Abstract
Transgenic tobacco plants expressing fused, tandem, inverted-repeat, double-stranded RNAs derived either from the three viruses [potato virus Y (PVY), potato virus A (PVA), and potato leafroll virus (PLRV)] or the five viruses [PVY, PVA, PLRV as well as tobacco rattle virus (TRV), and potato mop-top virus (PMTV)] were generated in this study to examine whether resistance could be achieved against these three viruses or five viruses, respectively, in the same plant. The transgenic lines were engineered to produce 600- or 1000-bp inverted hairpin transcripts with an intron, in two orientations each, which were processed to silencing–inducing RNAs (siRNAs). Fewer lines were regenerated from the transformants with either 1000-bp inverted hairpin transcripts, or a sense–intron–antisense orientation versus antisense–intron–sense orientation. Resistances to PVA and two strains of PVY (-O and -N) were achieved in plants from most of lines examined, as well as resistance to co-infection by a mixture of PVY-O and PVA, applied to the plants by either rub inoculation or using aphids. This was regardless of the orientation of the inserted sequences for the 600-bp insert lines, but only for one orientation of the 1000-bp insert lines. The lines containing the 1000-bp inserts also showed resistance to infection by TRV inoculated by rub inoculation and PMTV inoculated by grafting. However, all the lines showed only low-to-moderate (15–43%) resistance to infection by PLRV transmitted by aphids. The resistances to the various viruses correlated with the levels of accumulation of siRNAs, indicating that the multiple resistances were achieved by RNA silencing.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Genetically modified plants have been engineered to express resistance largely to one or two viruses within the same plant of several species, although predominantly in tobacco (Nicotiana tabacum) and N. benthamiana [1]. In addition, resistance in the same plant to three viruses, watermelon mosaic virus 2, zucchini yellow mosaic virus, and cucumber mosaic virus (CMV), was generated in transgenic cantaloupe [2], as well as in transgenic lines of squash [3], using multiple transgene cassettes expressing coat protein (CP) genes of these viruses. Using the strategy of expressing inverted, tandem repeat sequences separated by an intron, from a transformation vector, which provides a much higher percentage of resistant regenerated transformed lines [4], resistance to three viruses, tobacco mosaic virus, potato virus Y (PVY), and CMV, was obtained in transgenic tobacco [5]. The same strategy was utilized to obtain resistance against four tospoviruses: groundnut ringspot virus, tomato chlorotic spot virus, tomato spotted wilt virus, and watermelon silver mottle virus, in transgenic N. benthamiana [6]. In both of these studies, the transgenes consisted of fused fragments of 150 bp (tospoviruses), 200 bp (PVY and tobacco mosaic virus), or 250 bp (CMV) of sequences from the corresponding viral genomes, arranged in a linear order so that the homologous sense–antisense or antisense–sense sequences would base-pair to form inverted double-stranded (ds) RNA molecules after transcription [5, 6]. This illustrates that multiple virus resistance in a single plant can be obtained using a single construct and can, in principle, be extended to crop plants as well as model systems.
Potato (Solanum tuberosum) is an important horticultural crop worldwide, and is subject to infection by a number of viruses, the most important of which, in terms of effects on crop yields, are potato leaf roll virus (PLRV) and PVY, with potato virus X being of less importance, as its main effects on potato crops occurs only in combination with PVY or other potyviruses [7]. Potato virus A (PVA) and potato virus V are less important globally, but of high impact in certain areas. Similarly, in some areas, tobacco rattle virus (TRV) and potato mop-top virus (PMTV) cause recurrent problems, as they affect the quality of the tubers [7]. PVY and PVA are potyviruses transmitted by aphids in a non-persistent, non-circulative manner. PLRV is a polerovirus, also transmitted by aphids, but in a persistent, circulative manner; PLRV is also transmitted in seed tubers. TRV is soil-borne and transmitted by trichodorid nematodes, as well as in seed tubers. PMTV is soil-borne and transmitted by Spongospora subterranea, a plasmodiophorid, fungus-like vector [7]. Thus, different strategies need to be implemented to reduce infection of potato crops by these different viral pathogens. Natural resistance genes are not available for PMTV, and such genes have been used to only a limited extent in some potato cultivars against TRV, because of the severe hypersensitive response that occurs in tubers [7]. Transgenic tobacco, N. benthamiana, or potato plants have been constructed expressing sequences of these “potato viruses” to provide virus gene-mediated resistance to one virus alone [8–14] or two viruses [15, 16] at a time, but plants with resistance to more than two of these potato viruses have not been described. Here, we generated transgenic tobacco plants expressed sequences of fused, tandem repeat, virus-derived dsRNAs of 200 bp each for either three viruses (PVY-O, PLRV, and PAV), or five viruses (PVY-O, PLRV, PVA, TRV, and PMTV) and evaluated these for resistance against viruses introduced by mechanical inoculation (PVY-O, PVY-N, PVA, and TRV), agroinfection (PLRV), graft-inoculation (PMTV), or aphid transmission (PVY-O, PVY-N, PLRV, and PVA). We also examined the relationship between resistance and the accumulation of silencing–inducing RNAs (siRNAs) in the transgenic plants.
