Abstract
Cyanate and its derivatives are considered as environmental hazardous materials. Cyanate is released to the environment through many chemical industries and mining wastewater. Cyanase enzyme converts cyanate into CO2 and NH3 in a bicarbonate-dependent reaction. At low cyanate concentrations, the endogenous plant cyanases play a vital role in cyanate detoxification. However, such cyanate biodegradation system is probably insufficient due to the excess cyanate concentrations at contaminated sites. In this study, we have transferred the activity of the cyanobacterial cyanase into Arabidopsis thaliana plants in order to enhance plant resistance against cyanate toxicity. The enzyme was shown to be active in planta. Transgenic plants exposed to cyanate, either applied by foliar spray or supplemented in growth medium, showed less reduction in pigment contents, antioxidant enzymes, carbohydrate contents, and reduced levels of plant growth retardation. Plant growth assays under cyanate stress showed enhanced growth and biomass accumulation in cyanase overexpressors compared to control plants. Results of this study provide evidence for developing novel eco-friendly phytoremediation systems for cyanate detoxification.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
The toxic compound, cyanate, threatens both human health and ecosystems. It is produced by anthropogenic activities and industries that use electroplating, ore leaching, steal manufacturing, and synthetic fibers (Hamel 2011; Patil and Paknikar 2000; Sancho and Bellon 2005). Cyanate and its derivative compounds have been widely used for the production of herbicides and synthesis of polymers (Koshiishi et al. 1997; Kraus and Kraus 2001). It is also produced through degradation of a number of metabolites such as carbamoylphosphate and urea (Dirnhuber and Schutz 1948; Guilloton and Karst 1987). Moreover, cyanate is also released into the environment through spontaneous photo-oxidation of cyanide (Malhotra et al. 2005; Rader et al. 1995) as well as the oxidative treatment of cyanide-containing industrial wastes (Mekuto et al. 2016; Nowakowska et al. 2006).
The common detoxification methods for cyanate and cyanide compounds are mainly based on chemical treatments through oxidation or chlorination reactions (Akcil and Mudder 2003). However, these chemical methods are disadvantageous because of their high costs and/or production of toxic by-products (Srivastava and Muni 2010). Bioremediation using microorganisms or plants provides affordable and eco-friendly alternatives (Akcil and Mudder 2003). However, microbial biodegradation systems are seemingly inefficient due to accumulation of toxic microbial metabolites or overloading with excess pollutants (Ebbs 2004). Indeed, phytoremediation using vascular plants may be the preferable approach for detoxification of cyanate and cyanide pollutants (Bushey et al. 2006; Taebi et al. 2008; Yu et al. 2006).
Cyanase enzyme (EC 4.2.1.104) converts cyanate into carbon dioxide and ammonium in a bicarbonate-dependent reaction (Anderson 1980; Anderson and Little 1986; Johnson and Anderson 1987). The enzyme is encoded by cynS and was first identified in Escherichia coli where the enzyme was fully characterized in this bacterium (Anderson and Little 1986; Johnson and Anderson 1987; Sung et al. 1987; Taussig 1960; Walsh et al. 2000). It is also found in proteobacteria, some gram-positive bacteria, and fungi (Butryn et al. 2015; Elleuche and Poggeler 2008; Kamennaya and Post 2010). Cyanase activity was also shown to support the growth of some bacteria using cyanate as nitrogen and carbon sources (Kunz and Nagappan 1989; Luque-Almagro et al. 2008; Taussig 1960). The enzyme has also been discovered in cyanobacteria (Blank and Hinman 2016; Garcia-Fernandez and Diez 2004; Harano et al. 1997; Kamennaya et al. 2008; Miller and Espie 1994; Voigt et al. 2014) and plants (Aichi et al. 1998; Qian et al. 2011). The role of cyanase in cyanate detoxification is based mainly on cyanate toxicity at relatively low concentrations (Guilloton and Karst 1987; Kunz and Nagappan 1989). In living organisms, cyanase plays a vital role in the detoxification of cyanate and cyanide compounds (Ebbs 2004). Plant cyanase may be actively involved and thus plays an integral role in different physiological and biochemical pathways. It was observed that transcription level of a putative cyanase gene in Medicago truncatula is higher in roots compared to root nodules (Gyorgyey et al. 2000). In leaves of Suaeda aegyptica, cyanase gene transcription was found to be induced by salt stress (Askari et al. 2006). Seed germination and early seedling growth were inhibited by application of KCNO to Arabidopsis thaliana cyanase knockout mutants. However, transgenic A. thaliana plants overexpressing A. thaliana (AtCYN) or Oryza sativa (OsCYN) cyanase genes showed enhanced resistance to cyanate (Qian et al. 2011). So far, no significant research studies have been performed to test the efficacy of introducing prokaryotic cyanase into plants for cyanate detoxification purposes. In the current study, we have therefore cloned and transferred the activity of the cyanobacterial cyanase gene into A. thaliana plants. Transgenic plants showed enhanced tolerance to cyanate (KCNO) supplemented either in growth medium (up to 1.2 mM) or applied via foliar spray (2.5 mM). The study was extended to evaluate the biochemical response of transgenic plants under cyanate stress in vivo.
