Abstract
Transgenic potato plants of Solanum tuberosum cultivar Vales Sovereign were generated that expressed fused, tandem, 200 bp segments derived from the capsid protein coding sequences of potato virus Y (PVY strain O) and potato leafroll virus (PLRV), as well as the cylindrical inclusion body coding sequences of potato virus A (PVA), as inverted repeat double-stranded RNAs, separated by an intron. The orientation of the expressed double-stranded RNAs was either sense–intron–antisense or antisense–intron–sense RNAs, and the double-stranded RNAs were processed into small RNAs. Four lines of such transgenic potato plants were assessed for resistance to infection by PVY-O, PLRV, or PVA, all transmitted by a natural vector, the green-peach aphid, Myzus persicae. Resistance was assessed by the absence of detectable virus accumulation in the foliage. All four transgenic potato lines tested showed 100 % resistance to infection by either PVY-O or PVA, but variable resistance to infection by PLRV, ranging from 72 to 96 % in different lines. This was regardless of the orientation of the viral inserts in the construct used to generate the transgenic plants and the gene copy number of the transgene. This demonstrates the potential for using tandem, fused viral segments and the inverted-repeat expression system to achieve multiple virus resistance to viruses transmitted by aphids in potato.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Resistance to virus infection has been engineered into a number of transgenic plant species, against a range of individual viruses (rev. by [1, 2]). While most of these examples are in experimental hosts such as Nicotiana tabacum and Nicotiana benthamiana, which are also natural hosts for some of the targeted viruses, there are also many examples of crop plants engineered to express resistance to their viruses [2–4]. Since many crops are subject to infection by more than one virus, in only a limited number of situations would providing resistance to one virus protect the crop against viral infection or disease. Therefore, for many crops, it will be necessary to engineer them for resistance to multiple viruses. This was done first with potato expressing the coat protein (CP) coding sequences of potato virus X (PVX) and potato virus Y (PVY) and providing resistance to these viruses [5, 6], and then with squash [7] and cantaloupe [8], both conveying resistance to watermelon mosaic virus 2, zucchini yellows mosaic virus, and cucumber mosaic virus (CMV), all using multiple transgene cassettes expressing the CP coding sequences of these viruses. Subsequently, transgenic plants were generated that expressed fused viral segments from a single promoter and showed resistance to CMV and tomato leaf curl virus in tomato [9], tomato spotted wilt virus, and tomato leaf curl Taiwan virus in tomato [10], and CMV and watermelon mosaic virus in watermelon [11]. In other laboratories, the strategy of expressing inverted repeat sequences from a transformation vector of at first one [12, 13] and then multiple viruses [14–17] has come into vogue, as plants regenerated using this strategy showed a higher percentage of resistant lines. Among the latter studies, only one, to date, involved a food-crop plant, potato (Solanum tuberosum) [17].
Potato is an important food crop which is subject to a number of diseases, including those caused by about 40 viruses and two viroids. However, only nine viruses and one viroid are considered to have a significant economic impact on potato plants worldwide [18, 19]. Of these viruses, potato leafroll virus (PLRV) and PVY are considered the most important, with PVX of less importance, as its main effects are caused by synergistic interactions in combination with PVY or other potyviruses, such as potato virus A (PVA) and potato virus V (PVV). The latter two viruses have different global distributions with PVA being more prevalent, and PVV limited to Peru and parts of Western Europe [18, 19]. While PVX is transmitted mechanically, PLRV and the potyviruses are transmitted by aphid vectors; all are transmitted through seed tubers. Natural resistance genes to all of these viruses are known, but many such genes show differences in the extent of resistance, depending on the nature of the resistance, the challenge virus strain, and on the genetic background of the host. While a number of these resistance genes have been deployed in commercial cultivars, some of these genes cannot be transferred readily to such cultivars [18, 19]. Given these limitations, along with the time required to generate new potato cultivars that express new resistance phenotypes along with the other agronomic characteristics of the parent crop-plants, transgenic approaches to confer resistance against these viruses is considered a rapid and suitable alternative [2–4, 18, 19]. Therefore, in this study, we generated transgenic potato plants expressing segments of the CP coding sequences of PVY and PLRV, as well as the cylindrical inclusion body coding sequence of PVA, as tandemly arrayed, inverted-repeat sequences, and assessed lines of these transgenic potato plants for resistance to infection by PVY, PLRV, or PVA, transmitted by a natural aphid vector.
