Abstract
Cyanate and its derivatives are considered as highly dangerous materials that threaten human health and environment. Cyanate arises from both natural resources and anthropogenic activities including various chemical industries, herbicide production, and mining wastewater. Despite its toxicity, cyanate is considered as an important nitrogen (N) source in marine ecosystems. Cyanase (CYN) catalyzes the decomposition of cyanate into CO2 and NH3 in a bicarbonate-dependent reaction. In marine cyanobacteria, endogenous cyanases participate in detoxification of low concentrations of cyanate. However, this cyanate biodegradation system is seemingly inconvenient especially at contaminated sites due to high cyanate concentrations. In the current study, we have transferred the activity of the cyanobacterial enzyme cyanase into the micro-alga, Chlamydomonas reinhardtii, via Agrobacterium tumefaciens–mediated transformation method. The recombinant cyanase enzyme was shown to be active in transgenic C. reinhardtii lines. When variable concentrations of cyanate (up to 30 mM) is applied to growth medium, transgenic lines showed higher rate of NH3 release, reduced loss of pigmentation symptoms, decreased levels of induced antioxidant enzymes, and low percentage of growth retardation compared to wild-type controls. Results of this study provide an effective eco-friendly phytoremediation system for cyanate detoxification using micro-algae compared to previously reported plant systems.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Many researchers and research groups are now working on projects focusing on developing new technologies for the treatment of industrial and environmental hazardous wastes. Using biological treatment, systems received particular attention by many research groups. Aerobic and anaerobic microbial systems were applied successfully in the treatment and removal of such hazardous wastes including organic and inorganic compounds and metals (Mudder et al. 2001). Cyanide and its oxidation product cyanate are dangerous toxic chemicals produced through human activities and industries comprising ore leaching, electroplating, steal production, plastics, and synthetic fibers (Hamel 2011). Cyanate and its derivatives are being widely used for the manufacturing of a broad class of herbicides and in the synthesis of polymers (Koshiishi et al. 1997). Spontaneous photo-oxidation of cyanide as well as cyanide containing wastes are major sources for the release of the toxic cyanate into the environment (Malhotra et al. 2005; Mekuto et al. 2016). Moreover, cyanate is released into the environment through the biological breakdown of various metabolites such as urea and carbamoylphosphate (Guillotonm and Karst 1987). Chemical treatment methods such as oxidation or chlorination reactions are commonly applied for the detoxification of cyanate-containing compounds (Akcil and Mudder 2003). However, These chemical methods have some limitations such as high costs and production of hazardous byproducts (Srivastava and Muni 2010). Bioremediation systems based on the usage of plants or microorganisms are eco-friendly and more affordable alternatives (Akcil and Mudder 2003). In this regard, microbial degradation systems are probably inconvenient because of the accumulation of toxic metabolites and/or overloading the system with high amounts of the pollutant. This can negatively affect microbial growth and in turn the overall bioremediation process (Ebbs 2004). Phytoremediation using algal-based systems and/or vascular plant systems might become preferable alternatives for removal of cyanate-containing compounds and cyanide contaminants (Taebi et al. 2008).
Cyanase (EC 4.2.1.104) is able to degrade cyanate into CO2 and NH3 in the presence of bicarbonate (Johnson and Anderson 1987). The enzyme was first identified and fully characterized in Escherichia coli (Anderson and Little 1986; Taussig 1960; Walsh et al. 2000). Other Gram-positive bacteria and fungi were also shown to contain cyanase (Butryn et al. 2015; Kamennaya and Post 2010). Endogenous cyanase activity enables some bacteria to use cyanate as sole nitrogen and carbon source (Luque-Almagro et al. 2008; Taussig 1960). The enzyme was also isolated and characterized from cyanobacteria (Kamennaya et al. 2008; Voigt et al. 2014) and plants (Aichi et al. 1998; Qian et al. 2011). In living organisms, cyanases are important for degradation and/or removal of the toxic cyanide and cyanate compounds affecting their growth (Ebbs 2004). Moreover, cyanases are effectively involved in different biochemical and physiological pathways in plants. Application of KCNO to Arabidopsis thaliana cyanase knock-out mutants causes inhibition of seed germination and seedling development. On contrary, transgenic Arabidopsis thaliana plants overexpressing Oryza sativa (OsCYN) or A. thaliana (AtCYN) cyanases showed better growth performance and increased resistance under cyanate stress (Qian et al. 2011). Recently, overexpression of cyanobacterial cyanase in A. thaliana enable transgenic plants to tolerate higher levels of cyanate, relative to wild type plants (Kebeish and Al-Zoubi 2017). However, cyanate biodegradation using transgenic plants is seemingly limited to soil ecosystems and cannot therefore be applied to water ecosystems.
Chlamydomonas reinhardtii has attracted more attention as a model for studying biological systems because this organism is the most biologically characterized (Rosales-Mendoza et al. 2012). Research into recombinant protein production such as expression of enzymes, proteins, human growth factors, antibodies, and vaccine in Chlamydomonas reinhardtii has attracted increasing attention (Rasala et al. 2010; Rosales-Mendoza et al. 2012). The efficacy of introducing cyanobacterial cyanase into micro-algae for cyanate remediation purposes has not been implemented so far. In the present study, the cyanobacterial cyanase gene (CYN, gi16329170) was therefore genetically cloned and transferred into C. reinhardtii nuclear genome. Transgenic C. reinhardtii lines showed enhanced tolerance to cyanate (up to 30 mM) compared to wild types. The biochemical response of transgenic micro-algae under cyanate stress has been studied in vivo. Results of the present study provide effective eco-friendly solutions for CNO− remediation in aquatic systems.