Materials and methods
Plasmid constructs
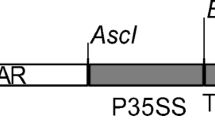
For bacterial expression, cyanase gene (CYN) coding sequence (gi16329170, slr0899) was firstly amplified by PCR using genomic DNA isolated from the cyanobacterial strain Synechococcus elongatus PCC6803 (kindly provided by Botany Department, Faculty of Science, Helwan University, Cairo, Egypt) as template and gene-specific primers (5′-ATGGCCATGGCTGGCACTGAAATTTC-3′ and 5′-GTCACTCGAGCCATTTCT TGTAGGGTAA-3′). The primers were designed with extensions for additional Nco I and Xho I sites, respectively. Amplified PCR fragments were then ligated into the bacterial expression vector pET22b(+) (Novagen, Darmstadt, Germany) in between Nco I and Xho I sites. This results in a C-terminal translational fusion with six histidines (His-tag). The structure of CYN bacterial expression cassette (pET-CYN) and CYN-PCR products is shown in Fig. 1a, c, respectively. For plant expression, the CYN gene coding sequence with the C-terminal His-tag was then PCR amplified using pET-CYN plasmid DNA as template, the same forward primer with Nco I extension (5′-ATGGCCATGGCTGGCACTGAAATTTC-3′), and a reverse primer with Xba I extension (5′-ATGTTCTAGATCAGTGGTGGTGGTGGTG-3′). The amplified CYN-His PCR product was then ligated into the binary plant expression vector pTRAK, a derivative of pPAM (gi13508478). The CYN-His gene expression cassette was flanked by the scaffold attachment region (SAR) of the tobacco RB7 gene (gi3522871), the 3′-UTR of CaMV 35S (pA35S), and 5′-UTR of the Cab22L tobacco leader peptide (TL). The nptII cassette of pPCV002 (Koncz and Schell 1986) was used for transgenic plant selection on kanamycin. Transcription of CYN gene was controlled by CaMV-35S promoter (Reichel et al. 1996). The detailed structure of this binary plant expression vector was previously described (Kebeish et al. 2015). The structure of plant CYN expression cassette is also shown in Fig. 1b.
Schematic representations of Synechococcus elongatus PCC6803 cyanase (CYN) gene expression cassettes and CYN gene PCR amplification products. The bacterial expression vector pET22b(+) with T7-P/T7-T promoter/termination signal of the bacterial T7 phage was used for the expression of cyanase (CYN, gi16329170, E.C.4.2.1.104) in Esherichia coli bacteria BL21 (DE3) (Novagen, Darmstadt, Germany) (a). The binary plant expression vector pTRA-K (gi13508478) (b), with a constitutive CaM promoter (p35SS), 5′-UTR of the Cab22L tobacco leader peptide (TL), and 3′-UTR of CaMV 35S (pA35S). CYN was amplified by PCR using S. elongatus PCC6803 genomic DNA as template and gene-specific primers containing extensions for Nco I and Xho I sites (for bacterial expression) and primers with Nco I and Xba I extensions for plant expression. One band in the expected size range (453 bp) appears after a 1% (w/v) agarose gel electrophoresis in 1× TAE for 45 min at 100 V (c). Amplified PCR products of CYN gene (lanes 1–4). PCR control without template (-ve)
Plant transformation and growth conditions
Wild-type A. thaliana ecotype Columbia (Col-0) was transformed with the respective constructs (i.e., pTRAK and pTRAK-CYN) following the Agrobacterium tumefaciens (GV3101)-mediated floral dip transformation protocol as described by Clough and Bent (1998). Plants used for physiological, biochemical, and cyanate stress experiments were grown at short-day growth conditions (8-h light and 16-h dark) in growth chambers at 22–25 °C with a light intensity of 100 μmol m−2 s−1.
SDS-PAGE and western blot analyses
Total proteins were extracted from A. thaliana plant leaves by grinding 200 mg fresh leaf tissues in liquid nitrogen. One hundred microliters of 4× Laemmli sample buffer (Bio-Rad, Germany) were added. Samples were then boiled for 5 min and centrifuged for 10 min at 14,000×g. The presence of CYN protein in plant leaf extracts was tested on 12% SDS-PAGE followed by western blot analysis using anti-His-HRP conjugate antibody (Qiagen, Hilden, Germany) after protein transfer onto nitrocellulose membrane.
Real-time RT-PCR
Quantitative real-time RT-PCR was performed as described previously (Kebeish et al. 2015). RNA was extracted from Arabidopsis leaves based on the 1-bromo-3-chlorpropane (BCP) protocol (Chomczynski and Mackey 1995). Synthesis of first-strand complementary DNA (cDNA) was performed as described by Niessen et al. (2007). qRT-PCR was performed using an ABI PRISM1 7300 Sequence Detection System (Applied Biosystems, Darmstadt, Germany) following the manufacturer’s instructions. PCR amplifications were carried out using SYBR Green reagents (SYBR1 GreenERTM qPCR SuperMixes; Karlsruhe, Germany), while the primers were purchased from Intron Biotechnology Inc. (Kyungki-Do, South Korea). For the detection of CYN transcripts, primers 5′-GGGAATCACGTTTGCTGATTT-3′ and 5′-AAGTTTCTCCGCCTCATC AA-3′ were used. For ACTIN2 transcripts, primers 5′-GGTAACATTGTGCTCAGTGGTGG-3′ and 5′-GGTGCAACGACCTTAATCTTCAT-3′ were used. The used primer concentration in the reaction mixture was 200 nM. Amplification program used for both CYN and ACTIN2 was 10 min of initial denaturation at 95 °C, followed by 40 cycles each of 15-s denaturation at 95 °C and 1 min of combined annealing and extension at 60 °C.
Growth assay conditions
Two growth assay systems were used for cyanate stress. Regarding the determination of root growth and plant rosette diameter in seedlings under cyanate stress (plate assay), measurements were performed using A. thaliana plants that were grown for 4 weeks on vertically placed Murachige and Skoog (MS) agar plates supplemented with different concentrations of potassium cyanate (0.0, 0.1, 0.2, 0.4, 0.6, 0.8, 1.0, and 1.2 mM). For determination of rosette diameter, fresh weight, and dry weight (foliar spray assay), plants were grown in soil for 6 weeks. Foliar plant parts were then sprayed with 2.5 mM potassium cyanate (foliar spray) for 2 weeks in 3-day intervals.
Enzymatic assays
Recombinant protein was expressed in the bacterial strain BL21 (DE3) (Novagen, Darmstadt, Germany) as described by Kebeish et al. (2016). The empty vector pET22b(+) was served as negative control. After induction of protein expression by 1 mM IPTG for 1.5 h at 37 °C, cells were washed once in 10 mM potassium phosphate (pH 7.5) and resuspended in the same buffer (15–20% (v/v) cell suspension). Cells were then lysed in bacterial lysis buffer (20 mM Tris-HCl pH 7.5, 300 mM NaCl, 10% glycerol, 5 mM DTT, 0.1% Triton X-1005 , mg ml−1 lysozyme, and 2 U DNAse I) for 1 h on ice. The extract was then centrifuged for 30 min at 30,000×g at 4 °C.