Materials and methods
Plasmid constructs, plant transformation, and selection of transformants
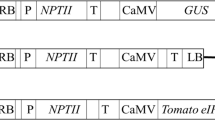
The construction of the plant transformation binary vector pFGC5941/600, containing two inverted copies of a 600 bp viral sequence cassette, was prepared as described previously [16]. Each 600 bp viral sequence cassette contained fused, tandem, 200 bp viral sequences derived from the coding regions of the PVY (strain O) CP coding sequence, the PLRV (Canadian isolate) CP gene, and the PVA (strain U) cylindrical inclusion body coding sequence. The same 600 bp cassette, in the intermediate gateway entry plasmid pDONR207, was used to introduce the 600 bp cassette twice, in opposite orientations, into the binary plant transformation vector pK7GWIWG2(I), using the primers given in Supplementary Table S1 and the Gateway recombination system, as described by the manufacturer (Invitrogen) (Supplementary Fig. S1). Following selection and characterization of the resulting plasmid pK7GWIWG2(I)/600, to verify both that it contained the double insert and the orientation of the inserts, the binary plasmid was transformed into Agrobacterium tumefaciens strain LBA4404 by the freeze–thaw method [20]. The binary vector pFGC5941/600 was introduced into A. tumerifaciens strain LBA4404 by the same method, as detailed previously [16]. Confirmation of repeated insertion of the 600 bp cassette into pFGC5941 and pK7GWIWG2(I) was done by the polymerase chain reaction (PCR), using the primer pairs given in [16] and in Supplementary Table S1, respectively.
Tissue culture plantlets of potato cv. Vales Sovereign were purchased from the Scottish Agricultural Scientific Agency (SASA; Edinburgh, UK). Internodal stem segments of 1 cm, and in some cases, leaf disks of the tissue culture plantlets, were used for plant transformation with Agrobacterium harboring either plasmid pFGC5941/600 or plasmid pK7GWIWG2(I)/600. The co-cultivation, transformation, and regeneration of transformed plants were done using the methods described in [21]. Verification of the presence of the transgenes in the transformed plants was done by both PCR using the primers given in Supplementary Table S1 and the presence of small-interfering RNAs (siRNAs).
Transgene DNA and siRNA analysis
Genomic DNA was extracted, digested with the restriction enzymes EcoRI or SacI, from plants transformed with pFGC5941/600 or pK7GWIWG2(I)/600, respectively, separated on 1.0 % agarose gels, blotted onto nitrocellulose membranes, hybridized with a 32P-labeled probe, and assessed for the presence of viral sequences, all as specified previously [16]. Total RNAs were extracted and analyzed by northern blot hybridization for the presence of viral siRNAs using the method of [22], with the modifications given in [16].
Plant inoculation and assessment of resistance
Cuttings (of 8 cm) were made from the stems of individual transgenic plant lines and were rooted in pots containing soil. Plants grown from the cuttings were inoculated using the green peach aphid (Myzus persicae) to transmit viruses from infected, non-transformed potato plants. The inocula for all three viruses were field isolates of PVY-O, PVA, and PLRV-infected plants obtained from SASA, as described previously [16]. Each plant was inoculated by aphids containing only one test virus. Ten aphids of the second or third instars were used for each plant. The duration of pre-acquisition starvation was 3–4 h for PVY and PVA, and no pre-acquisition starvation period was used for PLRV. The period of virus acquisition was 5 min for PVY and PVA, and 3 days for PLRV. The duration of feeding test plants was 24 h for PVY and PVA, and 3 days for PLRV. The aphids were then killed by fumigation overnight using nicotine shreds. Inoculated plants were maintained in a greenhouse with a 16 hr daylength, and day/night temperatures of 25 ± 5 °C. Plants were monitored for 30 days, and were assessed for infection by western blot analysis for PVY in the first instance and by reverse-transcription PCR (RT-PCR) in most cases; RT-PCR was used for analysis of infection by PLRV and PVA. The procedures for western blot analysis and RT-PCR analysis were as described previously [16].