2 Materials and Methods
2.1 Chlamydomonas reinhardtii Culture Conditions
The test organism, Chlamydomonas reinhardtii strains CC-124 (mt−), was kindly obtained from Prof. Dr. Mohammed Ismaeil (Botany Department, Faculty of Science, Mansoura University, Egypt). C. reinharditii was aseptically grown in Tris Acetate Phosphate (TAP) medium pH 7.4 (Gorman and Levine 1965). For selection of transformed C. reinhardtii colonies, solid TAP medium supplemented with 1.5% (w/v) agar and kanamycin (50 μg/ml) was used. The algal cultures were incubated in growth chamber at 25 ± 2 °C under long day conditions (16 h light/8 h dark) and kept under light intensity of 80 μmol m−2 s−1 with continuous shaking (75 rpm) in case of liquid cultures.
2.2 Gene Cloning and Plasmid Constructs
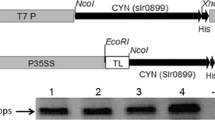
Cyanase gene (CYN gi16329170) was PCR-amplified using genomic DNA of Synechoccous elongatus PCC6803 as template (kindly obtained from Botany Department, Faculty of Science, Cairo University, Giza, Egypt). A forward primer with extension for NcoI site (5′- ATGGCCATGGCTGGCACTGAAATTTC-3′) and a reverse primer (5′-GTCACTCGAGCCATTTCTTGTAGGGTAA-3′) with extensions for XbaI site were used. Amplified CYN fragments were genetically cloned into the binary expression vector pTRAK (gi13508478). The resulted CYN gene expression cassette (Fig. 1) was flanked by 5′-UTR of the Cab22L tobacco leader peptide (TL), 3′-UTR of CaMV 35S (pA35S), and the scaffold attachment region (SAR gi3522871). The pPCV002 nptII gene was used for kanamycin selection of transgenic C. reinhardtii (Koncz and Schell 1986). CYN gene expression is transgenic lines is controlled by CaMV-35S promoter (Reichel et al. 1996).
Structure of cyanobacterial cyanase (CYN) gene expression cassette. Synechococcus elongatus PCC 6803 cyanase gene coding sequence (CYN, gi16329170, E.C.4.2.1.104) was cloned into the binary plant expression vector pTRA-K (gi13508478) in between NcoI and XbaI sites. CYN gene cassette is flanked by Scaffold attachment regions (SAR) of the tobacco RB7 gene (gi3522871). CYN gene expression is under the control of the constitutive CaM promoter (p35SS), 5′ UTR of the Cab22L tobacco leader peptide (TL), and 3′ UTR of CaMV 35S (pA35S). LB/RB: Left and right border sequences of Nopaline-Ti-plasmids pTiT37. pAnos: Polyadenylation signal of Nopaline synthetase gene from A. tumefaciens. NptII: Neomycin phosphotransferase type II that confers resistance to kanamycin. pnos: Promoter of nopaline synthase gene from A. tumefaciens
2.3 Transformation and Generation of Transgenic C. reinhardtii Lines
pTRAK-CYN construct was transformed into C. reinhardtii by the Agrobacterium tumefaciens (GV3101)-mediated co-cultivation transformation protocol as previously described (Kumar et al. 2004). single colony of wild type C. reinhardtii was inoculated into liquid TAP medium and was grown till reaching log phase. Cells were then plated on solid TAP medium and incubated for 48 h under continuous light until forming a lawn of cells. An Agrobacterium culture transformed with pTRAK-CYN plasmid was grown in liquid LB medium containing appropriate antibiotics (25 mg/l rifampicin and 50 mg/l kanamycin) at 28 °C till OD600 reaches 0.6. A. tumefaciens cells were then spun down (4000 ×g for 10 min at 4 °C) and resuspended in 250 μl liquid TAP medium containing 100 μM acetosyringone. The bacterial suspension was then co-cultivated with C. reinhardtii cells grown on the agar plates for 2 h at 28 °C followed by overnight incubation at 25 °C in dark. C. reinhardtii cells were harvested and washed twice with liquid TAP medium supplemented with 0.5 g/l cefotaxime (10 min each) in order to remove the remaining Agrobacterium. For selection of the transformed C. reinhardtii cells, the washed cells were cultivated on solid TAP agar plates containing 0.5 g/l cefotaxime and 50 μg/ml kanamycin and incubated at 25 °C in growth chamber for 8 days until the appearance of the transformed colonies (Fig. 2a). The presence of CYN transgene in the selected Chlamydomonas colonies was confirmed by colony PCR test using CYN gene–specific primers (2.2). Three independent transgenic C. reinhardtii lines were selected for further molecular and biochemical analyses. Presence of CYN transgene in transformed C. reinhardtii lines were confirmed by PCR test using genomic DNA isolated from transgenic lines as template prior to each assay.
Selection of transformed colonies of C. reinhardtii, qRT-PCR analysis of CYN transgene expression and cyanase enzymatic activity in transgenic lines of C. reinhardtii. a Photograph representing selection of the transformed colonies of C. reinhardtii and wild-type cells on solid TAP medium supplemented with kanamycin. b Results of qRT-PCR analysis of CYN transcript accumulation in three chosen transgenic colonies of C. reinhardtii. c Activity of cyanase enzyme measured from transgenic C. reinhardtii lines (Cr.CYN-1 to 3). Each data point is based on at least three independent measurements. Error bars indicate SE
2.4 qRT-PCR Analysis
Quantitative RT-PCR was performed as previously described (Kebeish and Al-Zoubi 2017). RNA was extracted from C. reinhardtii cells following the BCP (1-bromo-3-chlorpropane) protocol of Chomczynski and Mackey (Chomczynski and Mackey 1995). The protocol described by Niessen et al. (2007) was applied for the synthesis of first-strand cDNA. qRT-PCR analysis was performed on An ABI PRISM1 7300 (Applied Biosystems, Darmstadt, Germany) according to the manufacturer’s instructions. SYBR Green Reagents (Karlsruhe, Germany) were used for PCR amplifications. Oligonucleotides were purchased from Intron Biotechnology Inc. (Kyungki-Do, South Korea). For CYN transcripts, primers 5′-GGG AAT CAC GTT TGC TGA TTT-3′ and 5′-AAG TTT CTC CGC CTC ATC AA-3′ were used. Primers 5′-GCG ATG TGG ACA TCC GCA AG-3′ and 5′-GGG CCG TGA TCT CCT TGC TC-3′ were used for detection of ACTIN transcripts. In the reaction mixtures, final primer concentration was adjusted to be 200 nM. 10 min primary denaturation at 95 °C, followed by 40 cycles (15 s denaturation at 95 °C and 60 °C combined annealing and extension for 1 min) was applied as a PCR program for amplification of both CYN and ACTIN.