For in vitro cyanase assay from plant leaves, samples were taken from6 -week-old short-day grown plants and harvested 4 h after illumination. Frozen leaf material was extracted in a medium containing Hepes-KOH (pH 7.5), 10 mM MgCl2, 5 mM DTT, 1 mM EDTA, 20% glycerol, and 2% PVP. A Bradford protein assay was applied to measure the total protein concentration in the isolated protein samples (Bradford 1976). Cyanase activity in bacterial as well as plant extracts was determined by measuring the ammonia released in the reaction medium as described by Anderson (1980). The standard assay mixture was composed of 50 mM potassium phosphate buffer (pH 7.7), 3 mM sodium bicarbonate, and 2 mM potassium cyanate. The reaction was initiated by the addition of protein extracts and terminated by the addition of equal volumes of Nessler’s reagent after 1–10 min at 27 °C. The amount of ammonia released (micromole) was measured by monitoring the absorbance at 420 nm within 10 min after adding Nessler’s reagent based on a standard curve that was prepared using dilution series of ammonium chloride.
For the determination of antioxidant enzyme activities, 100 mg leaves were harvested from 6 to 7-week-old A. thaliana plant sprayed with 2.5 mM KCNO for 3× in 3-day intervals. Four hours after the third treatment, plant leaves were collected, ground in liquid nitrogen into fine powder, and homogenized in cold phosphate buffer (50 mM, pH 7.5). The extracts were centrifuged at 30,000×g for 20 min at 4 °C. Protein extracts in the supernatants were used for the determination of catalase, peroxidase, and superoxide dismutase activities as described by Xu et al. (2008).
Ammonia release assay, pigment contents, and total carbohydrate measurements
Ammonia release assay from detached leaves of transgenic A. thaliana plants with or without KCNO was carried out following the method of Rasco-Gaunt et al. (1999) with the modifications previously described by us (Kebeish et al. 2015). Three young Arabidopsis leaves were harvested and placed in a 1.5-ml reaction tube containing 1 ml of incubation medium (50 mM potassium phosphate pH 5.8, 2% sucrose, 0.1% Tween 20, 0.1 mg l−1 2,4-dichlorophenoxy acetic acid, 25 mg l−1 phosphinotricin, with or without 2 mM KCNO). Two reaction tubes containing only incubation medium were used as a blank. The tested samples were incubated under 100 μE light intensity for 16 h at 26 °C. One hundred microliters from each sample were transferred to a new 1.5-ml reaction tube containing 0.5 ml of reagent-I (0.21 M sodium salicylate, 0.085 M trisodium citrate, 25 g l−1 sodium tartrate, 0.4 mM sodium nitroprusside). Samples were mixed, and 0.5 ml of reagent-II (0.75 M NaOH, 2.3 mM sodium dichloroisocyanurate) was added. Mixtures were incubated in the dark at 37 °C and then room temperature for 15 min. The amount of ammonium ions in the tested samples was quantified by measuring the extinction at 655 nm compared to ammonium standards.
For determination of total soluble sugars, 100 mg leaves were harvested before and after 4 h of 2.5 mM KCNO application by foliar spray. Soluble sugars were extracted using 80% (v/v) boiling ethanol. The contents of soluble sugars in ethanolic extracts were then determined colorimetrically as described by Dubois et al. (1956) based on glucose standards.
Chlorophyll and carotenoid pigments were extracted from 6-week-old plants after 3× foliar spray application of 2.5 mM KCNO in 3-day intervals. Plant leaves were collected 24 h after the last application. Chlorophyll contents were measured as described by Nybom (1955). One hundred milligram leaves were harvested in a 2-ml reaction tube and immediately ground in liquid nitrogen followed by addition of 1 ml 80% acetone. Mixtures were vigorously vortexed and centrifuged at 30,000×g for 10 min. Extinctions at 663, 645, and 440 nm were measured, and pigment concentrations were calculated.
Statistical analysis
The data were represented as mean ± standard error (SE) of at least three independent experiments. Significance was determined according to Student’s t test using Excel software (Microsoft Corporation, USA). Two-sided tests were performed for homoscedastic matrices.
Results and discussion
Generation and establishment of transgenic lines
The aim of the current study was to test whether overexpression of cyanase (CYN, from S. elongatus PCC6803, gi16329170) could enhance cyanate resistance in Arabidopsis plants and whether the endogenously released products of cyanate degradation (i.e., NH3 and CO2) could be further involved in the basic plant metabolism and support plant growth or not. Clear differences in cyanase-mediated cyanate metabolism among species have been documented. In bacteria, cyanase has an important role in cyanate detoxification and cyanate-dependent production of NH3 as an alternative source for nitrogen (Anderson et al. 1990; Kunz and Nagappan 1989). However, in photosynthetic cyanobacteria, the products of cyanase activity, NH3, and CO2 are supposed to be utilized as nitrogen and carbon source (Espie et al. 2007). Based on sequence similarity, cyanase (cynS) was identified in Synechocystis sp. strain PCC6803, S. elongatus PCC7942 (Harano et al. 1997), and freshwater Synechococcus sp. strain PCC6301 (Espie et al. 2007). Cyanase gene has also been characterized and expressed in Arabidopsis (Aichi et al. 1998; Qian et al. 2011). Moreover, cyanase gene from S. elongatus PCC6803 was previously studied and well characterized (Harano et al. 1997). In the current study, we have decided to use the well-characterized cyanobacterial gene (CYN) for plant transformation in order to compare the activities of prokaryotic and eukaryotic cyanases in promoting cyanate resistance in planta. Wild-type Arabidopsis plants were transformed with the plant expression construct, pTRAK-CYN by A. tumefaciens-mediated floral dip transformation (Clough and Bent 1998). Transgenic lines were selected based on kanamycin resistance. Three independent CYN transgenic lines were generated. Accumulation of CYN gene mRNA transcripts was initially tested by quantitative RT-PCR in CYN transgenic lines (Fig. 2c). All the generated CYN lines show high expression levels of CYN gene. However, transgenic line, CYN-3, shows relatively higher expression level compared to CYN-1 and CYN-2 lines under the assay conditions. The expression of CYN in transgenic lines was further confirmed by western blot. Total protein extracts isolated from wild type and three CYN transgenic A. thaliana plants were subjected to 12% SDS-PAGE (Fig. 2a). The presence of CYN protein in plant extracts was then tested by western blot analysis using anti-His-HRP conjugate antibody (Qiagen, Hilden, Germany) after protein transfer onto nitrocellulose membrane. A specific protein band (∼16.5 kDa) corresponding to CYN protein was observed on the western blot membrane (Fig. 2b). The active E. coli cyanase is a homodecamer of five dimers comprising 17 kDa subunits (Walsh et al. 2000). The results of western blot analysis of the recombinant cyanobacterial cyanase confirm a size of about 16.5 kDa. Our results are in accordance with the data sheet of the cyanobacterial cyanase at BRENDA database (http://www.brenda-enzymes.org). The result of qRT-PCR and western blot assays (Fig. 3) indicates therefore that CYN gene is well expressed in planta.