Results
Transformation and regeneration of transgenic potatoes
The aim of this study was to express tandem, fused, 200 bp segments of three viruses (PVY, PLRV, and PVA) from a plant transformation vector in transgenic potato plants as inverted repeats (see Supplementary Fig. S1), such that the resulting RNA transcript would produce hairpin RNAs which would be targeted for degradation by the host RNA silencing system. This, in turn, was expected to produce plants that would show resistance, at some level to be ascertained, to the three viruses, as we had done previously in tobacco [16]. In addition, in the first instance, we also sought to use a plant transformation vector (pFGC5941) that facilitated selection of transgenic plants by the herbicide BASTA (ammonium glufosinate; phosphinotricine). However, we found that leaf disks and stem segments of potato cv. Vales Sovereign were highly sensitive to BASTA and very few would regenerate even in 10 μg/ml BASTA (Supplementary Table S2). The same was true for the effects of BASTA on further growth of shoots and rooting of shoots placed into medium containing BASTA on both cv. Vales Sovereign and also the cv. Maris Piper (data not shown). Therefore, we inserted the cassette of viral segments into the plant transformation vector pK7GWIWG2(I), which uses kanamycin resistance as the basis of selection (Supplementary Fig. S1), and transformed potato stem segments and/or leaf disks. The overall efficiency of regeneration of transgenic plants containing the cassette of viral sequences (as ascertained by selection on kanamycin and subsequent analysis by PCR) was about 1.3 % for cv. Vales Sovereign using plant transformation vector pK7GWIWG2(I)/600 (Supplementary Table S2) and about 0.2 % using plant transformation vector pFGC5941/600 (data not shown). Thus, we continued only with the regenerated Vales Sovereign potato plants transformed with pK7GWIWG2(I)/600.
There were two orientations of the viral sequences relative to the intron in the transformation vector, such that the RNAs in the transgenic plants would be expressed in either the sense–intron–antisense (IN) or the antisense–intron–sense (OUT) orientation. Initially, we obtained 11 IN lines and nine OUT lines, but several lines did not survive and/or were not assessed further. In addition, work was not continued with one vector construct [pK7GWIWG2(I)/600-4-11-IN], for which there were only four regenerated plants (Supplementary Table S2). Thereafter, all data mentioned were obtained using lines transformed with either vector pK7GWIWG2(I)/600-102-6-IN or vector pK7GWIWG2(I)/600-1-16-OUT. The corresponding line numbers will be abbreviated as 600/102-6-IN# or 600/1-16-OUT#.
Molecular characterization of the transgenic potato plants
The regenerated potato plants, which contained the sequences of all three viral segments, were analyzed by Southern blot hybridization to confirm the insertion of the transgene into the plant genome after digestion of extracted DNAs with SacI, for which there is only one site in the transferred vector DNA and none in the viral insert. Eight of the nine OUT lines and six of the seven IN lines showed the presence of viral sequences in the digested DNAs (Fig. 1 and data not shown). The transgenic IN Lines 600-102-6-IN38 and -IN39, as well as OUT Lines 600-1-16-OUT3 and -OUT21 each exhibit two independent insertion events (Fig. 1), while the other lines showed only the presence of a single band. The smallest band (from Line 600-1-16-OUT21) corresponded to about 8 kb, while the largest band (from the same line) corresponded to over 23 kb (the highest size marker band). The other bands were all between 10 and 23 kb (Fig. 1). The relative intensities of the bands suggested that most of the DNA fragments contained a single copy of the insert, while the intensities of the two bands in Line 600-102-6-IN39 suggested that the upper band might contain more than one insert (Fig. 1). This was not investigated further.
