Abstract
Newcastle disease (ND) is one of the most serious diseases affecting poultry worldwide. In 2022, we studied two strains of Newcastle disease virus (NDV) from pigeons and magpies identified by PCR and propagated in SPF chicken embryos. The whole genome of the virus was then expanded and its biological characteristics were studied. The results showed that NDV was isolated from pigeons and magpies. Virus present in the allantoic fluid could agglutinate red blood cells and could not be neutralized by serum positive for avian influenza. Sequencing showed that the gene length of the two isolates was 15,191 bp, had high homology and was located in the same branch of the phylogenetic tree, both belonging to genotype VI.1.1. The sequence of 112–117 amino acids in the F gene sequence was 112R-R-Q-K-R-F117, which constituted virulent strain characteristics. The HN gene contained 577 amino acids, which is also consistent with the characteristics of a virulent strain. The results from the study of biological characteristics revealed that the virulence of SX/TY/Pi01/22 was slightly stronger. There were only four different bases in the complete sequence of the two strains. Comprehensive analysis revealed that the G at 11,847 site of the SX/TY/Ma01/22 strain may change to T, leading to translation of amino acids from R to S, thereby weakening viral virulence. Therefore, NDV was transmitted from pigeons to magpies, indicating that the pathogen could be transmitted between poultry and wild birds.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Newcastle disease (ND) is an acute, febrile, and infectious disease caused by Newcastle disease virus (NDV) infection. The World Organization for Animal Health (WOAH) has required ND must be reported [1]. The Ministry of Agriculture and Rural Affairs of the People’s Republic of China has listed ND as a class two animal epidemic disease, which is extremely harmful to the poultry industry [2].
NDV is an avulavirus belonging to the family Paramyxoviridae. and genus Orthoavulavirus. NDV can be divided into two large groups: class I and class II. Class I is a single genotype (I.1.1, I.2.and I.1.2.1) [3] and Class II contains at least 20 genotypes, of which the main prevalent genotypes are V, VI, and VII. The main prevalent genotypes in chickens are VII, particularly VII.1.1 [4]. However, most of the NDV genotypes from pigeons are genotype VI, especially the subtype VI.1.1, which is termed pigeon paramyxovirus type 1 (PPMV-1) [5]. Virulence has been correlated with the amino acid sequence 112–117 in the fusion protein (F). The sequence of the virulent strain is 112K/R-R-Q-R/K-R-F117, whereas the sequence of the attenuated strain is usually 112E-R-Q-G/E-R-L117 [6]. It has also been reported that the virulence of pigeon NDV was also correlated with the NP, P, and L proteins [7].
NDV is known to infect over 250 avian species [8], due to a particularly high infection and carrying rate of wild birds. Although magpies are also susceptible [9,10,11], there have been few research reports on NDV infection in magpies worldwide, and the susceptible NDV genotype in magpies has not been previously reported.
Materials and methods
Sample collection
The disease materials of this study were obtained from carrier pigeons raised in a carrier pigeon farm in Taiyuan and a dead magpie near the road. A total of 134 carrier pigeons were raised in the farm, of which seven sick pigeons were found, but none were dead. Seven samples of throat and cloacal secretions of infected pigeons were collected with sterile flocking swabs [Hanwei (Changzhou) Biotechnology Co., Ltd., China] and placed into a collection tube containing non-inactivated preservation solution (main components: Hank’s buffer, phenol red, and BSA). The collection tube was placed in an incubator containing ice bags. The dead magpie on the roadside was placed into a sterile sealed bag and a heat preservation box with ice bag was used to bring it back to the laboratory. The magpie was dissected in the laboratory, and the heart, liver, spleen, lung, and brain tissues were obtained, placed into aseptic sealed bags, and stored in a − 20 °C freezer until further use.
Test animals
The 1-day-old and 42-day-old chicks were incubated by the purchased SPF eggs (Beijing Boehringer Ingelheim Vital Biotechnology Co., Ltd., China), layer chickens, and were reared in the BSL2 laboratory of the College of Veterinary Medicine, Shanxi Agricultural University. Feeding chicken pellet feed was obtained from Shanxi Shichuang Feed Co., Ltd.