Materials and methods
Virus sequence plasmid constructs
cDNA clones of various viral RNAs were used as templates for PCR to amplify sequences of 200 bp from each virus. The cDNA clones were of PVA (strain U [17]), PVY (strain O, SCRI-O isolate [18]), PLRV (Canadian isolate [19]), PMTV (Swedish isolate [20]), and TRV (isolate PpK20 [21]). The viral gene fragments amplified were as follows: PVY-O CP (Accession No. NC_001616, nucleotide 8894 to nucleotide 9093); PLRV CP (Accession No. AF453394, nucleotide 3831 to nucleotide 4030); PVA cylindrical inclusion (CI) (Accession No. Z21670, nucleotide 4472 to nucleotide 4671); TRV RNA1 replicase (REP) (Accession No. NC_003805, nucleotide 3,472 to nucleotide 3,626); and PMTV RNA1 REP (Accession No. NC_003723, nucleotide 3,411 to nucleotide 3,610).
The 600-bp and a 1000-bp fused, tandem viral sequences were prepared using overlapping PCR (Supplementary Fig. S1) with the primer pairs listed in Supplementary Table S1. The 600-bp fragment was prepared by fusing 200 bp each of the PVY CP-, PLRV CP-, and PVA CI-coding sequences. The 1000-bp fragment was prepared by fusing 200 bp each of the PVY CP-, PLRV CP-, PVA CI-, TRV REP- and PMTV REP-coding sequences. Amplification consisted of an initial denaturation at 94°C for 2 min, 5 cycles of 30-s denaturation at 94°C, 30-s annealing at 52°C, and 130-s elongation at 72°C, followed by 25 cycles of 20-s denaturation at 93°C, 20-s annealing at 59°C and 2-min elongation at 72°C. The various PCR products were then purified by Sepharose column chromatography (Sigma-Aldrich).
The 600- and the 1000-bp fragments were recombined into the Gateway entry plasmid pDONR207 using the Gateway recombination system (Invitrogen). This Gateway vector was then used to facilitate insertion of the viral DNA segments twice, in opposite orientations, into a Gateway-compatible version of the plant transformation binary vector pFGC5941 [22] according to the manufacturer’s instructions. The plasmids then were transformed into Escherichia coli DH5α competent cells (Invitrogen) using standard procedures [23].
Plant transformation and selection of transformants
Plasmids pFGC5941/600 and pFGC5941/1000 were transformed into Agrobacterium tumefaciens LBA4404 by the freeze–thaw method [24]. A colony of A. tumefaciens LBA4404 cells harboring pFGC5941/600 or pFGC5941/1000 was grown at 28°C in YEB media with 40 μg/ml kanamycin for 2 days. The bacterial cells were harvested by centrifugation at 4,500 rpm for 10 min and resuspended in MS media. The OD value of the Agrobacterium cells used for co-culture with disks cut from leaves of tobacco (N. tabacum cv. Samsun NN) was 0.7–1.0 at 600 nm. Transformation of 20 leaf disks per Agrobacterium construct was done as described by [25] with the modifications described by [26]. Calli were developed in the absence of glufosinate, which then was present during both shoot and root development.
The presence of the viral sequences in the transformed plants was confirmed by PCR, and siRNA analysis. Total plants RNAs were extracted using the RNeasy mini kit (Qiagen). The primer pairs used for the PCR confirmation of the insertion of 600 bp or 1000 bp into pFGC5941 and into the plant genome were based on the sequence of the 35S promoter, the chalcone synthase A intron (CHSA), and the octopine synthase (OCS) 3′ non-translated region. P35SF: 5′-AGAGGACACGCTCGAGTATA-3′; CHSAR: 5′-GAGCCAATTAAGATAAAACGTT-3′; CHSAF: 5′-ACTTACACTTGCCTTGGAGT-3′; OCSR: 5′-TAAGGATCTGAGCTACACATGT-3′. The siRNAs were analyzed as described previously [27] using the method of Llave et al. [28]. Synthesis of the probes and processing of the blot with digoxigenin-labeled probes were performed according to the manufacturer’s instructions using DIG RNA labeling mix (Roche diagnostics, Lewes, UK). The templates for the probes of PVY, PLRV, PVA, TRV, and PMTV were the respective 200-bp fragments of each virus previously cloned into pGEM-T easy (Promega). Plasmid DNA containing PVY and TRV was linearized by digestion with the restriction enzyme SalI and the RNA probe was synthesized in vitro with T7 RNA polymerase (Invitrogen), while PLRV, PVA, and PMTV were linearized with restriction enzyme NcoI, and the RNA probe was synthesized in vitro with SP6 RNA polymerase (Invitrogen).
Genomic DNA was extracted from young leaves of the T2-generation plants using the cetyltrimethylammonium bromide method [29]. Genomic DNA of these transgenic tobacco plants was digested with EcoRI, separated on 1.0% agarose gels and blotted to nitrocellulose membranes, as described [23]. The membrane was then hybridized with a 32P-labeled probe generated from the plasmid pYLRA (a cDNA clone containing the 600-bp insert) with the Rediprime DNA Labelling kit (Amersham Biosciences). Hybridization was done using Church’s buffer at 65°C [30]. Washing of the membrane and autoradiography were done by standard methods [23].
Plant inoculation and assessment of resistance
Transgenic seeds and control non-transgenic seeds of N. tabacum cv. Samsun NN were sown in sterilized soil. The plants were used for assessment of resistance when the first pair of leaves was fully expanded.
