GC spectra was compared with the standards, the sharp peak (Rt: 6.461 min) was identified as 2-HBP while the minor peaks at the Rt of 12.814 and 10.065 min were identified as DBT and DBTO2 respectively. The mass-spectrometry of detected peaks further validated the existence of 2-HBP, DBTO2 and DBT in the culture extract. The main ion peaks detected at the m/z of 184.1, 216.1, and 170.2 (Fig. 3B, C and D), correspond to the molecular masses of DBT, DBTO2, and 2-HBP respectively, when compared with MS Spectra in WILEY7n.L, and NIST02.L mass-spectral libraries and standards. The DBTO was not detected in the analysis which may be due to its instant conversion into DBTO2 or the hydrophilic nature that hinders its extraction with organic solvents from culture broth (Parveen et al. 2020; Su et al. 2018)
The enzymes of the “4S” pathway are encoded by the dszA, dszB, and dszC genes, which are found on the plasmid DNA in the form of an operon (Ferreira et al. 2017). In order to characterize the complete dszABC gene sequences in strain W3S5, initially the partial sequences of dszA (~ 350 bp), dszB (~ 600 bp) genes and almost the complete sequence of dszC (~ 1250 bp) gene were obtained by PCR amplification and sanger sequencing. The NCBI-BLASTn analysis of these sequences showed > 95% similarity with dsz genes of other desulfurizing bacteria. The RAST alignment search of partial dszA, dszB and dszC gene sequences against the genome sequence of W3S5 revealed the presence of dszABC genes on contig#22 in the form of an operon (Fig. 4A).
Characterization of complete dszABC gene sequences in strain W3S5 and determination of their evolutionary relationship with other related desulfurizing bacteria. A The structure of dsz operon located at contig#22, identified in W3S5 strain by searching the whole-genome sequence through RAST alignment tool. B The outcomes of BLASTn search displaying the closely related sequences to the complete dszABC gene sequences of W3S5. C Phylogenetic tree of dszABC genes of W3S5
Phylogenetic analysis offers a thorough understanding of evolutionary relationship among genus/species or genes (Gray et al. 1996; Kilbane II and Le Borgne, 2004; Pylro et al. 2012). The complete dszABC gene sequences (retrieved from RAST) of W3S5 showing > 88% homology with dszABC genes of other desulfurizing bacteria during NCBI-BLASTn analysis (Fig. 4B) were used to construct the phylogenetic tree using MEGA-X program. The phylogenetic trees in Fig. 4C show that the dszA, dszB, and dszC genes of W3S5 form a cluster (cluster I) with Gordonia sp. IITR100 (KC693733.1), Gordonia sp. WQ-01 (DQ448811.1), Gordonia alkanivorans RIPI90A (EU364831.1), and Gordonia alkanivorans 1B (AY678116.1) which shows a close evolutionary relatedness among their dszABC genes than with other species desulfurizing genes (see cluster II) making a separate cluster suggesting that the dsz genes of W3S5 strain are conserved and quite similar to the dsz genes of Gordonia species.
Based on the characterization of the DBT desulfurization metabolites i.e., DBT, DBTO2, 2-HBP and the identification of dszABC genes it is evident that the isolate W3S5 follows the same steps of “4S” pathway reported in the literature (Gunam et al. 2006; Parveen et al. 2020; Su et al. 2018). Keeping in view our findings we have proposed a basic schematic of “4S” pathway in W3S5 as shown in Fig. 3E.
Investigating the desulfurization of DBTs, BTs, and THs, both separately as well as a mixture by strain W3S5
The isolate W3S5 was interrogated for its ability to metabolize a variety of HDS resistant organic-sulfur compounds like DBTs, BTs, and THs. The line graph in Fig. 5A shows the cell growth of W3S5 cultivated in the presence of DBT, DBTS, 4-MDBT, 2,8-DMDBT, BT, 3-MBT, TH, 2-MTH, and 3-MTH. The isolate exhibited the highest growth in the presence of DBT, 2,8-DMDBT, 4-MDBT, DBTS, BT and 3-MBT. In addition to these compounds, a relatively less growth was also observed in the presence of TH and its derivatives, indicating the utilization of these compounds by W3S5 for its growth. The overall order of growth for all these compounds was: DBT > 2,8-DMDBT > BT > 3-MBT > DBTS > 4-MDBT > 3-MTH > 2-MTH > TH. The bar graph in the Fig. 5B represent the production of 2-HBP resulting from the desulfurization of DBT, DBTS, 4-MDBT, BT and 3-MBT within 144 h of incubation. A maximum 2-HBP production was observed after 96 h of incubation, indicating that the isolate W3S5 desulfurized DBT and DBTS into 2-HBP by ~ 95% and ~ 75% respectively, while 4-MDBT, BT, and 3-MBT were desulfurized into 2-HBPeq by ~ 50%. Overall, the order of desulfurization activity for these compounds was: DBT > DBTS > BT > 3-MBT > 4-MDBT. The formation of 2-HBP/2HBP equivalent was not detected in cultures treated with 2,8-DMDBT, TH, 2-MTH, and 3-MTH. This suggests their conversion into non-phenolic end products or that the produced phenolic end products did not react with the Gibbs reagent, that do not give a blue reaction with Gibb’s reagent. In addition to desulfurize DBTs, and BTs as a single-sulfur source, the isolate W3S5 was also investigated to desulfurize them simultaneously as a mixture as well. The Fig. 5C compares the growth and desulfurization activity (2-HBP production) by isolate W3S5 for the desulfurization of DBTs and BTs (DBT, 3-MDBT, DBTS, BT and 3-MBT) when supplied together in MG medium in equimolar ratios (0.1 mM). The isolate exhibited an increase in growth after 72 h of incubation, however, a sharp increase in growth and the production of 2-HBP was observed after 96 h of incubation and was found maximum at 192 h of cultivation. Although the lag-phase for growth was prolonged, the results indicate that the isolate W3S5 can effectively desulfurize DBTs, and BTs when supplied together as a mixture.