2.5 Growth Assay of Transgenic C. reinhardtii under Cyanate Stress
Three independent C. reinhardtii lines transgenic for cyanase gene in addition to wild type were used for evaluating the efficient detoxification of cyanate from the culture medium. Transgenic and wild-type C. reinhardtii (~ 5 × 103 cells) were inoculated into liquid TAP medium supplemented with 0, 5, 10, 15, 20, and 30 mM KCNO. After that cells were allowed to grow at long day growth conditions with continuous shaking. Cell densities at 665 nm (Robert 1979) were recorded each 24 h for 15 days. A growth response curve was generated and mid log phase of growth was observed after 7 days. The experiment was then repeated in trice and cell density for each genotype at all the applied KCNO concentrations were recorded after 7 days of growth where the percentage of growth reduction as a result of KCNO application was calculated based on the following equation: % reduction in growth = [(cell density of C. reinhardtii without KCNO − cell density at the desired KCNO concentration) / (cell density of C. reinhardtii without KCNO)] × 100.
2.6 Ammonia Release and Pigment Content Measurements
Ammonia released from transgenic C. reinhardtii at different concentrations of potassium cyanate was carried out according to the method described by Rasco-Gaunt et al. (Rasco-Gaunt et al. 1999) with modifications. Wild-type and transgenic C. reinhardtii cells were allowed to grow in liquid TAP medium for 7 days. Algal tissues were harvested and 0.05 g from each genotype were transferred into 2-ml micro tubes containing 1 ml of incubation medium [potassium phosphate pH 5.8 (50 mM), sucrose (2%), Tween 20 (1%), 2,4-dichlorophenoxy acetic acid (0.1 mg l−1), and phosphinotricin (25 mg/l)] supplemented with 0, 5, 10, 15, 20, and 30 mM KCNO. As blanks, three reaction micro tubes containing incubation medium without KCNO were used. The test samples were incubated for 16 h at 25 °C under 80 μmol m−2 s−1 light. After centrifugation (13,000 rpm/5 min), 100 μl from each sample were transferred into a new 1.5-ml reaction tube containing 0.5 ml of reagent-I (0.4 mM sodium nitroprusside, 25 g/l sodium tartrate, 0.085 M trisodium citrate, and 0.21 M sodium salicylate). Samples were mixed and 500 μl of reagent-II (2.3 mM sodium dichloroisocyanurate, 0.75 M NaOH) was added. The reaction mixtures were then left in the dark at 37 °C followed by incubation at room temperature for 15 min. Ammonium ions in the test samples were quantified by measuring the absorbance at 655 nm compared to standards of ammonium chloride.
Pigments, Chl. a, Chl. b, and carotenoids were extracted from wild-type and transgenic C. reinhardtii cells grown for 5 days in liquid TAP medium after application of different concentrations of KCNO (0, 5, 10, 15, 20, 30 mM) with 80% acetone. After homogenization and vigorous shaking, mixtures were centrifuged at 30.000 g for 10 min. Absorbance of the supernatants was measured at 664, 647, 630, and 452 nm. Pigment contents were calculated as described by Metzner et al. (1965) and Jeffrey and Humphrey (1975).
2.7 Protein Extraction and Enzymatic Assays
For total protein extraction from wild-type and transgenic C. reinhardtii cells, 100 mg fresh algal tissues was collected and directly frozen in liquid nitrogen. Frozen pellets were then homogenized and 500 μl of extraction buffer (50 mM sodium phosphate buffer (pH 7.4) containing 0.1% Triton X-100, 10 mM EDTA, 2% PVP, and 20% glycerol) were added. The homogenate was centrifuged twice at 30.000 ×g for 20 min at 4 °C. Supernatants were then collected and protein concentration was determined using Bradford method (Bradford 1976). These protein extracts were used for cyanase (CYN), catalase (CAT), peroxidase (POD), and superoxide dismutase (SOD) enzymatic assays based on the assay conditions.
Cyanase activity was measured in vitro from wild-type and transgenic C. reinhardtii cells following the protocol described by Anderson (P.M Anderson 1980). The reaction mixture composed of 50 mM potassium phosphate buffer (pH 7.7), 3 mM NaHCO3, and 2 mM KCNO. The reaction was started by addition of protein extract and stopped by addition of equal volumes of Nessler’s reagent after 1–10 min at 26 °C. Ammonium chloride dilution series were used for generating a standard curve. The amount of released ammonia (μmol) was estimated by measuring the optical density at 420 nm 10 min after addition of Nessler’s reagent compared to ammonium chloride standards. Cyanase activity was expressed as μmol ammonia released/mg protein/min.
For determination of antioxidant enzyme activities for wild-type and transgenic C. reinhardtii lines under cyanate stress, total proteins were extracted from 100 mg C. reinhardtii cells harvested after 5 days growth in liquid TAP medium supplemented with 0, 5, 10, 15, 20, and 30 mM KCNO. Extracted proteins from each genotype were used for quantification of catalase (Kar and Mishra 1976), peroxidase (Racusen and Foote 1965), and superoxide dismutase (Beauchamp and Fridovich 1971; Xu et al. 2008) activities at all potassium cyanate concentrations.