Southern blot hybridization analysis of T0 regenerated potato plants for the presence and size of the inserted transgene. Purified DNAs extracted from regenerated plants of Lines pK7GWIWG2(I)/600-102-6-IN31, -IN32, -IN34, -IN35, -IN38, and -IN39, plus Lines 600-1-16-OUT3, -OUT10, -OUT11, and -OUT21, as well as non-transformed plants (NT), were digested with the restriction enzyme SacI, fractionated by agarose gel electrophoresis, blotted onto nitrocellulose membranes, and hybridized with a 32P-labeled DNA probe. The size marker (M) was generated from phage λ DNA digested with the restriction enzyme HindIII. The position of the fragments of size 23.1, 9.4, and 6.5 kbp are shown
RNAs extracted from the above transgenic lines were assessed by northern blot hybridization for the presence of siRNAs against the three viruses used in the generation of the constructs. These transgenic lines all showed the presence of siRNAs derived from the inserted region of each virus (Fig. 2 and data not shown). Line 600-102-6-IN39 showed the presence of higher levels of siRNAs than the other lines, consistent with the presence of multiple copies of the inserted transgene (Fig. 2). Lines 600-102-6-IN34 and 600-1-16-OUT11, each of which showed the presence of a single band in the Southern blots (Fig. 1), showed similar levels of siRNA accumulation (Fig. 2), while Line 600-102-6-IN38, which exhibited two bands of equal intensities on the southern blot (Fig. 1), only showed a higher level of siRNA accumulation for PVY, but not for PLRV or PVA (Fig. 2). Nevertheless, the data support the conclusion that the transgenic plants contain transgenes expressing hairpin RNAs that are degraded in planta by the RNA silencing system into small RNAs of 21–24 nt.
Northern blot hybridization for the presence and level of accumulation of small RNAs accumulating in the transgenic plants. Purified small RNAs extracted from plants of Lines pK7GWIWG2(I)/600-102-6-IN34, -IN38, -IN39, and 600-1-16-OUT11, as well as from non-transformed (NT) plants, were fractionated by gel-electrophoresis, blotted onto nylon membranes, and hybridized with DIG-labeled RNA probes against the segments derived from the viruses indicated on the blot. The small RNAs were 21–25 nt in size, based on the mobility of the RNAs seen in stained gels
Assessment of resistance to natural infection
Although mechanical inoculation of potato by PVY and PVA can result in systemic infections, this is not the case for PLRV, for which mechanical inoculation would lead to only single cell infections of epidermal cells [23–25]. In addition, in our experience, infection with PVA was more difficult to establish in potato using mechanical inoculation (data not shown). Since infections by these viruses are established by aphids in the field, we chose instead to examine the extent of resistance to these three viruses after their transmission via the green peach aphid, M. persicae. Thus, ten aphids were used per plant for transmission of single virus species, with 20 or 25 plants of each line assessed for infection by each virus at 30 days post inoculation.
The four transgenic potato lines analyzed in Fig. 2 for their expression of siRNAs were used in the assessments of resistance to the PVY, PLRV, and PVA, since larger numbers of cuttings could be made from those lines to generate plants for the assessment or resistance. All the transgenic potato lines tested showed resistance to all three viruses (Table 1), although at different levels for PLRV versus PVY and PVA, as assessed by either western blot analysis (PVY) or RT-PCR (PLRV and PVA). The symptoms correlated with the presence of virus for PVY, with PVY-inoculated transgenic plants showing no symptoms and no virus accumulation, while PVY-inoculated non-transformed plants showed severe symptoms (Fig. 3 and data not shown); however, there was no such correlation for PLRV, where most of the infected plants, transgenic and non-transformed, showed no symptoms (Supplementary Fig. S2 and data not shown). In the case of PVA, mottle symptoms were clear on infected plants for those inoculations done in Scotland, but were not for those done in Korea. Thus, the data below refer to the detection of virus rather than the presence of symptoms. All four transgenic potato lines showed 100 % resistance to detectable infection by PVY, while 8.6 % of the control non-transformed plants remained uninfected (Table 1). All four transgenic potato lines also showed 100 % resistance to detectable infection by PVA (Supplementary Fig. S3 and data not shown); however, the non-transformed control plants were not efficiently infected by PVA, with only 36 % of those plants becoming infected (Table 1). The PVA used was a field isolate that also exhibited poor infection in non-transformed tobacco [16] when aphids were used for transmission, although in that case, two-thirds of the non-transformed control plants became infected. Only 9 % of non-transformed tobacco plants did not become infected with PLRV [16], while here, 24 % of non-transformed potato plants did not become infected with PLRV. However, the four transgenic potato lines demonstrated resistance to detectable infection by PLRV, although to various extents: Lines 600-102-6-IN34 and -IN38 showed over 90 % resistance, while Lines 600-102-6-IN39 and 600-1-16-OUT11 showed 72 and 84 % resistance to infection, respectively (Table 1). Thus, each of the four transgenic potato lines showed excellent resistance to PVA and PVY and good, but not complete resistance to PLRV.