Desulfurization of DBTs, BTs and THs by W3S5, as a single sulfur source and as a mixture. A Comparison of growth and B production of phenolic end-product i.e., 2-HBP/2-HBPequivalent by isolate W3S5, cultivated in MG media in the presence of 0.2 mM of DBT (Dibenzothiophene), DBTS (Dibenzothiophene Sulfone), 4-MDBT (4-Methyldibenzothiophene), 2,8-DMDBT (2,8-Dimethyldibenzothiophene), BT (Benzothiophene), 3-MBT (3-Methylbenzothiophene), TH (Thiophene), 2-MTH (2-Methylthiophene), and 3-MTH (3-Methylthiophene) respectively, as a single sulfur source. C A time-course study of cell growth and production of phenolic end-product i.e., 2-HBP/2-HBPequivalent for 192 h, when DBTs, and BTs (DBT, DBTS, 4-MDBT, BT, and 3-MBT) were supplied in MG medium together in equimolar ratio (0.1 mM)
Characterization of biosurfactants in W3S5 strain
Biosurfactants are amphiphilic surface-active compounds that help a bacterium in the biotransformation/biodegradation of water insoluble compounds by decreasing the surface tension of the medium (Mnif and Ghribi 2015). In order to check the production of biosurfactants by isolate W3S5 during DBT desulfurization, a qualitative estimation was carried out by performing the oil-displacement assay and calculating the emulsification index (% EI24) of the cell-free culture. A positive control (SDS, a strong amphiphilic compound) and a negative control (culture medium) were used to validate the result. In oil-displacement test, the cell-free culture created a clearing zone of 2.3 cm, while for positive control (SDS) it was 4 cm (see Supplementary Fig. 2A). The negative control did not show any oil displacement, which confirms that the clearing zone formed by cell-free culture was due to the presence of surface-active compounds (SAC) released in the culture medium during desulfurization of DBT. To further validate these results the emulsification index of the cell free culture, positive control and the negative control was also calculated. The EI24 for cell free culture was 53% and for positive control 65%, while negative control did not show any emulsification (see Supplementary Fig. 2B).
The crude biosurfactants were purified using Thin-Layer Chromatography (TLC). Yellow-colored spot appeared on the TLC plate with a purple-colored background when developed with potassium permanganate stain, indicating the glycolipid nature of the extracted biosurfactants. The TLC purified biosurfactants were subjected to FTIR analysis in the range of 4000–500 cm−1 and the resulting IR spectrum was interpreted by reading the IR spectrum chart/table from Sigma-Aldrich and previous reports on the characterization of glycolipids (De Smet et al. 2000; Wang et al. 2019). The Fig. 6A shows the FT-IR spectrum of the TLC purified biosurfactants. A wide band detected in the range of 3200–3500 cm−1 can be assigned to O-H bond vibrations, indicating the presence of an alcohol group (De Smet et al. 2000; Wang et al. 2019). A strong band detected at 2970–3007 cm−1 range exhibited the existence of unsaturated hydrocarbons and sharp bands observed at 2951–2850 cm−1 were designated to C–H stretching bands of CH2 and CH3 groups demonstrating the presence of saturated hydrocarbons (De Smet et al. 2000; Wang et al. 2019). A strong band at 1708 cm−1 was also observed, that is a characteristic band of esters denoting the presence of carbonyl groups in the extracted biosurfactans (De Smet et al. 2000; Wang et al. 2019). A medium sized band observed at 1464 cm−1 was ascribed to C–H stretching bands (De Smet et al. 2000; Wang et al. 2019). Moreover, the absorption bands observed at 1190–1456 cm−1 denote the existence of aliphatic chains in the analyzed compound (De Smet et al. 2000; Wang et al. 2019). The sharp bands detected in the range of 950–1155 cm−1 were attributed to C–O vibration bands, which are characteristic of the sugar rings (De Smet et al. 2000). These FT-IR findings strongly suggest that the extracted biosurfactants are glycolipid in nature and may belong to trehalose subclass.