PCR identification
Viral RNA extraction and amplification were performed according to the manufacturer’s instructions of the Newcastle disease virus universal RT-PCR detection kit (Beijing Anheal Laboratories Co., Ltd., China) for NDV.
Virus isolation and culture
The diseased tissue was repeatedly frozen and thawed three times. After grinding, PBS solution was added at a ratio of 1:5, after which penicillin and streptomycin [5000 U (µg)/mL] was added to the mixture. After reacting at 37 °C for 1 h and centrifuging at 598×g for 30 min, the supernatant was extracted using a sterile syringe and filtered through a 0.22-µm membrane filter. The filtrate was stored in a freezer at − 20 °C for further use. The preservation solution of the pharynx cloacal swab was directly added with penicillin and streptomycin for the reaction, filtered through a 0.22-µm membrane filter, and stored in a refrigerator at − 20 °C.
Next, 100-µL filtrate was inoculated in Martin Broth and Anaerobic Broth Liver Soup, respectively. We dipped the filtrate with the inoculation ring and inoculated it on a blood agar plate and Sabouraud Dextrose Agar bevel by scribing. The results were observed after culturing for 24 h at 37 °C.
Ten-day-old SPF eggs were inoculated with 0.2-mL supernatant filtrate into the allantoic cavity of the chicken embryo. Another two eggs were inoculated with sterilized saline as a negative control. The eggs were incubated at 37.6 °C and 60% humidity. The eggs were turned every 4 h and candled twice a day. Chicken embryos that died within 24 h were discarded. Allantoic fluid from dead embryos after 24 h and living chicken embryos after 120 h were collected aseptically As an antigen, the blood of the self-raised healthy rooster were collected, the red blood cells were separated, and the red blood cell solution was prepared with a 1% concentration with PBS solution for hemagglutinin (HA) and Avian influenza-positive serum (Harbin Weike Biotechnology Development Company, Harbin, China) was used for antibodies to achieve hemagglutination inhibition (HI) [12]. If the allantoic fluid could agglutinate red blood cells and could not be neutralized by avian influenza antibodies, it was judged as NDV positive. The allantoic fluid was frozen and stored in the freezer at − 20 °C for future used, and the chicken embryo was dissected and observed.
Genome amplification and sequencing
A total of 12 pairs of primers were designed to amplify the whole-gene sequence of NDV by referring to the full-genome sequence of pigeon NDV published on GenBank. The sequences are shown in Table 1.
The total viral RNA was extracted according to the manufacturer instructions of the NDV RNA extraction kit (Beijing Anheal Laboratories Co., Ltd., China).
According to the One-step RT-PCR kit [Takara Biomedical Technology (Beijing) Co., Ltd] instructions, the reaction system was as follows: PrimeScript 1 Step Enzyme Mix 2 μL, 2 × 1 Step Buffer 25 μL, forward and reverse primer 1 μL each, Template RNA 1 μL, and RNase-Free dH2O 20 μL. The reaction conditions were as follows: reverse transcription at 50 °C for 30 min, predenaturation at 94 °C for 2 min, denaturation at 94 °C for 30 s, annealing at 55 °C for 30 s, extension at 72 °C for 2 min, 35 cycles from the third step to the fifth step, and the reaction ended after extension at 72 °C for 10 min. A 5-μL sample of the PCR product was electrophoresed on agarose gel at a concentration of 1%. The PCR product was recovered and purified using a Gum recovery kit (Omega Biotech, Norcross, GA, USA). The purified product was connected to the cloning vector using a PEASY-Blunt cloning vector kit (Beijing TransGen Biotech Co., Ltd, Beijing, China), then transformed into Trans109-competent cells (Beijing TransGen Biotech Co., Ltd., China), and cultured in large numbers. The plasmids were extracted for PCR identification and the recombinant plasmid sent for sequencing [Bgi Tech Solutions (Beijing Liuhe, China) Co., Ltd. and Sangon Biotech (Shanghai) Co., Ltd.].
Sequence analysis
We used DNAMAN and DNAStar software to splice and compare the sequencing results. MEGA software was used to construct the phylogenetic tree.