SDS-PAGE, western blot, and CYN mRNA transcript accumulation in transgenic A. thaliana plants expressing the cyanobacterial cyanase gene. Total plant protein extracts isolated from wild type (WT) and three independent A. thaliana plants transgenic for cyanase gene (CYN-1, CYN-2, CYN-3) were analyzed on 12% SDS-PAGE and visualized by gel staining with Coomassie brilliant blue (a). Same protein samples were analyzed by western blot after transfer onto nitrocellulose membrane using anti-His-HRP conjugate antibodies (Qiagen, Germany) (b). Page ruler plus protein marker (Thermo Scientific, USA) (lane M). c CYN mRNA transcripts relative to Actin2 transcripts in three independent Arabidopsis lines overexpressing the CYN construct were analyzed by qRT-PCR. Samples were collected from the youngest fully expanded leaf 6 h after onset of light
Analysis of cyanase activity from bacterial (a) and plant extracts (b). Total protein extracts isolated from E. coli cultures overexpressing Synechococcus elongatus PCC6803 cyanase (pET-CYN), E. coli cultures transformed with the empty bacterial expression vector (pET22b(+)), or isolated from wild type (WT) or transgenic A. thaliana plants transformed with the empty plant expression vector (pTRA-K) or pTRA-K-CYN construct (CYN-1, CYN-2, and CYN-3) were tested for cyanase activity in vitro. Each data point is based on data obtained from at least three independent experiments. Vertical bars represent standard errors
Primary cyanase enzymatic assays were performed using bacterial protein extracts isolated from BL21 (DE3) bacteria transformed either with the empty bacterial expression vector pET22b(+) or with pET-CYN construct to ensure the functionality of the recombinant cyanobacterial enzyme before proceeding to plants. Fourfold increase in cyanase activity over background was observed (Fig. 3a). After generation of CYN transgenic Arabidopsis lines, we test the activity of CYN in transgenic plant extracts. As controls, protein extracts isolated from wild-type (WT) Arabidopsis plants and plants transformed with the empty plant expression vector (pTRA-K) were used. Approximately threefold to fourfold increase in cyanase activity over background (i.e., in WT and pTRA-K) was observed for transgenic lines. CYN-2 and CYN-3 lines show relatively higher cyanase activity compared to CYN-1 line (Fig. 3b). The background activity observed for pET22b(+) bacterial extracts and for WT and pTRA-K control plants may be attributed to the activity of the endogenous cyanases in both bacteria and plants (Aichi et al. 1998; Anderson and Little 1986). The enzymatic assay results indicate that the cyanobacterial CYN is active in transgenic plants.
Ammonia release and total carbohydrate contents measured from CYN transgenic lines under cyanate stress
Because the products of cyanase activity is NH3 and CO2, NH3 release was therefore measured from leaf disks of transgenic plants that were incubated in medium supplemented with 2 mM KCNO or without (−KCNO). This assay is based on the inhibition of ammonia fixation/re-fixation in the glutamine synthetase/glutamine oxoglutarate aminotransferase (GS/GOGAT) cycle in the presence of phosphinotricin (Rasco-Gaunt et al. 1999). As controls, leaves from azygous plants that were derived from CYN transformant’s segregation and transgenic plants that have been transformed with the empty plant expression vector (pTRA-K) were used. Figure 4a shows the results of this assay. In the absence of KCNO, NH3 was released from azygous and pTRA-K leaf samples in relatively similar levels, whereas transgenic CYN lines showed relative but not significant increase in the amount of NH3 released (gray bars in Fig. 4a). By addition of KCNO (black bars in Fig. 4a), NH3 release was not significantly affected in control samples. However, a significant twofold to threefold increase in NH3 release was observed for CYN transgenic lines. This indicates that CYN is able to induce NH3 production from the exogenously applied cyanate to transgenic plant leaves and CYN enzyme is therefore functionally active in planta. The released NH3 from the exogenously applied cyanate is expected to be re-assimilated by plant primary nitrogen assimilation pathway in the GS/GOGAT cycle. However, additional energy and reducing equivalents would be required for the re-fixation process (Peterhansel et al. 2010).