Characterization of biosurfactants production in W3S5 during DBT desulfurization, by Fourier-transform infrared spectroscopy (FTIR) and whole-genome sequence analysis. A The FT-IR spectrum of TLC purified biosurfactants in the range of 4000–500 cm−1. B A schematic of TreYZ, OtsAB and TreP pathways of trehalose biosynthesis identified in strain W3S5
Identification of trehalolipid-biosurfactant genes in W3S5 strain by genetic analysis
In total, five trehalose synthesis pathways have been reported in bacteria of which the OtsAB, TreYZ, and TreS are the three major pathways that are often present in more than one copies for some specific bacteria, depending on the physiological requirements in the environment (Cardoso et al. 2007; Ruhal et al. 2013). The existence of a certain metabolic pathway within an organism is generally inferred by recognizing the respective enzyme-coding genes. The trehalose-related metabolic pathways in W3S5 were identified by analyzing its whole genome sequence via different online genome annotation programs such as ClassicRAST, BlastKOALA, BV-BRC, and NCBI-PGAP. Moreover, the KEGG and MetaCYS pathway analysis were also performed and the proposed pathway in W3S5 were drawn on the basis of results obtained from all these studies. The trehalose-related genes predicted by the four annotations and their potential functions are presented in Table 1. All four annotations generated different results with respect to the number of genes identified and their copy number. ClassicRAST and BlastKOALA predicted 10 genes, whereas BV-BRC and NCBI-PGAP predicted 5 and 3 genes respectively. The results indicate that nearly all annotations predicted the presence of otsA, otsB, treY, treZ and treP genes (Table 1) in the genome that confirms the presence of OtsAB and TreYZ and TreP pathways in W3S5 strain (Fig. 6B). The OtsAB pathway, is the most prevalent pathway for stress response in bacteria that starts with glucose-6-phosphate and UDP-glucose to form trehalose-6-phosphate, catalyzed by trehalose-6-phosphate synthase (otsA) (Avonce et al. 2006), which is then dephosphorylated by trehalose-6-phosphate phosphatase (otsB) to release a free trehalose molecule (Avonce et al. 2006). Whereas, in TreYZ pathway, the glycogen or oligomaltodextrin is transformed into maltooligosyl-trehalose by transglycosylation of terminal maltosyl residue into a trehalosyl group by maltooligosyl-trehalose synthase (treY) and subsequently the trehalose is liberated from maltooligosyl-trehalose by the action of maltooligosyl-trehalose trehalohydrolase (treZ) (Avonce et al. 2006; Takayama et al. 2005). The TreP is another pathway found in bacteria, which consists of a single reversible reaction to synthesis trehalose from d-glucose and β-d-glucose 1-phosphate, catalyzed by trehalose phosphorylase (treP) (Takayama et al. 2005). Furthermore, the trehalose-monocorynomycolates (TMM) transporter gene (mmpL3) and trehalose O-mycolyltransferase gene (fbp) were also detected by BlastKOALA and BV-BRC annotations respectively (Table 1), indicating the synthesis of TMM lipids and their transport across the cell membrane (Li et al. 2019; Luong et al. 2018).
Certain genes associated with trehalose transport such as IpqY, SugA, and SugB were predicted by the BlastKOALA annotation (Table 1). Trehalose acyltransferase (papA3) and 2-sulfotransferase (stf0) genes that contribute to the biosynthesis of trehalose-based cell wall glycolipids were also detected by BlastKOALA annotation (Table 1). Beta-phosphoglucomutase (pgmB) and Glucose/mannose:H+ symporter (GlcP) genes involved in the uptake and utilization of trehalose and the trehalose phosphorylase (treP) gene implicated in trehalose catabolism/anabolism were identified by ClassicRAST, BlastKOALA, and NCBI-PGAP annotations (Table 1) indicating that isolate W3S5 can use trehalose as carbon/energy sources or as a solute for maintaining the osmotic pressure of the cell under stress conditions. Furthermore, the genes for 1,4-alpha-glucan (glycogen) branching enzyme (glgB) and putative glucanase (glgE) were also recognized by ClassicRAST annotation. The glgB and glgE genes participate in glycogen metabolism and are interconnected with trehalose biosynthesis as glycogen is used as a substrate for trehalose biosynthesis during the TreYZ pathway and therefore influences trehalose biosynthesis at these nodes.