F gene virulence locus and HN gene study
According to the amino acid sequence encoded by the F gene, the 112–117 amino acid sequence was identified to judge its virulence. Amino acids encoded by the HN gene were counted to judge the virulence.
Biological characteristics
The National Standards of the People’s Republic of China [13] was used to measure the three indexes of the mean death time (MDT), intracerebral pathogenicity index (ICPI), and intravenous pathogenicity index (IVPI) to determine pathogen virulence.
MDT: The virus allantoic fluid was diluted 10 times in sterile PBS buffer from 100 to 10−10. The 10-day-old SPF chicken embryos were inoculated with virus diluent at a dilution of 10−6–10−10 via the allantoic cavity. Each dilution was inoculated into 5 chicken embryos, and each embryo was inoculated with 0.1 mL. The death of chicken embryos was observed daily and the time of death was recorded until 96 h. The MDT value of NDV was the average time of total death of inoculated chicken embryos under the highest dilution multiple of the virus.
ICPI: The virus allantoic fluid was diluted with sterile PBS buffer, and 1-day-old SPF chickens were infected through intracerebral inoculation. Each chicken was inoculated with 50-μL virus fluid. A total of 10 SPF chickens were inoculated, and the chickens were permitted to eat and drink freely and were observed once daily at a fixed time for a total of 8 days. At the same time, the clinical manifestations of each chicken were recorded with reference to the scoring standard. The score was 0 for normal chickens, 1 for lameness, paralysis, and other clinical symptoms, and 2 for dead chickens. The ICPI value of NDV was the average of the cumulative total score of chickens divided by the cumulative total number of normal, diseased, and dead chickens.
IVPI: Fresh allantoic fluid infected with NDV with an HA titer higher than 24 was collected and diluted with sterile isotonic saline at a ratio of 1:10. SPF chickens aged 6 weeks were intravenously inoculated into 10 chickens and each chicken was inoculated with 0.1 mL. The chickens were observed once daily for 10 consecutive days and scored each time. Normal chickens were recorded as 0, sick chickens as 1, paralyzed chickens or other neurological symptoms as 2, and dead chickens as 3 (each dead chicken was still recorded as 3 in the daily observation after death). The IVPI value of NDV was the average of the cumulative total score of the chickens divided by the cumulative total number of normal, diseased, and dead chickens.
Results
RT-PCR identification results
The results showed that there were bands of the same size as the standard positive control in lane 1 and lane 2, which could confirm the presence of Newcastle disease virus in these disease, as shown in Fig. 1. The viruses were named China/Pigeon/Shanxi (Taiyuan)/Pi01/2022 (SX/TY/Pi01/22) and China/Magpie/ Shanxi (Taiyuan)/Ma01/2022 (SX/TY/ Ma01/22).
Results of virus isolation and culture
The allantoic fluid of dead chicken embryos was collected for hemagglutination and hemagglutination inhibition testing. It could be observed that it can agglutinate 1% chicken erythrocytes with an agglutination value of 6log2–7log2, and it could not be neutralized by avian influenza-positive serum, indicating that the isolated virus was NDV. The SPF chicken embryos inoculated with the supernatant filtrate at an age of 10 days died within 48–72 h. Bleeding was observed over the entire body, especially in the head and neck shown in Fig. 2.
Genome amplification results
According to the primer sequence in Table 1, 12 sequence fragments of SX/TY/ PI01/22 and SX/TY/ MA01/22 were amplified, and the fragment size was consistent with the design. The results are presented in Figs. 3 and 4.
Sequence analysis results
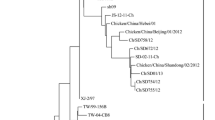
The splicing of the sequencing results showed that the whole-genome length of SX/TY/Pi01/22 and SX/TY/Ma01/22 was 15,191 bp. The similarity between the two was 99.84%, and four different loci, 1333, 4601, 10,237, and 11,847, were identified, respectively. Both viruses were located on the same branch on the phylogenetic tree and had the closest relationship. Moreover, both strains belong to the genotype of VI.1.1 and were in the same large branch with the pigeon NDV strains, MW147626.1 and MW147625.1 isolated in Beijing and Tianjin in 2017 and 2018, and had a close genetic relationship. The results are presented in Fig. 5.