Virus-infected plants were maintained in a greenhouse with a 16-h daylength, and day/night temperatures of 25 ± 5°C. The transgenic, control non-transgenic, and virus-source non-transgenic plants were inoculated mechanically using sterilized cotton swabs. Source plants for mechanical inoculation of viruses to control non-transgenic and transgenic tobacco plants were PVA-infected N. tabacum cv. Samsun NN, PVY-O- and PVY-N-infected N. tabacum cv. Samsun NN, PMTV-infected N. benthamiana, and TRV-infected N. benthamiana. Source plants for aphid transmission were PVY-O- and PVY-N-infected tobacco plants obtained from the Scottish Agricultural Scientific Agency (SASA), as well as PVA-infected tobacco and PLRV-infected Physalis floridana. PVA and PLRV source plants for aphid transmission were obtained from B. Fenton, whereas PVA, PMTV, PVY-O, PVY-N, and TRV inocula for mechanical transmission were obtained from T. Canto, G. Cowan, and S. MacFarlane, all of the James Hutton Institute (formerly the Scottish Crop Research Institute). Source plants of all the viruses were maintained in a glasshouse and renewed every month.
For transmission of virus, approximately five green peach aphids (Myzus persicae) of the second or third instars were used for each plant. The duration of pre-acquisition starvation was 3–4 h for PVY and PVA, and no pre-acquisition starvation period was used for PLRV. The period of virus acquisition was 5 min for PVY and PVA, and 3 days for PLRV. The duration of feeding on test plants was 24 h for PVY and PVA, and 3 days for PLRV. Insects were killed by fumigation with nicotine shreds.
The primers used for the reverse transcription-PCR (RT-PCR) verification of virus resistance are given in Supplementary Table S2. The RT reaction was carried out in 10 μl containing 1 μl of 10 μM specific primer and 1 μl of total plant RNA. The mixture was heated at 70°C for 10 min and then placed immediately on ice onto which 2 μl of 10× PCR buffer, 2 μl of 25 mM MgCl2, 2 μl of 10 mM dNTPs, and 1 μl of MuLV reverse transcriptase (Roche, USA) were added. The RT reaction was incubated at 37°C for 1 h. The reaction volume of the PCR was 30 μl, containing 5 μl of RT product, 2 μl of 25 mM MgCl2, 3 μl of 10× PCR buffer, 1 μl of 10 mM dNTPs, 1 μl of 10 μM each primer, and 0.5 μl of Taq DNA polymerase (Roche, USA). The PCR was then performed as described above.
For protein analysis, 0.3 g of leaf tissue was ground in 300 μl of 1× PBS buffer and centrifuged in an Eppendorf microcentrifuge. The supernatant was mixed with 5× sample buffer (60 mM Tris-HCl pH 6.8, 25% glycerol, 2% SDS, 14.4 mM 2-mercaptoethanol, and 0.1% bromophenol blue) and boiled for 5 min before loading onto a 12.5% polyacrylamide gel containing 0.1% SDS for separation of the protein samples. The samples were run at 100 V for 1.5 h. The protein was transferred to nitrocellulose membranes at 250 mM for 1 h, after which the membranes were blocked, probed with antiviral primary antisera, followed by probing with a secondary antiserum, which then was detected using the NBT/BCIP system (Sigma) for color development—all these according to standard procedures [23]. Rabbit antisera against viral CPs were obtained from the American Type Culture Collection (ATCC) for PVA (PVAS-266T), PLRV (PVAS-659), and TRV (PVAS-820), from the Plant Virus GenBank (Seoul Women’s University, Korea) for PVY (PVAS-50A), and from J.P.T. Valkonen (University of Helsinki) for PVA. A mouse monoclonal antiserum against PLRV (PVAS-649) was obtained from the ATCC. The secondary antiserum used was either an anti-rabbit IgG conjugated to alkaline phosphatase for polyclonal antibody or an anti-Mouse IgG conjugate (Promega, USA).
Results
Strategy for generation of plants expressing tandem, multi-virus hairpin dsRNAs
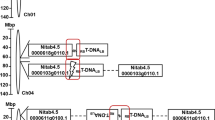
Plasmid vectors were constructed that contained tandem insert sequences derived from either three or five viruses. The three viruses were PVY-O, PVA, and PLRV. The five viruses were PVY, PVA, PLRV, PMTV, and TRV. Sequences of 200 bp from each viral genome cDNA clone were amplified by PCR and then joined by overlapping PCR to generate either 600-bp fragments containing sequences of PVY, PVA, and PLRV, or 1000-bp fragments containing sequences of all the five viruses (Supplementary Fig. S1). The particular viral sequences chosen for amplification were based on regions showing the least variation among isolates in the GenBank or in the case of PVY-O, the region showing the least variation between PVY-O and PVY-N (Supplementary Table S3). This included sequences derived from the CP-coding sequence of PVY, and PLRV, the CI-coding sequence of PVA, and the REP genes of PMTV and TRV (see Supplementary Fig. S1). The CP-coding regions of PVY and PLRV have been used successfully in sense RNA-expressing constructs [11, 15, 16, 31, 32]. The REP regions of PMTV and TRV were chosen, since infection with these viruses can still occur in the absence of the RNA segment encoding the CP gene [20, 33–35]. These PCR products were then inserted into the Gateway-compatible version of the plant transformation vector pFGC5941, at two sites flanking an intron in opposite orientation (Fig. 1a, b), such that hairpin RNAs would be produced upon transcription in planta from the upstream cauliflower mosaic virus 35S RNA promoter (Fig. 1c). The viral sequences were inserted in two orientations relative to the introns such that they would be expressed in the hairpin structure in either the sense–intron–antisense orientation (IN), or the antisense–intron–sense orientation (OUT) (Fig. 1a, b).