Discussion
Biodesulfurization of organic-sulfur compounds present in petro-fuels has attracted attention on a global scale due to the lack of economic and environmentally favorable methods for the removal of organic sulfur as opposed to inorganic sulfur removal (Davoodi-Dehaghani et al. 2010; Kalita et al. 2022; Al-khazaali and Ataei 2023; Silva et al. 2023). Certain microbes can obtain sulfur from complex organic-sulfur compounds for growth and other essential functions, and therefore can be used to remove organic-sulfur from petro-fuels with no negative environmental impact (Sousa et al. 2020). For the past several decades, DBT has been utilized as a model compound to isolate and characterize the organic-sulfur compound desulfurizing bacteria, since it is prevalent in ptero-fuels and challenging to remove using traditional methods (Davoodi-Dehaghani et al. 2010; Pavlopoulos et al. 2010; Xia 2018). Here, we are providing a detailed study on an efficient DBT desulfurizing strain, W3S5, isolated from an oil-contaminated soil sample that can desulfurize ~ 99% of DBT (0.2 mM) within 48 h of incubation. When grown in MG medium containing only DBT, the isolate did not show prominent growth, however, when both DBT and glucose were provided, a very rich growth was obtained indicating that the isolate utilizes DBT as a sole sulfur source and not as a carbon source (Fig. 1C). Through TYGS based phylogenomic analysis the strain has been identified as Gordonia rubripertincta, as it makes a cluster with Gordonia rubripertincta type strains in the phylogenomic tree by sharing similar genome features (Fig. 2). The TYGS has been applied in this study because it is one of the best tools for genome-based classification of species as it contains a large-scale database of type strains genomes having legally published names and utilizes trustworthy whole-genome based classification and phylogeny methods (Meier-Kolthoff et al. 2013; Meier-Kolthoff and Göker 2019). Our findings of GC-MS analysis of extracted metabolites and genetic studies of dsz genes revealed the production of core metabolites DBT, DBTO2, and 2-HBP indicating that W3S5 follows the “4S” pathway (Fig. 3) of DBT desulfurization encoded by dszA, dszB, and dszC genes present on contig# 22 in the form of an operon spanned by some mobile/transposable elements (Fig. 4A). In literature, two major pathways the ring destructive “Kodama pathway” and sulfur specific “4S pathway” have been reported for DBT desulfurization by aerobic bacteria (Gunam et al. 2006; Parveen et al. 2020; Su et al. 2018). In “4S” pathway the sulfur is specifically removed through enzymes by cleaving the C–S bond instead of C–C bond without breaking the carbon-skeleton, that preserves the calorific value of the fuel (Ferreira et al. 2017; Gunam et al. 2006; Parveen et al. 2020; Su et al. 2018). Our studies showed that W3S5 also follows the “4S” pathway of DBT desulfurization, and hence able to preserve the calorific value of the fuel when applied as a biocatalyst. The phylogenetic analysis of dszABC genes showed a close evolutionary relatedness with Gordonia species suggesting that the dsz genes of W3S5 strain are conserved and quite similar to the dsz genes of other desulfurizing Gordonia species (Fig. 4C).
Majority of BDS studies reported in literature could applied either DBTs or BTs, however only a few bacteria have been recorded to desulfurize both DBTs and BTs (Chen et al. 2021; Mohamed et al. 2015; Raheb and Hajipour 2011). We find that W3S5 can effectively metabolize all three classes of compounds i.e., DBTs, BTs, and THs when fed in MG medium separately as well as in a mixture. The overall growth order for all these compounds was: DBT > 2,8-DMDBT > BT > 3-MBT > DBTS > 4-MDBT > 3-MTH > 2-MTH > TH, and desulfurization activity order for compounds showing Gibb’s positive test was: DBT > DBTS > BT > 3-MBT > 4-MDBT (Fig. 5A and B). In case of methylated thiophenic compounds, the strain W3S5 showed a little more growth with double methylated compound (like 2,8-dimethyl DBT) as compared to single methylated compounds (Fig. 5A). In cultures treated with 2,8-DMDBT, TH, 2-MTH, and 3-MTH, the 2-HBP/2-HBP equivalent formation was not detected, however, the increase in growth in the presence of these compounds suggests that the isolate W3S5 have utilized these compounds to obtain sulfur for its growth and might have converted them into some non-phenolic end products that are non-reactive with Gibb’s reagent. Furthermore, there is a possibility that the produced phenolic end product did not react with the Gibbs reagent, which does not yield a blue reaction. For instance, 2,8-DMDBT may be converted into 3′,5-dimethyl-2-hydroxybiphenyl by the bacterium, a phenolic end product that does not give a blue reaction with Gibb’s reagent. Gibb’s reagent specifically reacts with phenols lacking substituents in the ortho position. Determining the formation of end products from the use of these phenolic compounds will require further studies through GC–MS/LC–MS/NMR in the future.
There could be several reasons for these variable effects of derivatizations; varying degrees of microbial toxicity of these compounds, the bacterium might be using various metabolic pathways for the more complicated compounds, or the more complex compounds may have a stronger affinity for the transport proteins. Collectively, the results indicate that as compared to the previous reported desulfurizing bacteria, the W3S5 strain can not only effectively desulfurize DBTs but also BTs as well as THs that are the highly recalcitrant organic-sulfur compounds present in petro-fuels (Chen et al. 2021; Mohamed et al. 2015; Raheb and Hajipour 2011).
In addition to desulfurize DBTs and BTs as a single-sulfur source, the isolate W3S5 was also investigated to desulfurize them together in equimolar ratios (0.1 mM). As compared to single-sulfur compounds (Fig. 5C), a delayed onset of log-phase was observed for the mixture of compounds (DBT, DBTS, 3-MDBT, BT and 3-MBT) after 96 h of incubation instead of 24 h, which might be due to the need for the cells to adjust to an environment rich in multiple organic-sulfur compounds and time for producing the respective BDS enzymes. In addition, there may be a competition (gradation in affinity) among different organic sulfur compounds to attach with the receptors/transporters responsible for the uptake of these compounds into the cell which can cause a delay in the onset of the log phase. So, even if one compound can be metabolized optimally when supplied individually, it may have a different behavior in uptake and metabolism in the presence of multiple sulfur compounds. After adaptation, the isolate successfully desulfurized 36% of the total amount of mixed-compounds within 192 h of cultivation, suggesting its potential to utilize multiple organic-sulfur compounds simultaneously. Overall, this is the first study on the desulfurization of DBTs as well as BTs, as single-sulfur source and a mixture of compounds by Gordonia rubripertinca W3S5.