Infection of potato cv. Vales Sovereign (VS) with PVY-O transmitted by aphids. Transgenic potato, Lines pK7GWIWG2(I)/600-1-16-OUT11 and non-transformed VS, were either mock-inoculated or inoculated with PVY-O using ten aphids per plant and were assessed for infection 30 days post-inoculation. The PVY-inoculated, non-transformed VS plant was positive for PVY by western blot analysis, was stunted, and showed deformed and necrotic leaves, as well as stem necrosis; the transgenic plant did not show any symptoms and did not contain detectable virus
Discussion
Transgene-mediated resistance to infection by multiple viruses has been achieved by several means, including expression of single virus segments from different promoters in transformation vectors containing multi-transcription units, or by co-transformation of plants by more than one vector each expressing a different transgene [5–8, 26, 27]; as fusions of viral segments expressed as single-stranded RNAs from the same promoter in one vector [9–11, 28]; and as tandem fusions of segments of multiple viruses expressed from a single plasmid vector as inverted hairpin RNAs [14–17]. All have achieved some degree of resistance to the corresponding viruses. Resistance to multiple viruses in potato has been described previously only for PVX and PVY [5, 6, 17]. Here, we report the establishment of transgenic potato plants expressing resistance to the two most important potato viruses (PLRV and PVY) along with resistance to a third potato virus that is becoming more important globally, PVA [18, 19], all of which are transmitted naturally by aphids.
Previously, we obtained excellent resistance to PVY, PVA, and a mixture of PVY and PVA in transgenic tobacco expressing the same cassette of three viral sequences, although we used a different plant transformation vector [16]. Here, we also obtained excellent resistance to PVA and PVY, with no virus detected in those plants within 30 days after inoculation (Table 1). However, in transgenic tobacco, we obtained less resistance against PLRV, with the single OUT line showing 43 % resistance and the single IN line showing only 15 % resistance [16]. In contrast to the situation in transgenic tobacco, the three transgenic potato IN lines all showed much better resistance (72–96 %), while the single transgenic OUT line also showed good resistance (84 %) (Table 1). We cannot explain why the level of resistance to PLRV is consistently less than that for PVY or PVA. However, it may relate to differences in either the extent to which RNA silencing operates in phloem companion cells and phloem parenchyma cells, or the ability of the PLRV RNA silencing suppressor to overcome hairpin RNA-mediated resistance versus the RNA silencing suppressors of PVA and PVY. Alternatively, the siRNAs generated from the segment of the PLRV CP gene, we used, might not efficiently bind to their viral genome targets, due to local secondary structure in the PLRV CP gene. It is worth noting that in two studies where a 449 bp fragment from part of the PLRV replicase gene was used to generate transgenic potato plants, resistance to infection occurred to varying extents in different lines, with none of the lines giving complete resistance to virus accumulation [29, 30], while several lines of transgenic potato generated earlier using the complete replicase gene gave complete resistance to PLRV accumulation [29–31]. This also could be interpreted as due to fewer, different siRNAs being available from the PLRV replicase fragment to target different regions of the PLRV replicase gene, although there may be other variables at play. Similarly, in another study, where transgenic potato plants of two other cultivars (Desiree and Kuroda) expressing an inverted-repeat hairpin containing 300 bp sequences of PVX (ORF2), PVY (HCPro), and PLRV (CP) were assessed for resistance to these viruses, the plant lines showing the strongest resistance (by lack of foliar symptoms) still showed the presence of the three viruses, although with considerably reduced titers [17]. In that case, those plants did not show differences in the extent of suppression of accumulation of PLRV versus PVX or PVY in plants of the various resistant transgenic lines, which would argue against differential suppression of the RNA silencing by the viral-encoded RNA silencing suppressor proteins.