Constructs used for transformation of tobacco to express inverted tandem repeat viral-derived dsRNAs. The plasmid inserts contained 200-bp fused segments of three viruses (a) or five viruses (b) inserted into the Gateway-compatible vector pFGC5941, twice, in opposite orientations (c), such that the transgene would produce a 600-bp dsRNA (a) or a 1000-bp dsRNA (b), both flanking an intron. The viral sequences were derived from the coat protein (CP)-coding sequences of PVY-O and PLRV, the cylindrical inclusion (CI)-coding sequences of PVA, and the replicase (REP)-coding sequences of TRV and PMTV. Upon transcription the orientation of the inserted viral sequences flanking the intron was either sense–intron–antisense (IN) or antisense–intron–sense (OUT). The vector containing the fused viral sequences (1200 bp or 2000 bp) (c) was used for transformation sequentially of Agrobacterium and then tobacco
After characterization for the presence and orientation of the inserts (not shown), the four plasmids (pFGC5941/1000-1-2-OUT, pFGC5941/1000-101-3-IN, pFGC5941/600-3-OUT, and pFGC5941/600-4-IN) were then transformed into A. tumefaciens that in turn were utilized to transform tobacco leaf disks. Twenty leaf disks were co-cultivated with Agrobacterium containing each of the four plasmids. The number of regenerated plants obtained was highly variable. Between 3 and 20 plants were regenerated from each set of 20 leaf disks (Supplementary Table S4), with the two plasmids containing viral sequences in the OUT orientation generating fewer plants than the corresponding plasmids containing viral sequences in the IN orientation. After further selection by resistance to the herbicide glufosinate (encoded by the BAR gene in the plasmid pFGC5941), only two and four lines of herbicide-resistant tobacco were obtained from the constructs pFGC5941/1000-1-2-OUT and pFGC5941/1000-101-3-IN, respectively, while five and 13 lines of tobacco were obtained from the constructs pFGC5941/600-3-OUT and pFGC5941/600-4-IN, respectively (Supplementary Table S4). The transgenic lines and plants will subsequently be referred to without the name of the transformation vector (pFGC5941).
Screening of transgenic lines: resistance to PVY-O and TRV
Plants lines of the T1-generation were screened initially by PCR for the presence of the transgene (Supplementary Table S5). Three lines did not contain the transgene and were not considered further. T1-generation progeny of the remaining 21 T0 lines were collected and were screened for the presence of the transgene by PCR and for the presence of siRNAs against each virus sequence contained in the transgene cassette. The number of T1 lines positive by both tests varied from zero to 23, with some T1 lines still segregating (Supplementary Table S5). T2-generation seeds were collected from various pods on the T1 plant lines that were positive for the presence of the transgene and siRNAs, and these were screened for resistance to infection by PVY-O and TRV introduced by mechanical inoculation (Table 1). T2 progeny plants derived from the two tobacco T0 lines 1000-1-2-OUT produced seeds resistant to infection by both viruses, while only T2 plants derived from one of the four T0 tobacco lines 1000-101-3-IN showed resistance to both viruses. In addition, T2 plants derived from four of the five T0 lines 600-3-OUT were resistant to infection by both viruses, while T2 plants derived from eight of the 13 tobacco T0 lines 600-4-IN showed resistance to both viruses (Table 1). Examples of T2 transgenic plants from several lines showing resistance or susceptibility to infection by PVY-O are shown in Supplementary Fig. S2. These include two resistant 1000-OUT lines, two resistant 600-OUT lines, and one still segregating 600-OUT line. Further characterization was then done using T2 plants from one line each of the four types of constructs. These lines were 1000-1-2-OUT-A1, 1000-101-3-IN-C3, 600-3-OUT-B2, and 600-4-IN-A1.
Characterization of the T2 lines
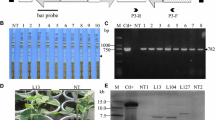
Individual T2-generation plants of the above regenerated lines were examined by Southern blot hybridization for the presence of the transgene cassette, after digestion of the plant DNA with the restriction endonuclease EcoRI (Fig. 2a), which occurs once in the T-DNA (Fig. 1c) and never in the inserts. All the T2 plants examined from the line 1000-1-2-OUT-A1 showed the presence of a band larger than 10 kb, while eight of 13 plants examined also showed the presence of a second band of about 9.4 kb (Fig. 2a and data not shown), indicating that some plants were still segregating a second independent insertion event from the common insertion event. All the T2 plants examined from the line 1000-101-3-IN-C3 showed the presence of a single band of less than 6 kb, as did the T2 plants from the line 600-3-OUT-B2. By contrast, all the T2 plants examined from the line 600-4-IN-A1 showed the presence of a single band of about 9.4 kb (Fig. 2a).
Hybridization analysis of T2-generation plants of transgenic tobacco lines. Individual plants of lines 1000-1-2-OUT-A1, 1000-101-3-IN-C3, 600-3-OUT-B2, and 600-4-IN-A1 were examined by (a) Southern blot hybridization for the presence and size of the inserted transgene, and (b) northern blot hybridization for the presence and levels of siRNAs accumulating in the transgenic plants. In (a), the position of size markers are indicated on the left and individual plant numbers are indicated across the top. In (b), the left panel shows the loading control of low mol. wt. RNAs, and the right panel shows the presence of siRNAs hybridizing against a mixture of RNA probes. NC = Negative Control: small RNAs extracted from non-inoculated plants; PC = Positive Control: small RNAs extracted from plants infected by the corresponding virus. Note that no small RNAs were detectable for PVA- or PLRV-infected plants
The presence of siRNAs derived from the transgenes containing sequences of each of the three or five viruses used in the corresponding construct was confirmed in T2-generation plants for each of the four constructs. Examples of each are shown in Fig. 2b and in Supplementary Fig. S2. In comparison to the levels of siRNAs in the other three transgenic lines, the level of siRNAs was much lower for all five viruses in T2 plants of the line 1000-101-3-IN-C3 (Fig. 2b and data not shown). Interestingly, the positive controls (lower molecular weight RNAs from plants infected by the corresponding viruses) did not show detectable levels of siRNAs in the plants infected by PLRV or PVA, and only a weak signal after infection by TRV, whereas the transgenic plants all showed the presence of such siRNAs. All T2 transgenic plants in the resistant lines showed the accumulation of siRNAs against the target viruses, while all the transgenic plants of T2 susceptible lines showed accumulation of lower levels of such siRNAs. In addition, one line still segregating for resistance showed detectable accumulation of siRNAs in the resistant plants and not in the susceptible plants (Supplementary Fig. S2 and data not shown).