Restricted mass transfer of substrate between oil and the aqueous phase is another main bottleneck in the large-scale application of BDS (Alves et al. 2015; Parveen et al. 2020; Hasanbeik et al. 2022). Biosurfactants are amphiphilic surface-active compounds that aid in enhancing the substrate mass transfer in biphasic systems by lowering the surface tension of the medium (Mnif and Ghribi 2015). We have found that strain W3S5 produces trehalose-surfactants during the biodesulfurization of DBT, as confirmed by TLC and FTIR analyses. The FTIR analysis of the TLC purified biosurfactants in the range of 4000–500 cm−1 confirmed that the produced biosurfactants are trehalose-surfactants (Fig. 6A). Trehalolipids are one of the best-studied subclasses of glycolipids, that have wide range industrial applications including microbial enhanced oil recovery, because of their adaptive structural and functional groups that give them the desired high surface activity (Arguelles 2000). Several bacterial species have been reported to produce trehalolipids, of which Rhodococcus genus is well reported to produce various types of trehalolipids such as trehalose-monocorynomycolates (TMM), trehalose-dicorynomycolates (TDM), and trehalose-tricorynomycolates (TTM), including succinoyl trehalose lipids and trehalose tetraesters (Franzetti et al. 2010; Kretschmer et al. 1982; Luong et al. 2018). However, no study has been reported for the production of trehalolipids by a desulfurizing Gordonia specie (Luong et al. 2018). We are first time reporting the production of trehalose-surfactants by a desulfurizing Gordonia rubripertincta W3S5 strain and have identified the trehalose-surfactant related genes by analyzing its whole genome sequence using several online genome annotation programs i.e., ClassicRAST, BlastKOALA, BV-BRC, and NCBI-PGAP (Table 1). We have identified three potential trehalose biosynthesis pathways, OtsAB, TreYZ, and TreP in strain W3S5 based on data obtained from these annotations (Fig. 6B). Additionally, few genes related to the biosynthesis of trehalose-monocornymycolate (TMM), a precursor for the synthesis of various types of trehalolipids were also identified. The results also suggest that it is best to perform a genomic analysis using multiple annotations as each tool has certain limitations such as differences in variances, the limited knowledge available in a database to correctly identify or annotate certain enzymes, or the sequences that could not meet the threshold of that database. A thorough understanding of biosurfactants produced by DBT desulfurizing bacteria would be helpful in studies intended in improving the desulfurization rate by increasing the mass transfer of the organic-sulfur compounds with the help of biosurfactants.
Conclusion
In conclusion, our study provides in-depth insights into the remarkable biodesulfurization potential of the strain W3S5, which was isolated from oil-contaminated soil. Our findings reveal that strain W3S5 follows the “4S” pathway of DBT desulfurization and can metabolize a variety of HDS-resistant organic-sulfur compounds, including DBTs, BTs, and THs separately as well as in a mixture. Moreover, it produces trehalose-surfactants during DBT desulfurization, enabling the bacterial cells to efficiently uptake the DBT from the medium by reducing the surface tension. Furthermore, we present the first comprehensive analysis of the whole-genome sequence of the desulfurizing strain W3S5 using different annotation programs, which identified three potential trehalose synthesis pathways, namely OtsAB, TreYZ, and TreP, as well as trehalose-monomycolate lipid synthesis genes. The identification of various trehalose-related genes in the strain W3S5 provides new insight into the mechanisms governing sulfur removal, which opens up new possibilities for optimizing this process. Overall, these findings suggest that the isolated strain Gordonia rubripertincta W3S5 has the potential for sustainable and efficient biodesulfurization of wide range of organic-sulfur compounds and could be a highly effective candidate in advanced biodesulfurization research aimed at addressing environmental issues related to sulfur-emissions.
Data availability
The whole genome shotgun sequence of isolate W3S5 is available at DDBJ/ENA/GenBank under the accession NZ_VLNS00000000 (BioProject: PRJNA555169; BioSample: SAMN12302752), version VLNS00000000.1. The identified complete sequences of dszA, dszB and dszC genes were submitted in GenBank database at NCBI (www.ncbi.nlm.nih.gov) and were assigned the following accession numbers: OP413448, OP413449, and OP413450 respectively.
