Abstract
A few amino acid differences in the viral protein VP2 account for important antigenic and biological changes among feline parvovirus (FPV), canine parvovirus (CPV-2) and CPV-2 variants 2a and 2b. Several pieces of evidence suggest that CPV-2 is still evolving as additional amino acid changes occurred within the main antigenic regions of CPV-2 capsid, altering the antigenic profile of the virus and stressing the need for implementing the diagnostic assays.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Letter
With interest we read the article by Wang et al. [1] published on Virus Genes. The authors describe the detection of CPV-2 isolates in Taiwan and their characterization by using a PCR-based typing assays published previously [2, 3]. Most of the strains turned to be CPV type-2b and only 2 strains were type-2a.
However, a major point arises by this work and requires to be addressed in detail. CPV-2 emerged during the early 1970s worldwide [4–7]. The virus was shown to be closely related, genetically and antigenically, to feline parvovirus (FPV) and to FPV-like parvoviruses from wild carnivores [8, 9] from which presumably originated by host species shift and subsequent rapid adaptation [10]. In the 1980s, two antigenic variants of CPV-2, distinguishable using monoclonal antibodies (MoAbs), arose almost simultaneously and were termed CPV type-2a and type-2b [11, 12]. Currently, the antigenic variants of CPV have completely replaced the original type-2, and are variously distributed in the canine population world-wide [13–19]. There are at least six or seven amino acid (aa) changes between FPV and CPV-2, and at least five or six aa changes between the variants 2a/b and the original type-2 (Table 1). Such few aa differences in the VP2 of FPV, CPV-2 and CPV-2a/b account for the important antigenic and biological changes observed, setting CPV as an important model to study virus evolution [8, 20–22].
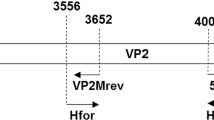
Additional mutations affecting important residues of the capsid protein of CPV, such as residues 297, 300 and 426, have been recognized recently (Table 1), suggesting that CPV is still evolving [16, 23–25]. The mutation Ala-297 was not present in the ‘old’ CPV-2 strains, while it is currently detected in most recent CPV-2 strains, irrespective of the antigenic type [19]. The mutation Asp-300 has been detected only in CPV-2a/b strains isolated from domestic or wild felids in Southern Asia [24]. The mutation 300-Asp is likely the expression of a further adaptation of CPV-2 variants a/b to replicate in the feline host, since replacement of 300-Gly by Asp in a natural mutant selected by serial passages in feline cells [26] and in a mutant created by site-directed mutagenesis has been shown to abolish the ability to replicate in the canine host [27]. The mutation Glu-426 was initially detected in 2000 in Southern Italy [16, 23] and subsequently in Southern Asia [25]. While the mutations Ala-297 and Asp-300 affect the antigenic region located over the shoulder region, the mutation Glu-426 affects the major antigenic region, located over the three-fold spike of CPV-2 capsid. Monoclonal antibodies have been developed that recognize the antigenic variant Asp-300 [28] and Glu-426 [25]. Therefore it is possible to characterize antigenically the variants 2a, 2b, Asp-300 and Glu-426, by assessing the reactivity to a panel of monoclonal antibodies in inhibition of haemagglutination. In addition, sequence analysis of recent CPV-2a isolates has revealed a reversion at position 555 to the sequence of FPV/CPV-2, Ile to Val (Table 1). This mutation restricts the differences among the antigenic variants 2a, 2b and Glu-426 to only one aa at position 426, that is Asn in CPV-2a, Asp in CPV-2b and Glu in the Glu-426 mutant. Most CPV-2 strains spreading currently in Italy differs only in this residue. The evolution observed in the VP2 of CPV-2 has important consequences for the PCR-based genotyping assay described by Pereira [2]. Due to the limited number of nt variations between CPV-2a and -2b, the 2b-specific primers were selected taking advantage of two single nt polymorphisms (SNPs), 4062-A→T and 4449-G→A, that determine the replacement of Asn by Asp at position 426 and of Ile by Val at position 555, in reference CPV-2a and -2b strains, respectively (Fig. 1 and Table 1). Each primer was selected to have one such mutation at the very 3′ end, as nt mismatches that occur at the 3′ end of a primer are highly detrimental to primer extension and strongly decrease PCR amplification. However, most new CPV-2a strains have the mutation 555 Val, due to the nt mutation 4449-G→A. Such mutation is present in CPV-2a strains spreading in Southern Asia, as also evidenced by Wang et al. [1]. Therefore, the PCR-based genotyping system developed by Pereira et al. [2] is no longer able to discriminate between type-2a and type-2b strains, as almost all the novel CPV-2a strains (555-Val) will go mischaracterized as type-2b. In addition, the genotyping system is not able to identify the Glu-426 antigenic variant, that is currently epidemic in Italy, and that has been also described sporadically in Southern Asia [25]. The nt change responsible for the mutation Glu-426 has occurred 2 nt downstream the primer binding region and thus cannot be detected in this PCR-based genotyping system. To address this point, it is necessary to adopt molecular assays able to recognize SNPs. For instance, restriction enzyme digestion of short PCR fragments of CPV-2 genome proved to be useful to discriminate between CPV-2b and Glu-426 strains [23]. In the effort to speed up routine analysis, we have now developed two minor groove binder (MGB) assays able to recognize the SNPs observed in the variants of CPV-2 (Decaro et al., submitted). Altogether, it is likely that in the epidemiological survey by Wang et al. [1] a number of strains that were typed as 2b are type-2a, and that the prevalence of the type-2b variant is overestimated. Also, other antigenic variants could have gone undetected.