2.8 Statistical Analysis
Data were represented as mean ± standard error (SE) of at least three independent measurements. Student’s t test using Excel software (Microsoft Corporation, USA) was applied to determine significance.
3 Results and Discussion
3.1 Gene Cloning and Generation of Transgenic C. reinhardtii Lines
The target of the present study was to test whether overexpression of the cyanobacterial cyanase (CYN, gi16329170) could support the growth and cyanate remediation capacity of C. reinhardtii or not. Among species, different pathways for cyanase-dependent cyanate metabolism have been documented. In bacteria, cyanase was shown to have an active role in the production of NH3 as an alternative source for nitrogen from cyanate (P.M. Anderson et al. 1990). Products of cyanase activity in photosynthetic cyanobacteria are used as carbon and nitrogen sources (Espie et al. 2007). The gene coding for cyanase has been identified, characterized, and genetically expressed in Arabidopsis (Aichi et al. 1998; Qian et al. 2011). Cyanase (cynS) was also characterized in Synechocystis sp. strain PCC7942, PCC6803, and PCC6301 (Harano et al. 1997). In the current study, cyanobacterial gene, CYN gi16329170, was used for the transformation of Chlamydomonas reinhardtii in order to evaluate the efficacy of this cyanobacterial cyanase in promoting cyanate remediation capacity of C. reinhardtii cells. Wild-type C. reinhardtii was transformed with the binary expression construct, pTRAK-CYN (Fig. 1) via Agrobacterium tumefaciens−mediated co-cultivation method (Kumar et al. 2004).
Transgenic Chlamydomonas lines were selected on solid TAP medium supplemented with kanamycin antibiotic (Fig. 2a). Three independent lines of transgenic Chlamydomonas reinhardtii named Cr.CYN-1, Cr.CYN-2, and Cr.CYN-3 were randomly selected and used for further molecular and biochemical analyses.
3.2 RT-PCR and Cyanase Enzymatic Assays
CYN gene mRNA transcripts were initially measured in CYN transgenic lines by qRT-PCR (Fig. 2b). High levels of CYN transcripts were observed for all the tested CYN transgenic C. reinhardtii lines compared to ACTIN transcripts. Transgenic line Cr.CYN-3 shows relatively higher levels of cyanase transgene expression followed by Cr.CYN-1 then Cr.CYN-2 lines. Variable endogenous promoters controlling gene expression in C. reinhardtii have been reported. The small subunit of the ribulose bisphosphate carboxylase, RbcS2 promoter was the most widely used for induction of transgene expression in C. reinhardtii (Cerutti et al. 1997). However, other constitutive promoters as PsaD, TubA1, Hsp70A, AtpC, and β-tub (Rosales-Mendoza et al. 2012) were shown to induce high transgene expression in chimeric constructs used for C. reinhardtii transformation. Moreover, Cauliflower mosaic virus 35S promoter (CaMV 35S) has also been shown to induce high gene expression in this micro-alga (Tang et al. 1995). It was previously reported that high expression of transgenes in the model organism C. reinhardtii could be enhanced when using genes comprising high GC content (Heitzer et al. 2007). Since GC content of the introduced cyanobacterial cyanase gene is approximately 47%, the observed expression levels of this transgene are therefore reasonable (Fig. 2b). However, codon usage adaptation of the introduced transgene is necessary for this micro-alga to induce higher expression levels.
To test the functionality of the expressed cyanase in transgenic lines, the activity of cyanase transgene was also tested on protein level in extracts of WT and transgenic C. reinhardtii lines in vitro. The results of cyanase activity assay are shown in Fig. 2c. Threefold to fourfold increase in the activity of cyanase was obvious for transgenic lines compared to WT controls. CYN-1 and CYN-3 lines show more activity for cyanase when compared to line CYN-2 (Fig. 2c). The background activity observed for WT control samples may be due to the action of endogenous cyanases and other nitrogen assimilating enzymes (Aichi et al. 1998; Anderson and Little 1986). It can be therefore concluded that the prokaryotic cyanase is active in transgenic C. reinhardtii lines.
3.3 Ammonia Release Assay from Transgenic C. reinhardtii
Cyanase converts cyanate into CO2 and NH3 in the presence of bicarbonate. Therefore, the amount of ammonia released from wild-type and transgenic C. reinhardtii lines in the presence of variable concentrations of cyanate was measured. This assay relies on suppression of ammonia fixation/re-fixation in the GS2/Fd-GOGAT cycle by phosphinotricin (Rasco-Gaunt et al. 1999). Wild-type and transgenic C. reinhardtii fresh cells were incubated in medium containing 2 mM NaHCO3 and variable concentrations of KCNO (i.e., 0.0, 5, 10, 15, 20, and 30 mM). Results of this assay are shown in Fig. 3. In the absence of KCNO, NH3 release was not significantly affected in both transgenic and wild-type control samples. A gradual decrease in the amount of NH3 was observed for wild-type C. reinhardtii by increasing KCNO concentrations. On the contrary, significant KCNO concentration-dependent (p ≤ 0.001) increases in the amount of NH3 released was observed for all the three transgenic lines compared to the corresponding wild-type controls. Again, line Cr.CYN-3 was superior compared to Cr.CYN-2 and Cr.CYN-1 lines. Reduction of ammonia release observed for wild type control samples may be due to the limited capacity of the endogenous cyanase to overcome the excess cyanate applied to the reaction mixture (Qian et al. 2011). In microbes, induction of cyanase gene transcription can be achieved through the application of exogenous cyanate (Luque-Almagro et al. 2008). However, induction of cyanase gene transcription in Arabidopsis thaliana and Oryza sativa was only observed under salt stress but not cyanate stress (Askari et al. 2006; Qian et al. 2011). Results of NH3 release assay indicate that CYN transgenic C. reinhardtii is able to produce more ammonia from exogenously applied cyanate and this, in turn, proves the functionality of the cyanobacterial cyanase enzyme in the green micro-alga, Chlamydomonas reinhardtii concerning cyanate remediation. This supports the significance of using CYN transgenic lines in cyanate detoxification.