References
Aggarwal S, Karimi IA, Ivan GR (2013) In silico modeling and evaluation of Gordonia alkanivorans for biodesulfurization. Mol Biosyst 9:2530–2540. https://doi.org/10.1039/c3mb70132h
Akhtar N, Ghauri MA, Anwar MA, Akhtar K (2009) Analysis of the dibenzothiophene metabolic pathway in a newly isolated Rhodococcus spp,. FEMS Microbiol Lett. https://doi.org/10.1111/j.1574-6968.2009.01797.x
Akhtar N, Ghauri M, Anwar M, Heaphy S (2014) Phylogenetic characterization and novelty of organic sulphur metabolizing genes of Rhodococcus spp. (Eu-32). Biotechnol Lett 37:837–847. https://doi.org/10.1007/s10529-014-1736-6
Akhtar N, Ghauri MA, Akhtar K (2016a) Dibenzothiophene desulfurization capability and evolutionary divergence of newly isolated bacteria. Arch Microbiol. https://doi.org/10.1007/s00203-016-1209-5
Akhtar N, Ghauri MA, Akhtar K (2016b) Exploring coal biodesulfurization potential of a novel organic sulfur metabolizing Rhodococcus spp. (Eu-32): a case study. Geomicrobiol J 33:468–472. https://doi.org/10.1080/01490451.2015.1052119
Akhtar N, Akhtar K, Ghauri MA (2018) Biodesulfurization of thiophenic compounds by a 2-Hydroxybiphenyl-resistant Gordonia sp. HS126-4 N carrying dszABC genes. Curr Microbiol 75:597–603. https://doi.org/10.1007/s00284-017-1422-8
Akhtar N, Ghauri MA, Akhtar K, Parveen S, Farooq M, Ali A et al (2019) Comparative analysis of draft genome sequence of Rhodococcus sp. Eu-32 with other rhodococcus species for its taxonomic status and sulfur metabolism potential. Curr Microbiol 76:1207–1214. https://doi.org/10.1007/s00284-019-01737-1
Al-khazaali WMK, Ataei SA (2023) Optimization of biodesulfurization of sour heavy crude oil. PLoS One 18:e0283285. https://doi.org/10.1371/journal.pone.0283285
Alves L, Paixão SM, Pacheco R, Ferreira AF, Silva CM (2015) Biodesulphurization of fossil fuels: energy, emissions and cost analysis. RSC Adv 5:34047–34057. https://doi.org/10.1039/C4RA14216K
Arguelles J (2000) Novel bifunctional trehalose synthase. Arch Microbiol 174:217–224
Avonce N, Mendoza-Vargas A, Morett E, Iturriaga G (2006) Insights on the evolution of trehalose biosynthesis. BMC Evol Biol 6:109. https://doi.org/10.1186/1471-2148-6-109
Bhanjadeo MM, Rath K, Gupta D, Pradhan N, Biswal SK, Mishra BK et al (2018) Differential desulfurization of dibenzothiophene by newly identified MTCC strains: influence of operon array. PLoS One 13:e0192536. https://doi.org/10.1371/journal.pone.0192536
Boniek D, Figueiredo D, dos Santos AFB, de Resende Stoianoff MA (2015) Biodesulfurization: a mini review about the immediate search for the future technology. Clean Technol Environ Policy 17:29–37. https://doi.org/10.1007/s10098-014-0812-x
Cardoso FS, Castro RF, Borges N, Santos H (2007) Biochemical and genetic characterization of the pathways for trehalose metabolism in Propionibacterium freudenreichii, and their role in stress response. Microbiology 153:270–280. https://doi.org/10.1099/mic.0.29262-0
Chen S, Zang M, Li L, Chen J, Liu Q, Feng X et al (2021) Efficient biodesulfurization of diesel oil by Gordonia sp. SC-10 with highly hydrophobic cell surfaces. Biochem Eng J 174:108094. https://doi.org/10.1016/j.bej.2021.108094
Davoodi-Dehaghani F, Vosoughi M, Ziaee AA (2010) Biodesulfurization of dibenzothiophene by a newly isolated Rhodococcus erythropolis strain. Bioresour Technol 101:1102–1105. https://doi.org/10.1016/j.biortech.2009.08.058
De Smet KAL, Weston A, Brown IN, Young DB, Robertson BD (2000) Three pathways for trehalose biosynthesis in mycobacteria. Microbiology 146:199–208. https://doi.org/10.1099/00221287-146-1-199
Feng S, Yang H, Zhan X, Wang W (2016) Enhancement of dibenzothiophene biodesulfurization by weakening the feedback inhibition effects based on a systematic understanding of the biodesulfurization mechanism by Gordonia sp. through the potential 4S pathway. RSC Adv. https://doi.org/10.1039/C6RA14459D
Ferreira P, Sousa SF, Fernandes PA, Ramos MJ (2017) Improving the catalytic power of the DszD enzyme for the biodesulfurization of crude oil and derivatives. Chemistry 23:17231–17241. https://doi.org/10.1002/chem.201704057
Franzetti A, Gandolfi I, Bestetti G, Smyth TJP, Banat IM (2010) Production and applications of trehalose lipid biosurfactants. Eur J Lipid Sci Technol 112:617–627. https://doi.org/10.1002/ejlt.200900162
Glekas PD, Martzoukou O, Mastrodima ME, Zarkadoulas E, Kanakoglou DS, Kekos D et al (2022) Deciphering the biodesulfurization potential of two novel Rhodococcus isolates from a unique Greek environment. AIMS Microbiol 8:484–506. https://doi.org/10.3934/microbiol.2022032
Gray KA, Pogrebinsky OS, Mrachko GT, Xi L, Monticello DJ, Squires CH (1996) Molecular mechanisms of biocatalytic desulfurization of fossil fuels. Nat Biotechnol 14:1705–1709. https://doi.org/10.1038/nbt1296-1705