Schematic representation of CPV2 genome. The sequence and position of the primers binding regions are shown. The sequence of primer Pbas [2] is reversed and complemented to simplify sequence comparison
In conclusion, in our hands the PCR-based genotyping assay proposed by Pereira [2] failed to predict the antigenic type of field CPV-2 strains, as the strategy developed to amplify selectively type-2b CPVs was affected by the genetic variations that accumulated in CPV-2 genome. Antigenic analysis with monoclonal antibodies, sequence analysis of the VP2 or detection of SNPs by restriction enzyme analysis or by MGB assays are required to characterize the antigenic variants of CPV-2.
References
H.C. Wang W.D. Chen S.L. Lin J.P.W. Chan M.L. Wong (2005) Virus Genes 31 171–174 Occurrence Handle16025242 Occurrence Handle1:CAS:528:DC%2BD2MXmtFyjurw%3D Occurrence Handle10.1007/s11262-005-1791-0
C.A. Pereira T.A. Monezi D.U. Mehnert M. D’Angelo E.L. Durigon (2000) Vet. Microbiol. 75 127–133 Occurrence Handle10889403 Occurrence Handle1:CAS:528:DC%2BD3cXks12lsLw%3D Occurrence Handle10.1016/S0378-1135(00)00214-5
M. Senda C.R. Parrish R. Harasawa K. Gamoh M. Muramatsu N. Hirayama O. Itoh (1995) J. Clin. Microbiol. 33 110–113 Occurrence Handle7699026 Occurrence Handle1:CAS:528:DyaK2MXjsVGns7w%3D
M.J.G. Appel W.F. Scott L.E. Carmichael (1979) Vet. Rec. 105 156–159 Occurrence Handle516347 Occurrence Handle1:STN:280:Bi%2BD1czhs1Q%3D
G. Burtonboy F. Coignoul P.P. Pastoret N. Delferriere (1979) Arch. Virol. 61 1–11 Occurrence Handle316320 Occurrence Handle1:STN:280:Bi%2BD1cnpslI%3D Occurrence Handle10.1007/BF01320586
R.H. Johnson P.B. Spradbrow (1979) Aust. Vet. J. 55 151
W.R. Kelly (1978) Aust. Vet. J. 54 593 Occurrence Handle753224 Occurrence Handle1:STN:280:CSaB3s7ivFM%3D
J.S.L. Parker W.J. Murphy D. Wang S.J. O’Brien C.R. Parrish (2001) J. Virol. 75 3896–3902 Occurrence Handle11264378 Occurrence Handle1:CAS:528:DC%2BD3MXisVSmsL0%3D Occurrence Handle10.1128/JVI.75.8.3896-3902.2001
U. Truyen A. Gruneberg S.F. Chang B. Obermaier P. Veijalainen C.R. Parrish (1995) J. Virol. 69 4702–4710 Occurrence Handle7609035 Occurrence Handle1:CAS:528:DyaK2MXmvFWhu7k%3D
L.A. Shackelton C.R. Parrish U. Truyen E.C. Holmes (2005) Proc. Natl. Acad. Sci. USA 102 379–384 Occurrence Handle15626758 Occurrence Handle1:CAS:528:DC%2BD2MXptFSksQ%3D%3D Occurrence Handle10.1073/pnas.0406765102
C.R. Parrish P.H. O’Connel J.F. Evermann L.E. Carmichael (1985) Science 230 1046–1048 Occurrence Handle4059921 Occurrence Handle1:CAS:528:DyaL28XivFGqtw%3D%3D
C.R. Parrish C.F. Aquadro M.L. Strassheim J.F. Evermann J.Y. Sgro H.O. Mohammed (1991) J. Virol. 65 6544–6552 Occurrence Handle1942246 Occurrence Handle1:CAS:528:DyaK3sXitV2jt7c%3D
D. Buonavoglia A. Cavalli A. Pratelli V. Martella G. Greco M. Tempesta C. Buonavoglia (2000) New Microbiol. 23 93–96 Occurrence Handle10946411 Occurrence Handle1:STN:280:DC%2BD3cvhvVCktQ%3D%3D
R.R. Ybanez ParticleDe C. Vela E. Cortes L. Simarro J.I. Casal (1995) Vet. Rec. 136 174–175 Occurrence Handle7762129