Ammonia release assay from wild-type and CYN transgenic C. reinhardtii under KCNO stress. Amount of NH3 released from wild type (Cr. WT) and three cyanase transgenic C. reinhardtii lines (Cr.CYN1-3) was measured from Chlamydomonas reinhardtii cells incubated with different concentrations of KCNO (0.0, 5, 10, 15, 20, and 30 mM) for 4 h. Each data point represents the average of at least three independent measurements ± SE. P value ˂ 0.001 for transgenic lines compared to wild type.
3.4 Growth Assays of Transgenic C. reinhardtii Under Cyanate Stress
In order to evaluate the efficacy of the cyanobacterial transgene cyanase in cyanate detoxification, a growth assay was performed using CYN transgenic C. reinhardtii lines. Similar amounts of Cr.CYN-1, Cr.CYN-2, Cr.CYN-3, and wild-type cells were inoculated into liquid TAP medium supplemented with 0.0, 5, 10, 15, 20, and 30 mM KCNO and grown for 7 days. Algal cell densities were then measured at 665 nm and the percentage of growth reduction was calculated. The results of this assay are shown in Fig. 4 and Table 1. A concentration-dependent inhibition of Chlamydomonas growth was obvious for wild type as well as CYN transgenic lines. However, growth of transgenic lines is highly significant compared to wild type at all potassium cyanate concentrations applied for this assay (Fig. 4). A sharp decrease in growth of wild-type cells was also remarkable at KCNO concentrations above 10 mM (59% and 88% reduction in growth at 15 and 20 mM KCNO, respectively) leading to a complete cell death at 30 mM KCNO (Table 1). On contrary, transgenic lines showed better growth performance and a decreased percentage of growth reduction compared to wild-type cells and growth of Cr.CYN-1 was relatively higher compared to Cr.CYN-2 and Cr.CYN-3 lines (Table 1). These growth assay results under cyanate stress indicate that expression of the cyanobacterial cyanase transgene in Chlamydomonas enhances the ability of this micro-alga to metabolize large amounts of the toxic cyanate from the culture medium (i.e., up to KCNO concentration of 20 mM) and consequently used its degradation products (i.e., CO2 and NH3) for supporting algal growth and biomass production. The increased reduction in growth of wild type starting from concentration of 10 mM KCNO may be attributed to the inefficient capacity of the endogenous cyanate metabolizing enzymes to remove the applied cyanate from the growth medium (Qian et al. 2011; Askari et al. 2006). It is important to compare the current growth assay results obtained for CYN transgenic C. reinhardtii with our previously reported results for CYN transgenic A. thaliana plants (Kebeish and Al-Zoubi 2017). A similar growth retardation phenomenon was observed for wild-type A. thaliana plants when cyanate concentration of 2.5 mM was applied to the foliar parts of the plant. Additionally, 0.4 mM cyanate in the culture medium caused a total inhibition of root hair formation in wild-type Arabidopsis plants. However, in both growth assays, expression of the cyanobacterial enzyme in A. thaliana plants enhance plant resistance to the exogenously applied cyanate up to 1.2 mM. Indeed, the enhanced growth of CYN transgenic C. reinhardtii at higher cyanate concentrations (i.e., 15, 20, and 30 mM KCNO) compared to wild-type cells support the usage of CYN transgenic C. reinhardtii as an efficient bio-system for detoxification of the harmful cyanate from the environment, especially from the contaminated water sites.
3.5 Effect of KCNO on Pigment Contents in Wild-Type and Transgenic C. reinhardtii Cells
Initial growth assays with wild-type C. reinhardtii showed that cyanate application in growth media leads to growth retardation accompanied by a loss of pigmentation symptoms. Therefore, chlorophyll and carotenoid pigments were measured from Cr.CYN transgenic lines and compared to wild-type controls under KCNO stress. Results of this assay are shown in Table 2. All the measured values for pigment contents were significantly higher in cyanase overexpressors compared to wild-type controls under the assay conditions. The observed values for chl.a, chl.b, and carotenoids in Cr.CYN-1, Cr.CYN-2, and Cr.CYN-3 transgenic lines at KCNO concentration of 20 mM reached about 1.9-, 1.8-, and 2.0-fold, respectively, compared to their wild-type counterparts. It was also observed that 30 mM KCNO cause a complete death of wild-type C. reinhardtii cells. However, transgenic C. reinhardtii can survive this high concentration of cyanate with weak signs of growth retardation symptoms. The decrease in pigment contents observed for wild-type C. reinhardtii may be due to inhibition of vital enzymes contributing to metabolic processes related to pigment biosynthesis and viability by the applied cyanate (Samuilov et al. 2006; Solomonson 1981). The mitochondrial cytochrome c oxidase that is actively involved in the respiratory electron transport chain was reported to be inhibited by cyanide and its oxidative product cyanate (Way 1984). Ribulose-1,5-bisphosphate carboxylase/oxygenase (Wishnik and Lane 1969) and stress-relieving enzymes involved in ROS scavenging mechanisms have been shown to be inhibited by cyanides (Karuppanapandian et al. 2011). Additionally, cyanide and cyanate are also able to inactivate important metalloenzymes involved in many biological processes such as respiration, carbon, and nitrate assimilation (Leavesley et al. 2008; Solomonson 1981). The observed results for pigment contents in Cr.CYN transgenic lines (Table 2) are in accordance with our previous results reported for A. thaliana plants transgenic for the cyanobacterial enzyme cyanase (Kebeish and Al-Zoubi 2017) or for the bacterial enzyme cyanidase (Kebeish et al. 2015). Transgenic A. thaliana plants overexpressing the cyanase or cyanidase showed significant increase in chlorophyll a, chlorophyll b, and carotenoids contents under cyanate or cyanide stress, respectively. The observed results in this assay indicate therefore that KCNO-dependent loss of pigmentation symptoms was ameliorated through the activity of the recombinant cyanobacterial enzyme cyanase in transgenic C. reinhardtii lines.