Table 2 shows that the two isolated NDV strains only differed in four loci compared with other isolates at home and abroad, with the mutation of bases at 1333 bp and 11,847 bp being relatively special. The base of SX/TY/Pi01/22 was T at position 1333 and all others were C. The base of SX/TY/Ma01/22 was T at position 11,847 and all others were G. However, the bases at the positions of 4601 bp and 10,237 bp were irregular.
Table 3 shows that the amino acids of the four base difference sites differed, with glutamine appearing at position 445 and arginine changing to serine at position 3949.
Virulence characteristics of the F and HN genes
By analyzing the F gene sequence of NDV, the two viruses were found to have the same base sequence located in the virulence site: AGGAGGCAGAAGCGCTTCATAGGTGCC and the amino acid sequence was 112R-R-Q-K-R-F117. This sequence completely conformed to the characteristics of a virulent strain. Through a sequence analysis of the HN gene, it was observed that 571 amino acids were encoded, which was consistent with the characteristics of virulent strains [5].
Identification results of biological characteristics
Table 4 shows that SX/TY/Pi01/22 belonged to the NDV moderately virulent strain, and SX/TY/Ma01/22 was a less virulent strain.
Conclusion
The results of this study showed that NDV isolated from pigeon and magpie belonged to gene VI.1.1 of the class II group. Moreover, because both strains belonged to the same branch on the developmental evolutionary tree, their homology was very high. Based on the amino acid sequence analysis of F and HN genes, they belonged to the virulent strain. Although only four base sites differed between the two strains, the determination of biological characteristics showed that SX/TY/Pi01/22 was a moderately virulent strain and SX/TY/Ma01/22 was a less virulent strain.
Discussion
ND in pigeons first occurred in Iraq in 1977 [14] and subsequently spread to all parts of the world. Pigeon-derived NDV is considered to be the main cause of the second and third ND pandemics [15, 16]. NDV was also isolated from pigeons in Hong Kong, China in 1985 and occurred continuously in China. This occurrence was mainly dominated by genotype VI.1.1 [17, 18], particularly after 2010 [19]; however, it has also been reported that strains isolated from pigeons were genotype VII [20]. Moreover, the sequence of the F protein cleavage sites of pigeon NDV isolated in recent years was consistent with the characteristics of virulent strains. During this study, the strains of pigeon NDV collected in Taiyuan, Shanxi Province were consistent with the types occurring in China, which was genotype VI.1.1, which was also consistent with the genotype of the strain isolated by Li et al. [21] from the Shanxi pigeon farm. This finding showed that the virus responsible for pigeon ND in Shanxi Province was the same as that in other regions of China.
Magpie is a common wild bird of Corvidae and Pica. They are widely distributed around the world and prefer to live in places where humans gather, including cities and villages. Magpie is susceptible to ND. Tong et al. [9] first reported magpie ND and transmitted the pathogen to chickens in China. Later, there were also some reports about magpies infected with ND [10, 11, 22, 23]. In 2009, Wang et al. [23] isolated NDV from magpie disease material and experimentally confirmed that it was an infection with a virulent strain. It can be inferred that it was caused by NDV infection of local chickens. The NDV isolated from the magpie in this study was consistent with the virus isolated from the local pigeon population in its genotype, which was consistent with most NDV occurring in China and belonged to genotype VI.1.1. Recently, the NDV strains in local chicken flocks were primarily attenuated type II [24] and virulent type VII [25], while there have been no reports of genotype VI. The dominant genotype of NDV in China is type VII, few chickens are infected with genotype VI, and the proportion of type VI in chickens and waterfowl and other hosts is only between 0.48 and 0.55% [14], indicating that the incidence of this type is very low. However, Yang et al. [26] showed that most NDV isolated from wild birds were class I, and the genotype I and genotype II in class II, and inferred that the virus may be transmitted between domesticated birds and wild birds. This conclusion has also been achieved experimentally by Ren et al. [27]. Therefore, the findings of this study could also infer that there was a correlation between the incidence of magpie and pigeon, which was consistent with the conclusion reported by Tong [9] and Wang [23].