Resistance to infection by mechanical inoculation, agroinfection, or grafting
Fifty T2-generation plants of the four lines listed above were then assessed for resistance to infection after mechanical inoculation with PVY-O, PVY-N, PVA, or a mixture of PVY-O and PVA (Table 2). The plants were assessed for resistance during 15–20 days post-inoculation, by western blot analysis using antisera against the various virus CPs. Plants of line 1000-1-2-OUT-A1, expressing sequences of five viruses (PVY, PVA, PLRV, PMTV, and TRV), showed resistance in all the plants tested to PVY-O and PVA, as well as to PVY-N, while all 50 control, non-transgenic plants became infected by all the three viruses. In addition, all the tested plants of this line also were resistant to infection by a mixture of PVY-O and PVA. By contrast, the plants of line 1000-101-3-IN-C3, also expressing sequences of the same five viruses, showed no resistance to PVY-O or PVY-N, and only 38% resistance to PVA. Moreover, when PVY-O and PVA were co-inoculated, the plants still showed no resistance to PVY-O, but only 22% were resistant to PVA (Table 2). However, as 30% of the control, non-transgenic plants inoculated with PVY-O plus PVA did not become infected by PVA (although they all became infected by PVY-O), there may have been some interference of infection by PVA in the presence of PVY, rather than any measurable resistance to PVA in the doubly inoculated plants. The plants of lines 600-3-OUT-B2 and 600-4-IN-A1, expressing sequences of three viruses (PVY, PVA, and PLRV), all showed 100% resistance to PVY-O, PVY-N, PVA, and a mixture of PVY-O and PVA, regardless of whether the insert orientation was IN or OUT, while the control, non-transgenic plants showed no resistance to infection by the individual viruses (Table 2).
T2 plants of the transgenic lines 1000-1-2-OUT-A1 and, 1000-101-3-IN-C3 also were assessed for resistance to infection by TRV after mechanical inoculation. None of 50 inoculated plants from transgenic line 1000-1-2-OUT-A1 became infected systemically by TRV, while all the 50 control, non-transgenic plants became infected by TRV; however, 35 of the 50 inoculated plants from transgenic line 1000-101-3-IN-C3 became infected by TRV (Table 3). Thus, as was the case with PVA, plants of line 1000-101-3-IN-C3 only showed a partial resistance.
PLRV is not transmissible mechanically, except in the presence of umbraviruses [36]; however, PLRV can be introduced into plants cells by agroinfection [19]. Therefore, T2 plants of both lines, 1000-1-2-OUT-A1 and 1000-101-3-IN-C3, were assessed for resistance to systemic infection by PLRV inoculated to the plants by agroinfection. All the six control, non-transgenic plants became infected systemically by PLRV. Similarly, none of six plants tested of each transgenic line showed resistance to infection by PLRV when the virus was introduced by agroinfection (Table 3). Other transgenic lines were not tested using this method of inoculation.
Although PMTV had been reported to be transmissible mechanically to N. tabacum cv. Xanthi nc and produced a limited systemic infection during the winter months [37], we were unable to transmit PMTV by mechanical inoculation to N. tabacum cv. Samsun NN (data not shown). However, PMTV could be transmitted to N. benthamiana by rubbing inocula onto leaves and the virus infected the plants systemically, as described previously [37]. Therefore, scions of T2 transgenic tobacco plants were side-grafted onto PMTV-infected N. benthamiana plants to assess whether plants of the lines 1000-1-2-OUT-A1 and, 1000-101-3-IN-C3 were resistant to infection by PMTV transmitted through grafting. While three of the five non-transgenic plants so grafted were infected by PMTV, none of five grafted plants from either transgenic line became infected by PMTV (Table 3). Thus, there is some level of resistance to PMTV in these two transgenic lines.
Resistance to infection by viruses transmitted by aphids
PVY-O, PVY-N, PVA, and PLRV are all transmitted naturally by aphids. Therefore, we assessed whether plants of the various transgenic lines were resistant to infection by these viruses, when transmitted by the aphid Myzus persicae.
The T2 plants of line 1000-1-2-OUT-A1 inoculated with PVY-O using aphids showed no infection by either PVY-O or PVY-N (Table 4). Similarly, the tested T2 plants of this line showed no infection by PVA, although only two-thirds of the control, non-transgenic plants became infected when using aphids as vectors for transmission of PVA. Similarly, all of the T2 plants of line 1000-1-2-OUT-A1 inoculated using aphids, which had probed on plants infected with both PVY-O and PVA, did not become infected by either virus, although 90.9% and 79.5% of the control, non-transgenic plants inoculated with both viruses became infected with PVY-O and PVA, respectively (Table 4). In contrast to the situation with mechanical inoculation of these two viruses (Table 2), PVY-O did not appear to affect aphid-mediated infection by PVA, which itself was less efficiently transmitted by aphids than PVY-O when both viruses were inoculated separately (Table 4). While these transgenic plants showed high levels of resistance to PVY-O, PVY-N, and PVA, they showed much less resistance to PLRV introduced by the same aphid vector species, with 38.3% of the transgenic plants being not infected versus 8.8% of the control, non-transgenic plants (Table 4).