3.6 Effect of KCNO on Antioxidant Enzyme Activities in Wild-Type and Transgenic C. reinhardtii Cells
Algae and higher plants show variable biochemical responses to biotic and abiotic stress conditions. Among these biochemical responses is the production of antioxidant enzymes in order to reduce the endogenously generated reactive oxygen species (ROS) (Liu et al. 2018). Therefore, the antioxidant enzymes, catalase (CAT), peroxidase (POD), and superoxide dismutase (SOD) activities were measured in extracts of transgenic and wild-type lines (Table 3) at different concentrations of KCNO. Wild-type extracts show enhanced levels of antioxidant enzyme activities at all the applied cyanate concentrations in a relative concentration-dependent manner. However, significant reduction in antioxidant enzymes was recorded for CYN transgenic lines compared to their corresponding controls of wild type at both low and high concentrations of KCNO. At KCNO concentration of 20 mM, wild-type C. reinhardtii showed 3.4-fold increase for CAT, 2.4-fold increase for POD, and 5-fold increase for SOD compared to untreated cells. However, transgenic lines showed low induction of these enzymes (− 31% catalase, − 42% peroxidase, and − 48% superoxide dismutase) compared to wild-type controls at the same KCNO concentration (i.e., 20 mM). Similar reduction in antioxidant enzyme activities was also observed for CYN transgenic lines at 5 mM, 10 mM, and 15 mM KCNO. Cr.CYN-3 line was superior when compared to the other two transgenic lines (Table 3). The observed reduction of CAT, POD, and SOD activities in transgenic C. reinhardtii lines can be due to the activity of the recombinant cyanobacterial enzyme cyanase in detoxification of the applied cyanate. SOD is considered as the first key player in the defense mechanism against oxidation as it converts O2− to H2O2 and O2 (Bowler et al. 1992). Previous studies have shown significant increase in SOD activities under KCN stress in Chlorella vulgaris however at low concentrations (0.1–10 mg l−1), hence SOD contributed to the removal of superoxides. On contrary, by increasing KCN concentration to 10 mg l−1, a decline in SOD activity was observed (Liu et al. 2018). This may be due to the inhibition of SOD activity by excess cyanide. Many reports have indicated that cyanide and perhaps cyanate inhibit antioxidant enzyme activities (Oracz et al. 2009; Siegien and Bogatek 2006). However, our recently published studies with A. thaliana expressing either cyanidase or cyanase (Kebeish et al. 2015; Kebeish and Al-Zoubi 2017) indicates the effective role of cyanide and/or cyanate-degrading enzymes in reducing the negative impact of the toxic cyanide compounds on the antioxidant machinery in transgenic plants through the efficient removal and/or degradation of the toxic factors. Therefore, the observed decrease in the activities of antioxidant enzyme in C. reinhardtii transgenic for CYN may reflect the efficient role of the recombinant cynobacterial cyanase in the removal of the applied cyanate from the surrounding growth medium. Taken together, the observed biochemical assay data in this study provide a strong evidence for the significance of this novel micro-algal–based detoxification system for cyanate removal from the environment.
References
Aichi, M., Nishida, I., & Omata, T. (1998). Molecular cloning and characterization of a cDNA encoding cyanase from Arabidopsis thaliana. Plant & Cell Physiology, 39, S135–S135.
Akcil, A., & Mudder, T. (2003). Microbial destruction of cyanide wastes in gold mining: process review. Biotechnology Letters, 25, 520–527.
Anderson, P. M. (1980). Purification and properties of the inducible enzyme cyanase. Biochemistry, 19, 2882–2888.
Anderson, P. M., & Little, R. M. (1986). Kinetic properties of cyanase. Biochemistry, 25, 1621–1626.
Anderson, P. M., Sung, Y.-C., & Fuchs, J. A. (1990). The cyanase operon and cyanate metabolism. FEMS Microbiology Reviews, 7, 247–252.
Askari, H., Edqvist, J., Hajheidari, M., Kafi, M., & Salekdeh, G. H. (2006). Effects of salinity levels on proteome of Suaeda aegyptiaca leaves. Proteomics, 6, 2542–2554.
Beauchamp, C., & Fridovich, I. (1971). Superoxide dismutase: improved assays and an assay applicable to acrylamide gels. Analytical Biochemistry, 44, 276–287.
Bowler, C., Montagu, M. V., & Inze, D. (1992). Superoxide dismutase and stress tolernace. Annual Review of Plant Physiology and Plant Molecular Biology, 43, 83–116.
Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Analytical Biochemistry, 72(1–2), 248–254.
Butryn, A., Stoehr, G., Linke-Winnebeck, C., & Hopfner, K.-P. (2015). Serendipitous crystallization and structure determination of cyanase (CynS) from Serratia proteamaculans. Acta. Crystallographica. Section F: Structural Biology Communications, 71, 471–476. https://doi.org/10.1107/S2053230X15004902.
Cerutti, H., Johnson, A. M., Gillham, W. N., & Boynton, J. E. (1997). A eubacterial gene conferring spectinomycin resistance on Chlamydomonas reinhardtii: Integration into the nuclear genome and gene expression. Genetics, 145, 97–110.
Chomczynski, P., & Mackey, K. (1995). Substitution of chloroform by bromochloropropane in the single-step method of RNA isolation. Analytical Biochemistry, 225, 163–164.