The sequence of amino acids in the F protein cleavage site of the two NDV strains isolated in this study was 112R-R-Q-K-R-F117, and there were 577 amino acids in the HN protein, indicating that the two NDV had typical virulent characteristics. However, biological characteristics tests revealed that they were not virulent strains. This was consistent with the research conclusions of genotype VI pigeon NDV reported in China [28]. This finding indicated that the F protein cleavage site was not the only factor that determined the virulence of pigeon NDV [29]. It has even been reported that the virulence factor of pigeon NDV had little relationship with the F protein cleavage site [30]. However, the virulence of the two strains isolated in this study was slightly different. The virulence of SX/TY/Pi01/22 isolated from pigeon was a moderate strain, similar to that of pigeon NDV gene VI.1.1 isolated from China, whereas SX/TY/Ma01/22 was a less virulent strain. Comprehensive analysis showed that there was a base mutation at 11,847, which changed from G to T, causing the corresponding amino acid to change from arginine to serine. Therefore, this alteration may be responsible for the variation in virulence and requires further study.
References
World Organization for Animal Health (2022) Newcastle disease. WOAH. https://www.woah.org/en/disease/newcastle-disease/. Accessed 26 December 2022
Notice of the Ministry of Agriculture and Rural affairs of People’s Republic of China (2022) Lists A, B and C Diseases. Ministry of Agriculture and Rural affairs of the People’s Republic of China. https://www.springer.com/journal/11262/submission-guidelines. Accessed 26 December 2022
Chen Y, Fan L, Cai J, Xiang B, Liao M, Xu C, Ren T (2020) Research progress on the difference of pathogenicity of Newcastle disease virus infecting different poultry. Poult Husb Dis Control 9:33–36
Meng L, Pang H, Zhao Z, Zhao K, Yuan W (2021) Status on Newcastle disease vaccines. Prog Vet Med 42:107–111
Li M, Li Y, Yang B, Xu Z, Wang H, Qiu X, Sun Y, Tan L, Liao Y, Song C, Liu W (2020) Identification and whole genome sequencing of a genotype VII Newcastle Disease Virus isolated from pigeon. Chin J Vet Med 56:5–9+14
Selim KM, Selim A, Arafa A, Hussein HA, Elsanousi AA (2018) Molecular characterization of full fusion protein (F) of Newcastle disease virus genotype VIId isolated from Egypt during 2012–2016. Vet World 11:930–938
Dortmans J, Koch G, Rottier PJM, Peeters BPH (2009) Virulence of pigeon paramyxovirus type 1 does not always correlate with the cleavability of its fusion protein. J Gen Virol 90:2746–2750
Dimitrov KM, Ramey AM, Qiu X, Bahl J, Afonso CL (2016) Temporal, geographic, and host distribution of avian paramyxovirus 1(Newcastle disease virus). Infect Genet Evol 39:322–334
Tong Z, Cao L, Cao Y, Wang X, Cheng W, Ye L, Sun H, Sun Y, Guo Y (1998) Report on the diagnosis of a Newcastle disease transmitted from a magpie to chickens. Heilongjiang Anim Sci Vet Med 4:38
Yang J (2002) Magpie Newcastle disease. Chin J Vet Med 3:52
Wu G, Wang Z, Zhang G, Zhou G (2007) Magpie Newcastle disease cases. Chin J Vet Med 5:56
Ministry of Agiculture and Rural Affairs of the People’s Republic of China (2020) GB/T 16550–2020, Diagnostic techniques for Newcastle disease, pp 2–3
National Standards of the People’s Republic of China (2009) Diagnostic techniques for Newcastle disease. Issued by Standardization Administration of the People’s Republic of China
Kaleta EF, Alexander DJ, Russell PH (1985) The first isolation of the avian PMV-1 virus responsible for the current panzootic in pigeons? Avian Pathol 14:553–557
Alexander DJ (2000) Newcastle disease and other avian paramyxovirus. Rev Sci Tech 19:443–462
Saif YM (2012) Diseases of poultry. China Agric Press 12:65–92