T2 plants of transgenic line 1000-101-3-IN-C3 showed either incomplete, less, or no resistance to infection by these four aphid-transmitted viruses: 50% of the tested transgenic plants of this line showed no infection by PVY-O, while all of the tested transgenic plants of this line showed infection by PVY-N; 93% of the tested transgenic plants of this line were not infected by PVA, and 21% were not infected by PLRV (Table 4). Surprisingly, when aphids were used for inoculating transgenic plants of this line with both PVY-O and PVA, the number of plants infected by both viruses increased sharply, although still below the levels observed in non-transgenic plants infected by both viruses via aphids (Table 4).
As was the case with mechanical inoculation of T2 plants of lines 600-3-OUT-B2 and 600-4-IN-A1, T2 transgenic plants of these two lines inoculated with PVY-O, PVY-N, PVA, or PVY-O plus PVA using aphids did not contain any detectable infection. However, these plants were partially or even largely susceptible to infection by PLRV via aphids (Table 4); in particular, in plants of line 600-4-IN-A1, 85% of the plants were infected by PLRV, versus in plants of the line 600-3-OUT-B2, where 57% of the plants became infected. Thus, as with the transgenic lines expressing sequences of five viruses, the transgenic lines expressing sequences of three viruses showed differences between IN and OUT lines in the extent of resistance to PLRV, with no lines providing 100% resistance to PLRV.
Discussion
The results presented here show that multiple virus resistances can be achieved by the use of constructs containing tandem repeats of fused segments of 200 bp of each virus, either for three viruses or five viruses. The transgenic plants producing siRNAs against three viruses showed excellent resistance to two of the three viruses (PVY and PVA), but less effective resistance against PLRV (Tables 2, 3, and 4). This was not the case for the transgenic plants producing siRNAs against five viruses, where resistance against three of the viruses (PVY, PVA, and TRV) depended also on the transgenic line (Table 2, 3, and 4), although resistance against PLRV also was only partial in both lines. None of the inoculated plants showed early symptoms followed by a recovery from infection. Recovery from infection is not a general feature associated with the use of inverted hairpin constructs [4–6, 14].
Unfortunately, we could not thoroughly evaluate the resistance to PMTV, as this virus was not transmissible mechanically to the tobacco cultivar we use for the transformation. Thus, we were only able to demonstrate resistance to PMTV introduced by graft-inoculation. Previous study using transgenic plants expressing the CP of PMTV in transgenic N. benthamiana showed that while these plants were resistant to accumulation of PMTV RNA 3 (encoding the CP) after mechanical inoculation of the virus to leaves, they were not resistant to accumulation of viral RNA 3 in the roots after soil transmission of the virus by its plasmodiophorid vector [33]. Thus, the issue of whether resistance to PMTV introduced by graft inoculation would also lead to resistance to PMTV introduced through infective soil remains to be examined.
Resistance to infection by virus introduced via mechanical inoculation versus by using aphids was largely similar (Table 2 versus Table 4). For example, lines 1000-1-2-OUT-A1, 600-3-OUT-B2 and 600-4-IN-A1 all showed 100% resistance to infection by PVY-O, PVY-N, or PVA, as well as PVY-O plus PVA, and much less resistance to infection by PLRV. However, plants of line 1000-101-3-IN-C3 inoculated mechanically showed moderate (38%) resistance to PVA and no resistance to infection by PVY-O, PVY-N, or either PVY-O or PVA from a mixed inoculum of PVY-O plus PVA. By contrast, plants of the same line inoculated using aphids showed moderate (50%) resistance to PVY-O, no resistance to PVY-N, and good (93%) resistance to PVA, but little or no significant resistance to PVY-O mixed with PVA. Thus, plants of lines resistant to virus applied mechanically are also likely to be resistant to virus introduced by aphids, although this would need to be verified in the field under higher inocula pressure and various abiotic stresses.
The 200 nt of the CP-encoding sequences of PVY-O, which were chosen for preparing the constructs, showed no more than 18-nt differences from the various corresponding PVY-N sequences available from the GenBank for this region (Supplementary Table S2). These sequences also contained regions of 21-nt, 37-nt, and 39-nt contiguous sequences with no differences between PVY-O and PVY-N, which may account for the ability of the siRNAs generated from this region to engender resistance to both PVY-O and PVY-N in most of the lines tested. A different CP-encoding sequence of a PVY-N isolate, but also with high sequence identity to other PVY strains and isolates, was used previously to obtain resistance in potato to PVY strains O, N, and NTN by expressing a 605-bp hairpin structure [32].
We cannot easily explain the excellent resistance observed in most of the lines examined in detail to infection by PVY and PVA, but not to PLRV (Tables 2, 3, and 4). Previous studies involving transgenic plants expressing sequences of PLRV showed variation in the extent of resistance, leading to a reduction in either the level of virus or the incidence of infection [11, 16, 31, 38–40]. We also observed a reduction in the incidence of infection. Thus, there may be some differences in the extent to which either the infecting PLRV is able to overcome RNA silencing versus the other viruses, or RNA silencing occurs in phloem tissues.