Ammonia release assay, total carbohydrate contents, pigment accumulation, and antioxidant enzyme activity measured from transgenic A. thaliana plants. NH3 release (a) was measured from two detached plant leaves from 6 to 7-week-old transgenic A. thaliana plants in the absence (−KCNO) or presence (+KCNO) of 2.5 mM KCNO. Total carbohydrate contents (b) were measured from A. thaliana plants before and after foliar spray inoculation of transgenic Arabidopsis plants with 2.5 mM KCNO. Pigment accumulation (c) and antioxidant enzyme activity (d) were measured from transgenic Arabidopsis plants after foliar spray with 2 mM KCNO. Six-week-old plants were sprayed three times in 4-day intervals with 2.5 mM KCNO. Segregants from CYN plants that lost the transgene (Azygous), empty vector transformants (pTRA-K), plants transgenic for cyanase gene (CYN-1–CYN-3). Data are means from at least four independent plants for each genotype ±SE. Asterisks represent statistically significant differences compared to azygous control plants (*p < 0.05, **p < 0.01, ***p < 0.001)
The second product of cyanate degradation by the action of CYN, CO2 is expected to be re-fixed by the plant photosynthetic machinery resulting in enhanced levels of carbohydrate biosynthesis. To elucidate this hypothesis, we have measured total carbohydrate contents before and after foliar application of 2.5 mM KCNO. Before KCNO application, all test plants show relatively similar levels of total carbohydrate contents (gray bars in Fig. 4b). Application of KCNO results in a significant twofold increase in total carbohydrate contents measured from CYN transgenic plants compared to nontransgenic plants. However, carbohydrate biosynthesis was even diminished in azygous and pTRA-K plants upon KCNO application (black bars in Fig. 4b). The exact amount of CO2 release from cyanate is seemingly more difficult to be measured because of the re-fixation of CO2 in the chloroplast by the photosynthetic machinery. However, our total carbohydrate measurement data may provide evidence for the enhanced release of CO2 from cyanate in transgenic lines after foliar application of cyanate and its subsequent integration into the photosynthetic products, carbohydrates. The reduction in carbohydrate biosynthesis in control plants may be related to the negative effects of the toxic cyanate on Rubisco enzyme (Grossmann 1996). This may also be related to the low capability of the endogenous cyanase to cope with the exogenously applied cyanate (Qian et al. 2011). Exogenous cyanate was shown to induce cyanase gene transcription in microbes (Elleuche and Poggeler 2008; Luque-Almagro et al. 2008) and used as a sole carbon and nitrogen source providing energy and reductant for microbial growth (Kumar et al. 2016; Palatinszky et al. 2015). However, this phenomenon is almost absent in plants because endogenous cyanase gene transcription was observed to be only induced under salt stress (Askari et al. 2006; Qian et al. 2011). Results of NH3 release assay and total carbohydrate measurements together with the enhanced growth data observed for CYN transgenic Arabidopsis lines under cyanate stress (Figs. 5 and 6 below) indicate the incorporation of both cyanate degradation products into the basic plant metabolism in vivo as carbon and nitrogen source.
Growth assay of transgenic Arabidopsis plants and controls on MS agar plates supplemented with different concentrations of KCNO. a Representative photograph of transgenic and control Arabidopsis seedlings grown under cyanate stress. b Plant rosette diameter and c plant root length measured from transgenic and control Arabidopsis plants grown in vertical MS agar plates supplemented with different concentrations of KCNO (0–1.2 mM). Segregants from CYN plants that lost the transgene (Azygous). Plants transgenic for cyanase gene lines 2 and 3 (CYN-2 and CYN-3). Data are means from at least four independent plants for each genotype ±SE. Rosette diameter and root length values recorded for CYN-2 and CYN-3 lines are significantly different compared to azygous plants at all the applied KCNO concentrations (p < 0.001)
Growth assay of transgenic Arabidopsis plants and controls grown in soil and sprayed with 2.5 mM KCNO. Plant rosette diameter (a) before and after 2.5 mM KCNO foliar spray inoculation, total plant fresh weight (b), and total plant dry weight (c) measured from 8-week-old transgenic Arabidopsis plants and controls grown in soil after foliar spray with 2.5 mM KCNO at 3-day intervals for 2 weeks. d Representative photograph of transgenic Arabidopsis thaliana plants and control plants grown in soil after foliar spray with 2.5 mM KCNO for 2 weeks. Segregants from CYN plants that lost the transgene (Azygous), empty vector transformants (pTRA-K), plants transgenic for cyanase gene (CYN-1–CYN-3). Data are means from at least three independent plants for each genotype ±SE. Asterisks represent statistically significant differences compared to azygous control plants (**p < 0.01, ***p < 0.001)
Effect of exogenous cyanate application on total pigments and antioxidant enzyme activities in planta
Primary experiments with wild-type plants showed that cyanate application on Arabidopsis plant leaves (foliar spray) lead to a loss of pigmentation symptoms. This assay was used to test the efficacy of the cyanobacterial enzyme in ameliorating the negative effects of cyanate toxicity on plants. Chlorophyll and carotenoid contents of CYN transgenic lines were therefore measured in comparison to azygous and pTRA-K controls after three times application of 2.5 mM KCNO in 3-day intervals. The result of this assay is shown in Fig. 4c. Significant increase in all pigments was observed for CYN overexpressors compared to azygous and pTRA-K plants. This result indicates that KCNO-induced loss of pigmentation was alleviated by overexpression of CYN in planta.
To gain more information about the biochemical response of plants under cyanate stress, the work was extended to measure the antioxidant enzymes, catalase (CAT), peroxidase (POD), and superoxide dismutase (SOD), that were expected to be induced upon foliar cyanate application. The antioxidant enzyme activities were measured in extracts from leaves of CYN transgenic lines and control lines (Fig. 4d). Again, azygous and pTRA-K controls show increased levels of all the measured antioxidant enzymes. However, CYN transgenic lines showed reduced induction of these enzymes (−85% for superoxide dismutase, −75% for catalase, and −30% for peroxidase). This reduction in antioxidant enzyme activities in CYN overexpressors may be due to the efficient removal of the exogenously applied cyanate by the cyanobacterial cyanase enzyme. The biochemical assay data observed in the current study (Fig. 4) are in accordance with the results observed with transgenic Arabidopsis plants overexpressing the bacterial enzyme cyanidase (CYND) when cyanide was applied via foliar spray to CYND transgenic plants (Kebeish et al. 2015). CYND transgenic A. thaliana plants showed significant increase in pigment contents and reduced induction of CAT, POD, and SOD enzymes compared to control plants under cyanide stress.