Ebbs, S. (2004). Biological degradation of cyanide compounds. Current Opinion in Biotechnology, 15, 231–236.
Espie, G. S., Jalali, F., Tong, T., Zacal, N. J., & So, A. K.-C. (2007). Involvement of the cynABDS operon and the CO2-concentrating mechanism in the light-dependent transport and metabolism of cyanate by cyanobacteria. Journal of Bacteriology, 189, 1013–1024.
Gorman, D. S., & Levine, R. P. (1965). Cytochrome f and plastocyanin: their sequence in the photosynthetic electron transport chain of Chlamydomonas reinharditi. Proceedings of the National Academy of Sciences of the United States of America, 54(6), 1665–1669.
Guillotonm, M., & Karst, F. (1987). Isolation and characterization of Escherichia coli mutants lacking inducible cyanase. Microbiology, 133, 645–653.
Hamel, J. (2011). A review of acute cyanide poisoning with a treatment update. Critical Care Nurse, 82, 31–72.
Harano, Y., Suzuki, I., Maeda, S.-I., Kaneko, T., Tabata, S., & Omata, T. (1997). Identification and nitrogen regulation of the cyanase gene from the cyanobacteria Synechocystis sp. strain PCC 6803 and Synechococcus sp. strain PCC 7942. Journal of Bacteriology, 179, 5744–5750.
Heitzer, M., Eckert, A., Fuhrmann, M., & Griesbeck, C. (2007). Influence of codon bias on the expression of foreign genes in microalgae. In Transgenic microalgae as green cell factories (pp. 46–53). New York: Springer.
Jeffrey, S. W., & Humphrey, G. F. (1975). New spectrophotometric equations for determining chlorophylls a, b, c1 and c2 in higher plants, algae and natural phytoplankton. Biochemie und Physiologie der Pflanzen, 167, 191–194.
Johnson, W., & Anderson, P. (1987). Bicarbonate is a recycling substrate for cyanase. The Journal of Biological Chemistry, 262, 9021–9025.
Kamennaya, N. A., & Post, A. F. (2010). Characterization of cyanate metabolism in marine Synechococcus and Prochlorococcus spp. Applied and Environmental Microbiology, 77, 291–301. https://doi.org/10.1128/AEM.01272-10.
Kamennaya, N. A., Chernihovsky, M., & Post, A. F. (2008). The cyanate utilization capacity of marine unicellular cyanobacteria. Limnology and Oceanography, 53, 2485–2495.
Kar, M., & Mishra, D. (1976). Catalase, peroxidase, polyphenol oxidase activities during rice leaf senescence. Journal of Plant Physiology, 57, 315–319.
Karuppanapandian, T., Moon, J. H., Kim, C., Manoharan, K., & Kim, W. (2011). Reactive oxygen species in plants: their generation, signal transduction, and scavenging mechanisms. Australian Journal of Crop Science, 5(6), 709–725.
Kebeish, R., & Al-Zoubi, O. (2017). Expression of the cyanobacterial enzyme cyanase increases cyanate metabolism and cyanate tolerance in Arabidopsis. Environmental Science and Pollution Research International, 12(24), 11825–11835.
Kebeish, R., Aboelmy, M., El-Naggar, A., El-Ayouty, Y., & Peterhansel, C. (2015). Simultaneous overexpression of cyanidase and formate dehydrogenase in Arabidopsis thaliana chloroplasts enhanced cyanide metabolism and cyanide tolerance. Environmental and Experimental Botany, 110, 19–26. https://doi.org/10.1016/j.envexpbot.2014.09.004.
Koncz, C., & Schell, J. (1986). The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of agrobacterium binary vector. Molecular and General Genetics MGG, 204, 383–396.
Koshiishi, I., Mamura, Y., & Imanari, T. (1997). Cyanate causes depletion of ascorbate in organisms. Biochimica et Biophysica Acta (BBA)- General Subjects, 1336, 566–574.
Kumar, S. V., Misquitta, R. W., Reddy, V. S., Rao, B. J., & Rajam, M. V. (2004). Genetic transformation of the green alga-Chlamydomonas reinhardtii by Agrobacterium tumefaciens. Plant Science, 166, 731–738.
Leavesley, H. B., Li, L., Prabhakaran, K., Borowitz, J. L., & Isom, G. E. (2008). Interaction of cyanide and nitric oxide with cytochrome c oxidase: implications for acute cyanide toxicity. Toxicological Sciences, 101, 101–111.
Liu, Q., Zhang, G., Ding, J., Zou, H., Shi, H., & Huang, C. (2018). Evaluation of the removal of potassium cyanide and its toxicity in green algae (Chlorella vulgaris). Bulletin of Environmental Contamination and Toxicology, 100(2), 228–233.
Luque-Almagro, V. M., Huertas, M.-J., Sáez, L. P., et al. (2008). Characterization of the Pseudomonas pseudoalcaligenes CECT5344 cyanase, an enzyme that is not essential for cyanide. Assimilation. Applied and Environmental Microbiology, 74, 6280–6288.
Malhotra, S., Pandit, M., Kapoor, J., & Tyagi, D. (2005). Photo-oxidation of cyanide in aqueous solution by the UV/H2O2 process. Journal of Chemical Technology and Biotechnology, 80, 13–19.
Mekuto, L., Ntwampe, S., & Akcil, A. (2016). An integrated biological approach for treatment of cyanidation wastewater. Science of the Total Environment, 571, 711–720.
Metzner, H., Rau, H., & Senger, H. (1965). Untersuchungen zur synchronisierbarkeit einzelner pigment-mangel mutanten von Chlorella. Planta, 65, 186–194.
Mudder, T. I., Botz, M., & Smith, A. (2001). Chemistry and treatment of cyanidation wastes. London: Mining Journal Books.