Transgenic potato Lines 600-102-6-IN38 and -IN39 both contained two independent insertion events in the potato genome (Fig. 1), yet they did not show similar levels of siRNA accumulation derived from all three viral segments (Fig. 2). Moreover, the levels of accumulation of siRNAs to PLRV and PVA from Line 600-102-6-IN38 were similar to those observed from the single copy transgene Lines 600-102-6-IN34 and 600-1-16-OUT11 (Fig. 2). Not all transgenes are necessarily expressed, and when two copies are present, in some cases, one transgene can lead to inactivation of expression from the second copy [32]. However, this does not explain why transgenic potato Line 600-102-6-IN38 accumulated high levels of PVY siRNAs, but low levels of PLRV and PVA siRNAs, in comparison with Lines 600-102-6-IN34 and -IN39, respectively (Fig. 2).
Transgenic potato Line 600-102-6-IN39 showed a higher level of PLRV siRNA accumulation than did the other three lines tested (Fig. 2), yet this line showed the lowest level of resistance to PLRV (Table 1). Thus, while several studies have shown a correlation between the levels of accumulating siRNAs and the extent of resistance [14, 16, 33–35], this may not always be the case. Alternatively, the data may reflect a minimal threshold level of siRNAs required for RNA silencing, and there may be other factors that can affect the extent of RNA silencing once the threshold is reached. Moreover, very little is known about the various factors involved in RNA silencing in crop plants and their interactions and regulation, versus those in the model species Arabidopsis thaliana where the vast majority of work on RNA silencing has been done. Such work on specific crop plants may be necessary to fully characterize transgenic plants utilizing RNA silencing technology to confer resistance.
References
M. Prins, M. Laimer, E. Noris, J. Schubert, M. Wassenegger, M. Tepfer, Mol. Plant Pathol. 9, 73–83 (2008)
J. Gottula, M. Fuchs, Adv. Virus Res. 75, 161–183 (2009)
R.G. Dietzgen, N. Mitter, Austral. Plant Pathol. 45, 605–618 (2006)
D.V.R. Reddy, M.R. Sudarshana, M. Fuchs, N.C. Rao, G. Thottappilly, Adv. Virus Res. 75, 185–220 (2009)
W. Kaniewski, C. Lawson, B. Sammons, L. Haley, J. Hart, X. Delannay, N.E. Tumer, Nat. Biotechnol. 8, 750–754 (1990)
C. Lawson, W. Kaniewski, L. Haley, R. Rozman, C. Newell, P. Sanders, N.E. Tumer, Nat. Biotechnol. 8, 127–134 (1990)
D.M. Tricoli, K.J. Carney, P.F. Russell, J.R. McMaster, D.W. Groff, K.C. Hadden, P.T. Himmel, J.P. Hubbard, M.L. Boeshore, H.D. Quemada, Nat. Biotechnol. 13, 1458–1465 (1995)
M. Fuchs, J.R. McFerson, D.M. Tricoli, J.R. McMaster, R.Z. Deng, M.L. Boeshore, J.F. Reynolds, P.F. Russell, H.D. Quemada, D. Gonsalves, Mol. Breed. 3, 279–290 (1997)
S. Praveen, A.K. Mishra, G. Antony, Plant Cell, Tissue Organ Cult. 84, 47–53 (2006)
C.-Y. Lin, H.-M. Ku, W.-S. Tsai, S.K. Green, F.-J. Jan, Transgenic Res. 20, 261–270 (2011)