In general, the number of the recovered transgenic lines was fewer for the constructs that contained two 1000-bp inserts than for those containing two 600-bp repeats (Supplementary Table S4), which may indicate a limitation as to how much additional sequence (a) will be tolerated by this particular vector, (b) could be transferred during the integration process, (c) would remain stably integrated, or (d) would not succumb immediately to cis-mediated transcriptional gene silencing. Thus, it may be necessary to use individual virus segments of fewer than 200 bp. While such shorter segments have been utilized to either demonstrate RNA interference [41–45] or obtain resistant transgenic plants [6, 46–49], using very short segments is fraught with the risk of the small RNA (siRNA or miRNA) being less able to interact with the small target, due presumably to local secondary structure in the target RNA [48], becoming ineffectual because of mutation in the viral target, especially at the 5′ end of the small RNA [50], and/or off-target effects of the small RNA [44], despite the small size of these RNAs.
We also saw fewer transgenic lines generated using constructs with the viral segments in the OUT orientation than the IN orientation (Supplementary Table S4), although the significance of this is unknown. Surprisingly, the two IN lines used in the most of the study performed here showed some degree of less resistance to virus infection than the two OUT lines used here (Tables 2, 3, and 4). While we cannot conclude from these data that IN lines, in general, may show less resistance than OUT lines, this was the general conclusion from the study of Bucher et al. [6] with transgenic plants expressing sequences of four tospoviruses in tandem. Further study using different transformation vectors, larger numbers of independent transgenic lines, and different plant species would be needed to establish the validity of this conclusion.
As was the case with previous studies [6, 14, 51, 52], we also observed a correlation between the presence of high levels of siRNAs in the transgenic plants and resistance. However, we also saw less or no resistance in a line that accumulated a lower level of siRNAs (Tables 2, 3, and 4 and Fig. 2b), consistent with previous studies [6, 14, 51, 52]. Moreover, as described in [53], the use of constructs expressing hairpin RNAs is not sufficient to ensure the production of siRNAs. Thus, not all such lines will produce siRNAs and not all the lines producing siRNAs will show resistance to the corresponding target viruses.
References
M. Prins, M. Laimer, E. Noris, J. Schubert, M. Wassenegger, M. Tepfer, Mol. Plant Pathol. 9, 73–83 (2008)
M. Fuchs, J.R. McFerson, D.M. Tricoli, J.R. McMaster, R.Z. Deng, M.L. Boeshore, J.F. Reynolds, P.F. Russell, H.D. Quemada, D. Gonsalves, Mol. Breed. 3, 279–290 (1997)
D.M. Tricoli, K.J. Carney, P.F. Russell, J.R. McMaster, D.W. Groff, K.C. Hadden, P.T. Himmel, J.P. Hubbard, M.L. Boeshore, H.D. Quemada, Nat. Biotechnol. 13, 1458–1465 (1995)
N.A. Smith, S.P. Singh, M.-B. Wang, P.A. Stoutjesdijk, A.G. Green, P.M. Waterhouse, Nature 407, 319–320 (2000)
C.-X. Zhu, Y.-Z. Song, G.-H. Yin, F.-J. Wen, J. Phytopathol. 157, 101–107 (2009)
E. Bucher, D. Lohuis, P.M.J.A. van Poppel, C. Geerts-Dimitriadou, R. Goldbach, M. Prins, J. Gen. Virol. 87, 3597–3701 (2006)
H. Barker, M.F.B. Dale, in Natural Resistance Mechanisms of Plants to Viruses, ed. by G. Loebenstein, J.P. Carr (Springer, Dordrecht, The Netherlands, 2006), pp. 341–366
C. Hemenway, R.-X. Fang, W.K. Kaniewski, N.-H. Chua, N.E. Tumer, EMBO J. 7, 1273–1280 (1988)
C.M.P. Van Dun, J.F. Bol, Virology 167, 649–652 (1988)
A. Hoekema, M.J. Huisman, L. Molendijk, P.J.M. van den Elzen, B.J.C. Cornelissen, Nat. Biotechnol. 7, 273–278 (1989)
L.M. Kawchuk, R.R. Martin, J. McPherson, Mol. Plant Microbe Interact. 3, 301–307 (1990)
B. Reavy, M. Arif, S. Kashiwazaki, K.D. Webster, H. Barker, Mol. Plant-Microbe Interact. 8, 286–291 (1995)
E.I. Savenkov, J.P.T. Valkonen, J. Gen. Virol. 82, 2275–2278 (2001)
A. Germundsson, J.P.T. Valkonen, Virus Res. 116, 208–213 (2006)
C. Lawson, W. Kaniewski, L. Haley, R. Rozman, C. Newell, P. Sanders, N. Tumer, Nat. Biotechnol. 8, 127–134 (1990)
M. Arif, P.E. Thomas, J.M. Crosslin, C.R. Brown, Pak. J. Bot. 41, 945–954 (2009)
V. Paalme, E. Gammelgård, L. Järvekülg, J.P.T. Valkonen, J. Gen. Virol. 85, 739–747 (2004)