Gunam IBW, Yaku Y, Hirano M, Yamamura K, Tomita F, Sone T et al (2006) Biodesulfurization of alkylated forms of dibenzothiophene and benzothiophene by Sphingomonas subarctica T7b. J Biosci Bioeng 101:322–327. https://doi.org/10.1263/jbb.101.322
Hasanbeik NY, Pourmadadi M, Ghadami A, Yazdian F, Rahdar A, Kyzas GZ (2022) Biodesulfurization of dibenzothiophene by decorating Rhodococcus erythropolis IGTS8 using montmorillonite/graphitic carbon nitride. Catalysts 12:1450. https://doi.org/10.3390/catal12111450
Kalita M, Chutia M, Jha DK, Subrahmanyam G (2022) Mechanistic understanding of Gordonia sp. in biodesulfurization of organosulfur compounds. Curr Microbiol 79:82. https://doi.org/10.1007/s00284-022-02770-3
Kayser KJ, Bielaga-Jones BA, Jackowski K, Odusan O, Kilbane JJ (1993) Utilization of organosulphur compounds by axenic and mixed cultures of Rhodococcus rhodochrous IGTS8. Microbiology 139:3123–3129. https://doi.org/10.1099/00221287-139-12-3123
Kilbane IIJJ, Le Borgne S (2004) Petroleum biorefining: the selective removal of sulfur, nitrogen, and metals. Stud Surf Sci Catal 15:29–65. https://doi.org/10.1016/S0167-2991(04)80143-5
Kilbane JJ, Robbins J (2007) Characterization of the dszABC genes of Gordonia amicalis F.5.25.8 and identification of conserved protein and DNA sequences. Appl Microbiol Biotechnol 75:843–851. https://doi.org/10.1007/s00253-007-0895-y
Kilbane J, Stark B (2016) Biodesulfurization: a model system for microbial physiology research. World J Microbiol Biotechnol 32:8. https://doi.org/10.1007/s11274-016-2084-6
Kretschmer A, Bock H, Wagner F (1982) Chemical and physical characterization of interfacial-active lipids from Rhodococcus erythropolis grown on n-alkanes. Appl Environ Microbiol 44:864–870. https://doi.org/10.1128/aem.44.4.864-870.1982
Li L, Shen X, Zhao C, Liu Q, Liu X, Wu Y (2019) Biodegradation of dibenzothiophene by efficient Pseudomonas sp. LKY-5 with the production of a biosurfactant. Ecotoxicol Environ Saf 176:50–57. https://doi.org/10.1016/j.ecoenv.2019.03.070
Luong TM, Ponamoreva ON, Nechaeva IA, Petrikov KV, Delegan YA, Surin AK et al (2018) Characterization of biosurfactants produced by the oil-degrading bacterium Rhodococcus erythropolis S67 at low temperature. World J Microbiol Biotechnol 34:20. https://doi.org/10.1007/s11274-017-2401-8
Martínez I, El-Said Mohamed M, Santos VE, García JL, García-Ochoa F, Díaz E (2017) Metabolic and process engineering for biodesulfurization in gram-negative bacteria. J Biotechnol 262:47–55. https://doi.org/10.1016/j.jbiotec.2017.09.004
Meier-Kolthoff JP, Göker M (2019) TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat Commun 10:2182. https://doi.org/10.1038/s41467-019-10210-3
Meier-Kolthoff JP, Auch AF, Klenk H-P, Göker M (2013) Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform 14:60. https://doi.org/10.1186/1471-2105-14-60
Mnif I, Ghribi D (2015) Microbial derived surface active compounds: properties and screening concept. World J Microbiol Biotechnol 31:1001–1020. https://doi.org/10.1007/s11274-015-1866-6
Mohamed ME-S, Al-Yacoub ZH, Vedakumar JV (2015) Biocatalytic desulfurization of thiophenic compounds and crude oil by newly isolated bacteria, 2015. Front Microbiol. https://doi.org/10.3389/fmicb.2015.00112
Mohebali G, Ball AS (2008) Biocatalytic desulfurization (BDS) of petrodiesel fuels. Microbiololy 154:2169–2183. https://doi.org/10.1099/mic.0.2008/017608-0
Mohebali G, Ball AS (2016) Biodesulfurization of diesel fuels–past, present and future perspectives. Int Biodeterior Biodegrad 110:163–180. https://doi.org/10.1016/j.ibiod.2016.03.011
Parveen S, Akhtar N, Ghauri MA, Akhtar K (2020) Conventional genetic manipulation of desulfurizing bacteria and prospects of using CRISPR-Cas systems for enhanced desulfurization activity. Crit Rev Microbiol 46:300–320. https://doi.org/10.1080/1040841X.2020.1772195
Parveen S, Akhtar N, Akram J (2021) Genomic analysis provides insights into the Gordonia sp. W3S5 taxonomy and sulfur metabolism-related genes. 3 Biotech 11:300. https://doi.org/10.1007/s13205-021-02850-4
Pavlopoulos GA, Soldatos TG, Barbosa-Silva A, Schneider R (2010) A reference guide for tree analysis and visualization. BioData Min 3:1. https://doi.org/10.1186/1756-0381-3-1
Porto B, Maass D, Oliveira JV, de Oliveira D, Yamamoto CI, Ulson de Souza AA et al (2018) Heavy gas oil biodesulfurization using a low-cost bacterial consortium. J Chem Technol Biotechnol 93:2359–2363. https://doi.org/10.1002/jctb.5580
Prasoulas G, Dimos K, Glekas P, Kalantzi S, Sarris S, Templis C et al (2021) Biodesulfurization of dibenzothiophene and its alkylated derivatives in a two-phase bubble column bioreactor by resting cells of Rhodococcus erythropolis IGTS8. Processes 9(11):2064. https://doi.org/10.3390/pr9112064