C.-Y. Lin, H.-M. Ku, Y.-H. Chiang, H.-Y. Ho, T.-A. Yu, F.-J. Jan, Transgenic Res. 21, 983–993 (2012)
N.A. Smith, S.P. Singh, M.-B. Wang, P.A. Stoutjesdijk, A.G. Green, P.M. Waterhouse, Nature 407, 319–320 (2000)
S.V. Wesley, C.A. Helliwell, N.A. Smith, M.-B. Wang, D.T. Rouse, Q. Liu, P.S. Gooding, S.P. Singh, D. Abbott, P.A. Stoutjesdijk, S.P. Robinson, A.P. Gleave, A.G. Green, P.M. Waterhouse, Plant J. 27, 509–581 (2001)
E. Bucher, D. Lohuis, P.M.J.A. van Poppel, C. Geerts-Dimitriadou, R. Goldbach, M. Prins, J. Gen. Virol. 87, 3697–3701 (2006)
C.-X. Zhu, Y.-Z. Song, G.-H. Yin, F.-J. Wen, J. Phytopathol. 157, 101–107 (2009)
B.N. Chung, P. Palukaitis, Virus Genes 43, 454–464 (2011)
M. Arif, U. Azhar, M. Arshad, Y. Zafar, S. Mansoor, S. Asad, Transgenic Res. 21, 303–311 (2012)
H. Barker, M.F.B. Dale, in Natural Resistance Mechanisms of Plants to Viruses, ed. by G. Loebenstein, J.P. Carr (Springer, Dordrecht, 2006), pp. 341–366
P. Palukaitis, Plant Pathol. J. 28, 248–258 (2012)
M. Holsters, D. de Waele, A. Depicker, E. Messens, M. van Montagu, J. Schell, Mol. Gen. Genet. 163, 181–187 (1978)
Y.H. Joung, J.H. Jeon, K.W. Choi, H.S. Kim, H. Joung, Korean J. Plant Tissue Cult. 23, 77–81 (1996)
C. Llave, K.D. Kasschau, J.C. Carrington, Proc. Natl. Acad. Sci. USA 97, 13401–13406 (2000)
K.M. Nurkiyanova, E.V. Ryabov, U. Commandeur, G. Duncan, T. Canto, S.M. Gray, M.A. Mayo, M.E. Taliansky, J. Gen. Virol. 81, 617–626 (2000)
E.V. Ryabov, G. Fraser, M.A. Mayo, H. Barker, M. Taliansky, Virology 286, 363–372 (2001)
K.A. Peter, F. Gildow, P. Palukaitis, S.M. Gray, J. Virol. 83, 5419–5429 (2009)
M. Fuchs, D. Gonsalves, Nat. Biotechnol. 13, 1466–1473 (1995)
M. Prins, P. de Haan, R. Luyten, M. van Veller, M.Q.J.M. van Grinsven, R. Goldbach, Mol. Plant-Microbe Interact. 8, 85–91 (1995)
F.-J. Jan, C. Fagoaga, S.-Z. Pang, D. Gonsalves, J. Gen. Virol. 81, 2103–2109 (2000)
M. Arif, P.E. Thomas, J.M. Crosslin, C.R. Brown, Pak. J. Bot. 41, 945–954 (2009)
M. Arif, P.E. Thomas, J.M. Crosslin, C.R. Brown, Pak. J. Bot. 41, 1477–1488 (2009)
P.E. Thomas, E.C. Lawson, J.C. Zalewski, G.L. Reed, W.K. Kaniewski, Virus Res. 71, 49–62 (2000)
A. Dalakouras, M. Tzanopoulou, M. Tsagris, M. Wassenegger, K. Kalantidis, Transgenic Res. 20, 293–304 (2011)
K. Kalantidis, S. Psaradakis, M. Tabler, M. Tsagris, Mol. Plant-Microbe Interact. 15, 826–833 (2002)
Y.K. Chen, D. Lohuis, R. Goldbach, M. Prins, Mol. Breed. 14, 215–226 (2004)
A. Germundsson, J.P.T. Valkonen, Virus Res. 116, 208–213 (2006)
Acknowledgments
This study was supported by grant number RDA-PJ009568022013 from the Next Generation BioGreen21 Program of the Rural Development Administration, Republic of Korea.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Chung, B.N., Yoon, JY. & Palukaitis, P. Engineered resistance in potato against potato leafroll virus, potato virus A and potato virus Y. Virus Genes 47, 86–92 (2013). https://doi.org/10.1007/s11262-013-0904-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11262-013-0904-4