H. Barker, K.D. McGeachy, N. Toplak, K. Gruden, J. Zel, I. Browning, Am. J. Pot. Res. 86, 227–238 (2009)
K.M. Nurkiyanova, E.V. Ryabov, U. Commandeur, G.H. Duncan, T. Canto, S.M. Gray, M.A. Mayo, M.E. Taliansky, J. Gen. Virol. 81, 617–626 (2000)
E.I. Savenkov, A. Germundsson, A.A. Zamyatnin Jr., M. Sandgren, J.P.T. Valkonen, J. Gen. Virol. 84, 1001–1005 (2003)
H. Liu, B. Reavy, M. Swanson, S.A. MacFarlane, Virology 298, 232–239 (2002)
O. Nyamsuren, C. Firnhaber, N. Hohnjec, A. Becker, H. Küster, F. Krajinski, Plant Sci. 173, 84–95 (2007)
J. Sambrook, D.W. Russell, Molecular Cloning: A Laboratory Manual, 3rd edn. (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, 2001)
M. Holsters, D. de Waele, A. Depicker, E. Messens, M. van Montagu, J. Schell, Mol. Gen. Genet. 163, 181–187 (1978)
R.B. Horsch, J.E. Fry, N.L. Hoffman, D. Eichholz, S.G. Rogers, R.T. Fraley, Science 279, 1486–1487 (1985)
F. Rakhshandehroo, M. Takeshita, J. Squires, P. Palukaitis, Mol. Plant-Microbe Interact. 22, 1312–1318 (2009)
T. Canto, F. Cillo, P. Palukaitis, Mol. Plant-Microbe Interact. 15, 1137–1146 (2002)
C. Llave, K.D. Kasschau, J.C. Carrington, Proc. Natl Acad. Sci. USA 97, 13401–13406 (2000)
J.J. Doyle, E.E. Dickson, Taxon 36, 715–722 (1987)
E. Foster, D. Schneiderman, M. Cloutier, S. Gleddie, L.S. Robert, Plant J. 31, 477–486 (2002)
H. Barker, B. Reavy, A. Kumar, K.D. Webster, M.A. Mayo, Ann. Appl. Biol. 120, 55–64 (1992)
A. Missiou, K. Kalantidis, A. Bouta, S. Tzortzakaki, M. Tabler, M. Tsagris, Mol. Breed. 14, 185–197 (2004)
A. Germundsson, M. Sandgren, H. Barker, E.I. Savenkov, J.P.T. Valkonen, J. Gen. Virol. 83, 1201–1209 (2002)
K.D. McGeachy, H. Barker, Mol. Plant-Microbe Interact. 13, 125–128 (2000)
S.A. MacFarlane, Mol. Plant Pathol. 11, 577–583 (2010)
E. Ryabov, G. Fraser, M.A. Mayo, H. Barker, M. Taliansky, Virology 286, 363–372 (2001)
B.D. Harrison, R.A.C. Jones, Ann. Appl. Biol. 65, 393–402 (1970)
L.M. Kawchuk, R.R. Martin, J. McPherson, Mol. Plant Microbe Interact. 4, 247–253 (1991)
F. Van der Wilk, D.P.-L. Willink, M.J. Huisman, H. Huttinga, R. Goldbach, Plant Mol. Biol. 17, 431–439 (1991)
P.E. Thomas, E.C. Lawson, J.C. Zalewski, G.L. Reed, W.K. Kaniewski, Virus Res. 71, 49–62 (2000)
C.L. Thomas, L. Jones, D.C. Baulcombe, A.J. Maule, Plant J. 25, 417–425 (2001)
C. Lacomme, K. Hrubikova, I. Hein, Plant J. 34, 543–553 (2003)
S. Ossowski, R. Schwab, D. Weigel, Plant J. 53, 674–690 (2008)
S. Praveen, S.V. Ramesh, A.K. Mishra, V. Koundal, P. Palukaitis, Transgenic Res. 19, 45–55 (2010)
Y. Tang, F. Wang, J. Zhao, K. Xie, Y. Hong, Y. Liu, Plant Physiol. 153, 632–641 (2010)
Q.-W. Niu, S.-S. Lin, J.L. Reyes, K.-C. Chen, H.-W. Wu, S.D. Yeh, N.-H. Chua, Nat. Biotechnol. 24, 1420–1428 (2006)
J. Qu, J. Ye, R.-X. Fang, J. Virol. 81, 6690–6699 (2007)
C.-G. Duan, C.-H. Wang, R.-X. Fang, H.-S. Guo, J. Virol. 82, 11084–11095 (2008)
X. Zhang, H. Li, J. Zhang, C. Zhang, P. Gong, K. Ziaf, F. Xiao, Z. Ye, Transgenic Res. (2010). doi:https://doi.org/10.1007/s11248-010-9449-3
C. Simón-Mateo, J.A. García, J. Virol. 80, 2429–2436 (2006)
K. Kalantidis, S. Psaradakis, M. Tabler, M. Tsagris, Mol. Plant-Microbe Interact. 15, 826–833 (2002)
Y.K. Chen, D. Lohuis, R. Goldbach, M. Prins, Mol. Breed. 14, 215–226 (2004)
A. Dalakouras, M. Tzanopoulou, M. Tsagris, M. Wassenegger, K. Kalantidis, Transgenic Res. 20, 293–304 (2011)
Acknowledgments
The authors thank Brian Fenton, Stuart MacFarlane, and Graham Cowan of the James Hutton Institute for providing materials, aphids, transmission facilities, advice and/or training. The authors also thank Jari Valkonen of the University of Helsinki and Lilian Järvekülg of the Tallinn University of Technology as well as the Plant Virus GenBank, Seoul Women's University, for providing materials. This study was supported in part by a grant for an International Cooperative Research Project from the Korean Rural Development Agency. PP was supported by a special grant from the Seoul Women’s University in 2010.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Chung, B.N., Palukaitis, P. Resistance to multiple viruses in transgenic tobacco expressing fused, tandem repeat, virus-derived double-stranded RNAs. Virus Genes 43, 454–464 (2011). https://doi.org/10.1007/s11262-011-0655-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11262-011-0655-z