Raheb J, Hajipour MJ (2011) The stable rhamnolipid biosurfactant production in genetically engineered Pseudomonas strain reduced energy consumption in biodesulfurization. Energy Sources A: Recov Util Environ 33:2113–2121. https://doi.org/10.1080/15567030903503167
Raheb J, Hajipour MJ, Saadati M, Rasekh B, Memari B (2009) The enhancement of biodesulfurization activity in a novel indigenous engineered Pseudomonas putida. Iran Biomed J 13:207–213 doi -
Rangra S, Kabra M, Gupta V, Srivastava P (2018) Improved conversion of dibenzothiophene into sulfone by surface display of dibenzothiophene monooxygenase (dszc) in recombinant Escherichia coli. J Biotechnol 287:59–67. https://doi.org/10.1016/j.jbiotec.2018.10.004
Ruhal R, Kataria R, Choudhury B (2013) Trends in bacterial trehalose metabolism and significant nodes of metabolic pathway in the direction of trehalose accumulation. Microb Biotechnol 6:493–502. https://doi.org/10.1111/1751-7915.12029
Sadare OO, Obazu F, Daramola MO (2017) Biodesulfurization of petroleum distillates—current status, opportunities and future challenges. Environments 4:85. https://doi.org/10.3390/environments4040085
Santos SC, Alviano DS, Alviano CS, Pádula M, Leitão AC, Martins OB et al (2006) Characterization of Gordonia sp. strain F.5.25.8 capable of dibenzothiophene desulfurization and carbazole utilization. Appl Microbiol Biotechnol 71:355–362. https://doi.org/10.1007/s00253-005-0154-z
Silva TP, Alves L, Paixão SM (2020) Effect of dibenzothiophene and its alkylated derivatives on coupled desulfurization and carotenoid production by Gordonia alkanivorans strain 1B. J Environ Manage 270:110825. https://doi.org/10.1016/j.jenvman.2020.110825
Silva TP, Paixão SM, Alves L (2023) A new impetus for biodesulfurization: bypassing sulfate inhibition in biocatalyst production. Green Chem 25:6416–6431. https://doi.org/10.1039/D3GC01612A
Sousa JPM, Neves RPP, Sousa SF, Ramos MJ, Fernandes PA (2020) Reaction mechanism and determinants for efficient catalysis by DszB, a key enzyme for crude oil bio-desulfurization. ACS Catal 10:9545–9554. https://doi.org/10.1021/acscatal.0c03122
Su T, Su J, Liu S, Zhang C, He J, Huang Y et al (2018) Structural and biochemical characterization of BdsA from Bacillus subtilis WU-S2B, a key enzyme in the 4S desulfurization pathway. Front Microbiol 9:231. https://doi.org/10.3389/fmicb.2018.00231
Tabatabaee A, Assadi MM, Noohi A, Sajadian V (2005) Isolation of biosurfactant producing bacteria from oil reservoirs. J Environ Health Sci Eng 2:6–12. https://doi.org/10.3389/fmicb.2020.00064
Takayama K, Wang C, Besra GS (2005) Pathway to synthesis and processing of mycolic acids in Mycobacterium tuberculosis. Clin Microbiol Rev 18:81–101. https://doi.org/10.1128/cmr.18.1.81-101.2005
Tamura K, Nei M (1993) Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol 10:512–526. https://doi.org/10.1093/oxfordjournals.molbev.a040023
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729. https://doi.org/10.1093/molbev/mst197
Pylro VS, Vespoli LS, Duarte GF, Yotoko KSC (2012) Detection of horizontal gene transfers from phylogenetic comparisons. Int J Evol Biol https://doi.org/10.1155/2012/813015
Wang J, Butler RR III, Wu F, Pombert J-F, Kilbane JJ, Stark II (2017) Enhancement of microbial biodesulfurization via genetic engineering and adaptive evolution. PLoS One 12:e0168833. https://doi.org/10.1371/journal.pone.0168833
Wang Y, Nie M, Diwu Z, Lei Y, Li H, Bai X (2019) Characterization of trehalose lipids produced by a unique environmental isolate bacterium Rhodococcus qingshengii strain FF. J Appl Microbiol 127:1442–1453. https://doi.org/10.1111/jam.14390
Xia W (2018) A novel and effective method for removing organic sulfur from low rank coal. J Clean Prod 172:2708–2710. https://doi.org/10.1016/j.jclepro.2017.11.141
Funding
This work is supported by the Higher Education Commission (HEC), Pakistan (Grant No: NRPU-3884 and NRPU-11570) and International Foundation for Science (IFS), Sweden (Grant No: F-5379-2).
Author information
Authors and Affiliations
Contributions
SP contributed in Investigation, Methodology, Formal analysis, Data curation and Writing. NA contributed in Conceptualization, Resources, Methodology, Validation, Supervision, Writing—review & editing. TEK and AIH contributed in Formal analysis, Visualization, Data curation. RB and KA contributed in Writing—review & editing. All authors have approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interest
The authors declare no competing interests.
Ethical approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Parveen, S., Akhtar, N., E-kobon, T. et al. Biodesulfurization of organosulfur compounds by a trehalose biosurfactant producing Gordonia sp. isolated from crude oil contaminated soil. World J Microbiol Biotechnol 40, 103 (2024). https://doi.org/10.1007/s11274-024-03899-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11274-024-03899-y