Niessen, M., Thiruveedhi, K., Rosenkranz, R., Kebeish, R., Hirsch, H.-J., Kreuzaler, F., et al. (2007). Mitochondrial glycolate oxidation contributes to photorespiration in higher plants. Journal of Experimental Botany, 58, 2709–2715.
Oracz, K., El-Maarouf-Bouteau, H., Kranner, I., Bogatek, R., Corbineau, F., & Bailly, C. (2009). The mechanisms involved in seed dormancy alleviation by hydrogen cyanide unravel the role of reactive oxygen species as key factors of cellular signaling during germination. Plant Physiology, 150, 494–505.
Qian, D., Jiang, L., Lu, L., Wei, C., & Li, Y. (2011). Biochemical and structural properties of cyanases from Arabidopsis thaliana and Oryza sativa. PLoS One, 6(3), e18300.
Racusen, D., & Foote, M. (1965). Protein synthesis in dark grown bean leaves. Canadian Journal of Botany, 43, 817–824.
Rasala, B. A., Muto, M., Lee, P. A., Jager, M., Cardoso, R. M. F., Behnke, C. A., et al. (2010). Production of therapeutic proteins in algae, analysis of expression of seven human proteins 215 in the chloroplast of Chlamydomonas reinhardtii. Plant Biotechnology Journal, 8, 719–733.
Rasco-Gaunt, S., Riley, A., Lazzeri, P., & Barcelo, P. (1999). A facile method for screening for phosphinothricin (PPT)-resistant transgenic wheats. Molecular Breeding, 5, 255–262.
Reichel, C., Mathur, J., Eckes, P., Langenkemper, K., Koncz, C., Schell, J., et al. (1996). Enhanced green fluorescence by the expression of an Aequorea victoria green fluorescent protein mutant in mono-and dicotyledonous plant cells. Proceedings of the National Academy of Sciences of the United States of America, 93, 5888–5893.
Robert, R. (1979). Growth measurements. Division rate. Physiological methods. Culture methods and growth measurements (pp. 29–311). Cambridge: Cambridge University. Press.
Rosales-Mendoza, S., Paz-Maldonado, L. M. T., & Soria-Guerra, R. E. (2012). Chlamydomonas reinhardtii as a viable platform for the production of recombinant proteins: current status and perspectives. Plant Cell Reports, 31, 479–494.
Samuilov, V. D., Kiselevsky, D. B., Sinitsyn, S. V., Shestak, A. A., Lagunova, E. M., & Nesov, A. V. (2006). H2O2 intensifies CN-induced apoptosis in pea leaves. Biochemistry-Moscow, 71, 384–394.
Siegien, I., & Bogatek, R. (2006). Cyanide action in plants - from toxic to regulatory. Acta Physiologiae Plantarum, 28, 483–497.
Solomonson, L. P. (1981). Cyanide as a metabolic inhibitor. In B. Vennesland, E. E. Conn, C. J. Knowles, J. Westley, & F. Wissing (Eds.), Cyanide in biology (pp. 11–28). London: Academic Press.
Srivastava, A. C., & Muni, R. R. D. (2010). Phytoremediation of cyanide. In Plant adaptation and phytoremediation (pp. 399–426). Springer.
Taebi, A., Jeirani, K., Mirlohi, A., & Zadeh Bafghi, A. R. (2008). Phytoremediation of cyanide-polluted soils by non-woody plants. JWSS-Isfahan University of Technology, 11, 515–523.
Tang, D., Qiao, S.-Y., & Wu, M. (1995). Insertion mutagenesis of Chlamydomonas reinhardtii by electroporation and heterologous DNA. Biochemistry and Molecular Biology International, 36, 1025–1035.
Taussig, A. (1960). The synthesis of the induced enzyme, cyanase, in E. coli. Biochimica et Biophysica Acta, 44, 510–519.
Voigt, K., Sharma, C. M., Mitschke, J., Lambrecht, S. J., Voss, B., Hess, W. R., et al. (2014). Comparative transcriptomics of two environmentally relevant cyanobacteria reveals unexpected transcriptome diversity. The ISME Journal, 8, 2056–2068.
Walsh, M. A., Otwinowski, Z., Perrakis, A., Anderson, P. M., & Joachimiak, A. (2000). Structure of cyanase reveals that a novel dimeric and decameric arrangement of subunits is required for formation of the enzyme active site. Structure, 8, 505–514.
Way, J. L. (1984). Cyanide intoxication and its mechanism of antagonism. Annual Review of Pharmacology and Toxicology, 24, 451–481.
Wishnik, M. W., & Lane, M. D. (1969). Inhibition of ribulose diphosphate carboxylase by cyanide. The Journal of Biological Chemistry, 244, 55–59.
Xu, P., Zou, J., Meng, Q., Zou, J., Jiang, W., & Liu, D. (2008). Effects of Cd2+ on seedling growth of garlic (Allium sativum L.) and selected physiological and biochemical characters. Bioresource Technology, 99(14), 6372–6378.
Acknowledgements
This work is in part under the supervision of the Applied Scientific Research Center, Herbal and Medicinal Plants research group, Taibah University. We are thankful to Prof. Dr. Mohammed Ismaeil (Botany Department, Faculty of Science, Mansoura University, Egypt) for providing us with C. reinhardtii culture.
Declarations:
- All authors participate equally in this publication.
- We do not have any conflict of interest with other research institutions or any potential financial support that could be perceived to influence the outcomes of the research.
- No conflicts, informed consent, human or animal rights applicable.
- All authors agreed to the authorship and submission of the manuscript for peer review in its current form.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
El-Ayouty, Y., Ismaiel, M., Al-Badwy, A. et al. Overexpression of Cyanase in Chlamydomonas reinhardtii: a Promising Approach for Biodegradation of Cyanate in Aquatic Systems. Water Air Soil Pollut 230, 123 (2019). https://doi.org/10.1007/s11270-019-4175-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11270-019-4175-0