Evaluation of transgenic plant resistance to cyanate stress (growth assays)
Two growth assays were performed to test the degree of cyanate resistance in transgenic Arabidopsis plants. In the first assay, cyanate was supplied via the root system as would be expected on contaminated soil at polluted sites. A pool of azygous and two CYN transgenic lines (CYN-2 and CYN-3) were germinated on vertical MS agar plates containing different concentrations of KCNO (0.0, 0.1, 0.2, 0.4, 0.6, 0.8, 1.0, and 1.2 mM) at short-day growth condition. Primary root length and rosette diameter were measured after 4 weeks. Primary root length and rosette diameter of azygous and CYN transgenic lines were almost identical for both genotypes (i.e., transgenic and azygous control plants) in the absence of KCNO. However, a negative relationship was observed between external KCNO concentration and primary root elongation as well as rosette diameter of azygous plants (light gray bars in Fig. 5b, c). A clear induction in plant rosette diameter was obvious for CYN-2 and CYN-3 transgenic lines at KCNO concentration of 0.1 and 0.2 mM compared to untreated controls (Fig. 5b). This indicates that low concentrations of KCNO (up to 0.2 mM) promote plant growth, whereas KCNO concentrations above 0.2 mM KCNO have negative impact on both CYN transgenic plants and azygous controls. Interestingly, root hair formation was inhibited in azygous control plants at KCNO concentrations of 0.2 mM and thereafter, whereas this effect was observed at KCNO concentration above 0.6 mM in CYN overexpressors (see Fig. 5a). It is clear that primary root length and plant rosette diameter measurements of CYN transgenic lines are significantly higher compared to azygous controls at all the used KCNO concentrations (Fig. 5b, c). Previous research studies have shown that seedlings of the model plant A. thaliana are sensitive to cyanides and show growth retardation phenotypes even at low concentration of the pollutant (Garcia et al. 2010; Mc Mahon Smith and Arteca 2000; O’Leary et al. 2014). In contrast, CYN transgenic plants survived concentrations up to 0.6 mM cyanate without any visible signs of stress. Although soil contamination is considered as the major source of cyanide and cyanate toxicity to plants (Trapp et al. 2003), our initial growth assays were based on foliar spray application because a significant amount of the exogenously applied cyanate is not expected to be metabolized in plant roots, however ends up in shoots (Yu et al. 2012). Thus, in the second growth assay, 2.5 mM cyanate was applied via plant leaves (foliar spray) to 6-week-old plants for 2 weeks in 2-day intervals. Plant rosette diameter, fresh weight (FW), and dry weight (DW) were measured as indicators for KCNO tolerance (Fig. 6). Plant rosette diameter was recorded before and after KCNO application. Before KCNO application, CYN transgenic plants and controls showed very similar growth pattern (gray bars in Fig. 6a). However, KCNO application has negative effects on growth of azygous and pTRA-K control plants (Fig. 6a, d). A significant increase in plant rosette diameter was observed for CYN transgenic lines compared to controls (black bars in Fig. 6a). About 10% increase in plant rosette diameter was observed for azygous and pTRA-K controls. In contrast, 70–90% increase in plant rosette diameter was observed for CYN transgenic lines compared to untreated plants.
Total plant fresh weight and total plant dry weight were also measured from 8-week-old plants that were sprayed with 2.5 mM KCNO (foliar spray) for 2 weeks in 2-day intervals. Again, CYN transgenic lines show significant increase in both parameters compared to azygous and pTRA-K controls under the assay condition. Significant fourfold increase in FW (Fig. 6b) and significant threefold increase in DW (Fig. 6c) were observed for CYN transgenic lines compared to controls. Thus, CYN overexpressors perform superior when KCNO was applied to plant leaves. These results indicate that CYN expression enhances cyanate tolerance and promotes growth in Arabidopsis plants.
It is useful to compare the currently observed effects of cyanobacterial CYN expression with the previously published approach for the functionality of Arabidopsis and rice cyanases (Qian et al. 2011). The authors have overexpressed the cyanase gene from A. thaliana (AtCYN) and O. sativa (OsCYN) in Arabidopsis plants. Transgenic lines overexpressing AtCYN or OsCYN exhibited resistance to KCNO stress (up to 1 mM) when cyanate was applied to Arabidopsis seedlings in MS agar plates. However, our growth assay data showed that CYN overexpressors can withstand up to 1.2 mM KCNO under the same assay conditions (Fig. 5). Moreover, application of higher concentrations of KCNO (2.5 mM) to plant leaves via foliar spray resulted in a significant induction of CYN transgenic plant growth (Fig. 6). Control plants showed very weak or stunt growth phenotype upon KCNO application. This growth assay indicates that the endogenous cyanase is insufficient to detoxify the exogenously applied cyanate. This may be due to the fact that endogenous cyanase transcription is not induced by application of exogenous cyanate to Arabidopsis plants (Qian et al. 2011). The enhanced growth of CYN transgenic plants under cyanate stress might be due to (1) the efficient removal of the exogenous biotic stress factor cyanate and (2) the re-assimilation of the cyanase-dependent cyanate degradation products, NH3 and CO2, by the primary plant nitrogen and carbon assimilation pathways as shown above (Fig. 4a, b) although additional energy and reducing equivalents would be required for the re-assimilation process (Kebeish et al. 2007; Peterhansel et al. 2010).
Cyanate in soil may come from chemical compounds containing urea or cyanide, and anthropogenic activities released additional cyanate containing wastes to the environment (Dirnhuber and Schutz 1948; Malhotra et al. 2005; Rader et al. 1995; Sancho and Bellen 2005). Full cyanate tolerance has not been achieved in the current study. Thus, an effective bioremediation system has to be developed in order to release the toxic cyanide and cyanate from the environment. We suggest that a gene stacking approach incorporating effective microbial cyanases, mitochondrial and/or plastid targeted nitrilases, and stress-eliminating enzymes (e.g., glutathione-S-transferase (GST)) in combination with augmentation of the plant endogenous cyanate detoxification pathway would introduce new research opportunities for developing an effective environmental friendly cyanate remediation system through the use of genetically engineered plants, especially if implemented in a plant species with higher natural tolerance to CNO−. It remains to be shown in further studies whether this approach is more effective when transgenic plants are allowed to grow in a cyanate-contaminated soil harboring soilborne cyanate-resistant microorganisms.