Zhao Y, Zhao Y, Wu H, Yang Z, Liu H (2022) Analysis of epidemic genotypes of Newcastle Disease Virus isolated from poultry in China. Prog Vet Med 43:107–111
Luo Y, Wang J, Wang Y, Wei R, Lv Y, Zhao Y, Zheng D, Yu S, Zhang L, Liu H (2018) Distribution and molecular characteristics of 10 strains of pigeon paramyxovirus in some regions of China from 2014 to 2017. Chin J Vet Sci 38:1883–1886+1931
Qiu X, Meng C, Zhan Y, Yu S, Li S, Ren T, Yuan W, Xu S, Sun Y, Tan L, Song C, Liao Y, Ding Z, Liu X, Ding C (2017) Phylogenetic, antigenic and biological characterrizeation of pigeon paramyxovirus type 1 circulating in China. Virol J 14:186
Tian Y, Xue R, Yang W, Li Y, Xue J, Zhang G (2020) Characterization of ten paramyxovirus type 1 viruses isolated from pigeons in China during 1996–2019. Vet Microbiol 244:108661
Li T, Lu B, Ding S, Tang J, Wang C, Liu H (2021) Isolation and identification of an isolate of pigeon Paramyxovirus type 1 and sequence analysis of F gene. Heilongjiang Anim Sci Vet Med 24:51–56
Liu W, Wang X, Liu C, Li C, Tong Z (2003) Diagnostic report of Newcastle disease in chickens caused by magpie Newcastle disease. Heilongjiang Anim Sci Vet Med 8:80
Wang Z, Wu G, Zhang G, Liu J, Zheng C, Lv L (2009) Isolation, identification and biological characteristics of Newcastle disease virus in magpies. Heilongjiang Anim Sci Vet Med 5:92–93
Liu H, Li T, Tang J, Ding S, Lu B, Wang C (2021) Isolation, identification and virulence analysis of NDV isolated from Shanxi Province. Mod J Anim Husb Vet Med 11:43–47
Liu Y, Xue J, Ma X, Wang G, Wang Z, Zhang W (2016) 1 Isolation and identification of a virulent strain of Wu ji Newcastle Disease virus and genetic evolution analysis of F gene. J Econ Anim 20:13–17
Yang S, Huang Q, Cui N, Sun S, Xu C (2022) Molecular characterization and phylogenetic analysis of Newcastle disease virus isolates from healthy wild birds. Chin J Vet Sci 42:452–456
Ren S, Xie X, Wang Y, Tong L, Gao X, Jia Y, Wang H, Fan M, Zhang S, Xiao S, Wang X, Yang Z (2016) Molecular characterization of a Class I Newcastle disease virus strain isolated from a pigeon in China. Avian Pathol 45:408–417
Pei Y, Sun Y, Zhao Y, Zhang G, Xue J (2022) Genome sequencing and pathogenicity analyses of four isolates of the Newcastle Disease Virus in pigeons. Chin J Virol 38:402–414
Olszewska-Tomczyk M, Dolka L, Switon E, Smietanka K (2018) Genetic changes in pigeon paramyxovirus type-1 induced by serial passages in chickens and microscopic lesions caused by the virus in various avian hosts. J Vet Res 62:447–455
Heiden S, Grund C, HÖper D, Mettenleiter TC, RÖmer-OberdÖrfer A, (2014) Pigeon paramyxovirus type l variants with polybasic F protein cleavage site but strikingly different pathogenicity. Virus Genes 49:502–506
Funding
This study was funded by Innovation Team Programes of College of Veterinary Medicine, Shanxi Agricultural University (DY-CX005) and General Program of Shanxi Youth Science and Technology Research Fund (201701D221198).
Author information
Authors and Affiliations
Contributions
Huadong Liu wrote the main manuscript text,All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
All authors declare that they have no conflict of interest.
Research involving human participants and/or animals
All procedures involving animals were approved by the Institutional Animal Care and Use Committee of Shanxi Agricultural University (Permit Number: SXAU-EAW-2022C.AL.00603001) and conducted according to the guidelines for animal welfare by the World Organization for Animal Health.
Additional information
Edited by Nicola Decaro.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Liu, H., Li, T., Ding, S. et al. Complete genome sequence analysis and biological characteristics of Newcastle disease viruses from different hosts in China. Virus Genes 59, 449–456 (2023). https://doi.org/10.1007/s11262-023-01988-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11262-023-01988-y