N.M. Greenwood W.S.K. Chalmers W. Baxendale H. Thompson (1996) Vet. Rec. 138 495–496 Occurrence Handle8736503 Occurrence Handle1:STN:280:BymA2cjjs1A%3D
V. Martella A. Cavalli A. Pratelli G. Bozzo M. Camero D. Buonavoglia D. Narcisi M. Tempesta C. Buonavoglia (2004) J. Clin. Microbiol. 42 1333–1336 Occurrence Handle15004112 Occurrence Handle10.1128/JCM.42.3.1333-1336.2004
M. Mochizuki R. Harasawa H. Nakatani (1993) Vet. Microbiol. 38 1–10 Occurrence Handle8128593 Occurrence Handle1:CAS:528:DyaK2cXisVajsb8%3D Occurrence Handle10.1016/0378-1135(93)90070-N
A. Steinel E.H. Venter M. Vuuren ParticleVan C.R. Parrish U. Truyen (1998) Onderstepoort J. Vet. Res. 65 239–242 Occurrence Handle10192835 Occurrence Handle1:STN:280:DyaK1M3gvV2itw%3D%3D
U. Truyen A. Steinel L. Bruckner H. Lutz K. Mostl (2000) Schweiz. Arch. Tierheilkd. 142 115–119 Occurrence Handle10748710 Occurrence Handle1:STN:280:DC%2BD3c3ht1Wqug%3D%3D
K. Hueffer J.S.L. Parker W. Weichert R.E. Geisel J.Y. Sgro C.R. Parrish (2003) J. Virol. 77 1718–1726 Occurrence Handle12525605 Occurrence Handle1:CAS:528:DC%2BD3sXmtFygtA%3D%3D Occurrence Handle10.1128/JVI.77.3.1718-1726.2003
U. Truyen C.R. Parrish (1992) J. Virol. 66 5399–5408 Occurrence Handle1323703 Occurrence Handle1:STN:280:By2A28rht10%3D
U. Truyen J.F. Evermann E. Vieler C.R. Parrish (1996) Virology 215 186–189 Occurrence Handle8560765 Occurrence Handle1:CAS:528:DyaK28XmvFKrtg%3D%3D Occurrence Handle10.1006/viro.1996.0021
C. Buonavoglia V. Martella A. Pratelli M. Tempesta A. Cavalli D. Buonavoglia G. Bozzo G. Elia N. Decaro L.E. Carmichael (2001) J. Gen. Virol. 82 3021–3025 Occurrence Handle11714979 Occurrence Handle1:CAS:528:DC%2BD3MXovVCksro%3D
Y. Ikeda M. Mochizuki R. Naito K. Nakamura T. Myazawa T. Mikami E. Takahashi (2000) Virology 278 13–19 Occurrence Handle11112475 Occurrence Handle1:CAS:528:DC%2BD3cXosFGnsrc%3D Occurrence Handle10.1006/viro.2000.0653
M. Nakamura Y. Tohya T. Miyazawa M. Mochizuki H.T. Phung N.H. Nguyen L.M. Huynh L.T. Nguyen P.N. Nguyen P.V. Nguyen N.P. Nguyen H. Akashi (2004) Arch. Virol. 149 2261–2269 Occurrence Handle15503211 Occurrence Handle1:CAS:528:DC%2BD2cXoslGitr0%3D Occurrence Handle10.1007/s00705-004-0367-y
C.R. Parrish L.E. Carmichael (1986) Virology 148 121–132 Occurrence Handle3942033 Occurrence Handle1:CAS:528:DyaL28Xot1yksg%3D%3D Occurrence Handle10.1016/0042-6822(86)90408-3
J.S.L. Parker C.R. Parrish (1997) J. Virol. 71 9214–9222 Occurrence Handle9371580 Occurrence Handle1:CAS:528:DyaK2sXnsVCqurk%3D
M. Nakamura K. Nakamura T. Miyazawa Y. Tohya M. Mochizuki H. Akashi (2003) Clin. Diagn. Lab. Immunol. 10 1085–1089 Occurrence Handle14607871 Occurrence Handle1:CAS:528:DC%2BD3sXpslGns70%3D Occurrence Handle10.1128/CDLI.10.6.1085-1089.2003
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Martella, V., Decaro, N. & Buonavoglia, C. Evolution of CPV-2 and Implicance for Antigenic/Genetic Characterization. Virus Genes 33, 11–13 (2006). https://doi.org/10.1007/s11262-005-0034-8
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/s11262-005-0034-8