References
Aichi M, Nishida I, Omata T (1998) Molecular cloning and characterization of a cDNA encoding cyanase from Arabidopsis thaliana. Plant Cell Physiol 39:S135–S135
Akcil A, Mudder T (2003) Microbial destruction of cyanide wastes in gold mining: process review. Biotechnol Lett 25:445–450
Anderson PM (1980) Purification and properties of the inducible enzyme cyanase. Biochemistry 19:2882–2888
Anderson PM, Little RM (1986) Kinetic properties of cyanase. Biochemistry 25:1621–1626
Anderson PM, Sung Y-C, Fuchs JA (1990) The cyanase operon and cyanate metabolism. FEMS Microbiol Rev 7:247–252
Askari H, Edqvist J, Hajheidari M, Kafi M, Salekdeh GH (2006) Effects of salinity levels on proteome of Suaeda aegyptiaca leaves. Proteomics 6:2542–2554
Blank CE, Hinman NW (2016) Cyanobacterial and algal growth on chitin as a source of nitrogen; ecological, evolutionary, and biotechnological implications. Algal Res 15:152–163
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Bushey JT, Ebbs SD, Dzombak DA (2006) Development of a plant uptake model for cyanide. International Journal of Phytoremediation 8:25–43
Butryn A, Stoehr G, Linke-Winnebeck C, Hopfner K-P (2015) Serendipitous crystallization and structure determination of cyanase (CynS) from Serratia proteamaculans. Acta Crystallographica Section F: Structural Biology Communications 71:471–476. doi:10.1107/S2053230X15004902
Chomczynski P, Mackey K (1995) Substitution of chloroform by bromo-chloropropane in the single-step method of RNA isolation. Anal Biochem 225:163–164
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacteriumâ-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743
Dirnhuber P, Schutz F (1948) The isomeric transformation of urea into ammonium cyanate in aqueous solutions. Biochem J 42:628
Dubois M, Gilles KA, Hamilton JK, Rebers P, Smith F (1956) Colorimetric method for determination of sugars and related substances. Anal Chem 28:350–356
Ebbs S (2004) Biological degradation of cyanide compounds. Curr Opin Biotechnol 15:231–236
Elleuche S, Poggeler S (2008) A cyanase is transcriptionally regulated by arginine and involved in cyanate decomposition in Sordaria macrospora. Fungal Genet Biol 45:1458–1469
Espie GS, Jalali F, Tong T, Zacal NJ, So AK-C (2007) Involvement of the cynABDS operon and the CO2-concentrating mechanism in the light-dependent transport and metabolism of cyanate by cyanobacteria. J Bacteriol 189:1013–1024
Garcia I, Castellano JM, Vioque B, Solano R, Gotor C, Romero LC (2010) Mitochondrial β-cyanoalanine synthase is essential for root hair formation in Arabidopsis thaliana. Plant Cell 22:3268–3279
Garcia-Fernandez JM, Diez J (2004) Adaptive mechanisms of nitrogen and carbon assimilatory pathways in the marine cyanobacteria Prochlorococcus. Res Microbiol 155:795–802
Grossmann K (1996) A role for cyanide, derived from ethylene biosynthesis, in the development of stress symptoms. Physiol Plant 97:772–775
Guilloton M, Karst F (1987) Isolation and characterization of Escherichia coli mutants lacking inducible cyanase. Microbiology 133:645–653
Gyorgyey J, Dl V, Jimenez-Zurdo JI, Charon C, Troussard L, Kondorosi A, Kondorosi E (2000) Analysis of Medicago truncatula nodule expressed sequence tags. Mol Plant-Microbe Interact 13:62–71
Hamel J (2011) A review of acute cyanide poisoning with a treatment update. Crit Care Nurse 31:72–82
Harano Y, Suzuki I, Maeda S-I, Kaneko T, Tabata S, Omata T (1997) Identification and nitrogen regulation of the cyanase gene from the cyanobacteria Synechocystis sp. strain PCC 6803 and Synechococcus sp. strain PCC 7942. J Bacteriol 179:5744–5750
Johnson W, Anderson P (1987) Bicarbonate is a recycling substrate for cyanase. J Biol Chem 262:9021–9025
Kamennaya NA, Post AF (2010) Characterization of cyanate metabolism in marine Synechococcus and Prochlorococcus spp. Appl Environ Microbiol 77:291–301. doi:10.1128/AEM.01272-10
Kamennaya NA, Chernihovsky M, Post AF (2008) The cyanate utilization capacity of marine unicellular Cyanobacteria. Limnol Oceanogr 53:2485
Kebeish R et al (2007) Chloroplastic photorespiratory bypass increases photosynthesis and biomass production in Arabidopsis thaliana. Nat Biotechnol 25:593–599
Kebeish R, Aboelmy M, El-Naggar A, El-Ayouty Y, Peterhansel C (2015) Simultaneous overexpression of cyanidase and formate dehydrogenase in Arabidopsis thaliana chloroplasts enhanced cyanide metabolism and cyanide tolerance. Environ Exp Bot 110:19–26. doi:10.1016/j.envexpbot.2014.09.004
Kebeish R, El-Sayed A, Fahmy H, Abdel-Ghany A (2016) Molecular cloning, biochemical characterization, and antitumor properties of a novel L-asparaginase from Synechococcus elongatus PCC6803. Biochem Mosc 81:1173–1181. doi:10.1134/S000629791610014X
Koncz C, Schell J (1986) The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Mol Gen Genet MGG 204:383–396
Koshiishi I, Mamura Y, Imanari T (1997) Cyanate causes depletion of ascorbate in organisms. Biochimica et Biophysica Acta (BBA)-General Subjects 1336:566–574
Kraus LM, Kraus AP (2001) Carbamoylation of amino acids and proteins in uremia. Kidney Int 59:S102–S107
Kumar R, Saha S, Dhaka S, Kurade MB, Kang CU, Baek SH, Jeon B-H (2016) Remediation of cyanide-contaminated environments through microbes and plants: a review of current knowledge and future perspectives Geosystem Engineering:1–13
Kunz DA, Nagappan O (1989) Cyanase-mediated utilization of cyanate in Pseudomonas fluorescens NCIB 11764. Appl Environ Microbiol 55:256–258
Luque-Almagro VM et al (2008) Characterization of the Pseudomonas pseudoalcaligenes CECT5344 cyanase, an enzyme that is not essential for cyanide assimilation. Appl Environ Microbiol 74:6280–6288
Malhotra S, Pandit M, Kapoor J, Tyagi D (2005) Photo-oxidation of cyanide in aqueous solution by the UV/H2O2 process. J Chem Technol Biotechnol 80:13–19
Mc Mahon Smith J, Arteca RN (2000) Molecular control of ethylene production by cyanide in Arabidopsis thaliana. Physiol Plant 109:180–187
Mekuto L, Ntwampe S, Akcil A (2016) An integrated biological approach for treatment of cyanidation wastewater Science of The Total Environment
Miller AG, Espie GS (1994) Photosynthetic metabolism of cyanate by the cyanobacterium Synechococcus UTEX 625. Arch Microbiol 162:151–157
Niessen M, Thiruveedhi K, Rosenkranz R, Kebeish R, Hirsch H-J, Kreuzaler F, Peterhãnsel C (2007) Mitochondrial glycolate oxidation contributes to photorespiration in higher plants. J Exp Bot 58:2709–2715
Nowakowska M, Sterzel M, Szczubiałka K (2006) Photosensitized oxidation of cyanide in aqueous solutions of photoactive modified hydroxyethylcellulose. J Polym Environ 14:59–64
Nybom N (1955) The pigment characteristics of chlorophyll mutations in barley. Hereditas 41:483–498
O’Leary B, Preston GM, Sweetlove LJ (2014) Increased β-cyanoalanine nitrilase activity improves cyanide tolerance and assimilation in Arabidopsis. Mol Plant 7:231–243
Palatinszky M et al (2015) Cyanate as an energy source for nitrifiers. Nature 524:105–108
Patil Y, Paknikar K (2000) Development of a process for biodetoxification of metal cyanides from waste waters. Process Biochem 35:1139–1151
Peterhansel C, Horst I, Niessen M, Blume C, Kebeish R, Kürkcüoglu S, Kreuzaler F (2010) Photorespiration. Arabidopsis Book 8:e0123
Qian D, Jiang L, Lu L, Wei C, Li Y (2011) Biochemical and structural properties of cyanases from Arabidopsis thaliana and Oryza sativa. PLoS One 6:e18300
Rader WS, Solujic L, Milosavljevic EB, Hendrix JL, Nelson JH (1995) Photocatalytic detoxification of cyanide and metal cyano-species from precious-metal mill effluents. Environ Pollut 90:331–334
Rasco-Gaunt S, Riley A, Lazzeri P, Barcelo P (1999) A facile method for screening for phosphinothricin (PPT)-resistant transgenic wheats. Mol Breed 5:255–262
Reichel C et al (1996) Enhanced green fluorescence by the expression of an Aequorea victoria green fluorescent protein mutant in mono-and dicotyledonous plant cells. Proc Natl Acad Sci 93:5888–5893
Sancho J, Bellen F (2005) Developments of an alternative technology to remove cyanide from mining wastewater. In: I9th International Mine Water Congress, pp 413–417
Sancho J, Bellon F (2005) Developments of an alternative technology to remove cyanide from mining wastewater. In: I9th International Mine Water Congress, pp 413–417
Srivastava AC, Muni RRD (2010) Phytoremediation of cyanide. In: Plant Adaptation and Phytoremediation. Springer, pp 399–426
Sung Y-C, Parsell D, Anderson P, Fuchs J (1987) Identification, mapping, and cloning of the gene encoding cyanase in Escherichia coli K-12. J Bacteriol 169:2639–2642
Taebi A, Jeirani K, Mirlohi A, Zadeh Bafghi A (2008) Phytoremediation of cyanide-polluted soils by non-woody plants. JWSS-Isfahan University of Technology 11:515–523
Taussig A (1960) The synthesis of the induced enzyme, cyanase, in E. coli. Biochim Biophys Acta 44:510–519
Trapp S, Larsen M, Pirandello A, Danquah-Boakye J (2003) Feasibility of cyanide elimination using plants ejmp & ep. Eur J Miner Process Environ Prot 3:128–137
Voigt K, Sharma CM, Mitschke J, Lambrecht SJ, Voss B, Hess WR, Steglich C (2014) Comparative transcriptomics of two environmentally relevant cyanobacteria reveals unexpected transcriptome diversity. The ISME journal 8:2056–2068
Walsh MA, Otwinowski Z, Perrakis A, Anderson PM, Joachimiak A (2000) Structure of cyanase reveals that a novel dimeric and decameric arrangement of subunits is required for formation of the enzyme active site. Structure 8:505–514
Xu P, Zou J, Meng Q, Zou J, Jiang W, Liu D (2008) Effects of Cd 2+ on seedling growth of garlic (Allium sativum L.) and selected physiological and biochemical characters. Bioresour Technol 99:6372–6378
Yu X-Z, Zhou P-H, Yang Y-M (2006) The potential for phytoremediation of iron cyanide complex by willows. Ecotoxicology 15:461–467
Yu X-Z, Lu P-C, Yu Z (2012) On the role of β-cyanoalanine synthase (CAS) in metabolism of free cyanide and ferri-cyanide by rice seedlings. Ecotoxicology 21:548–556
Acknowledgments
This work was supported by the Deanship of Scientific Research, Taibah University, KSA (project no. 6925) and partially supported by the Egyptian Science and Technology Development Fund (STDF). We are thankful to Dr. Ahmed Saleh and Dr. Ahmed Khalil (Biology Department, Faculty of Science-Yanbu, Taibah University) for the valuable discussion and correction of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible editor: Yi-ping Chen
Rights and permissions
About this article
Cite this article
Kebeish, R., Al-Zoubi, O. Expression of the cyanobacterial enzyme cyanase increases cyanate metabolism and cyanate tolerance in Arabidopsis . Environ Sci Pollut Res 24, 11825–11835 (2017). https://doi.org/10.1007/s11356-017-8866-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11356-017-8866-z