Abstract
Tribological evaluation of biological materials, such as cells and tissues, present both opportunities and challenges to experimentalists. When working with living materials, maintaining homeostasis during testing in vitro or in vivo often requires appropriate control of the environment, selection of the testing time and duration, applied loads, and shear stresses. This manuscript provides much of the background and design information used in the development of a microtribometer that has been modified to perform biotribology measurements in vitro. The focus of this manuscript is on mammalian cells in monolayer, and a series of order-of-magnitude calculations are used to inform future instrument designs and considerations, including: sliding speed ranges from 10 nm/s to 100 mm/s, contact pressures less than 6 kPa, temperature ~ 37 °C, and contact areas on the order of 1,000’s of µm2. The design and development of these biotribology instruments enable in situ fluorescence microscopy and allow for statistically significant gene expression analyses such as quantitative reverse-transcription polymerase chain-reaction (RT-qPCR) and enzyme-linked immunosorbent assay (ELISA).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Biomedical implants make extensive use of soft materials for contact lenses, clamps, catheters, and soft tissue prostheses. A large number of soft materials are already approved for implantation, including hydrogels, silicones, and decellularized tissue matrix. The FDA generally relies on toxicological analyses to evaluate the “biocompatibility” of a material [1], but there is growing evidence that inflammatory responses to sliding against soft implants may cause discomfort or adverse effects for some patients [2,3,4,5,6]. In vitro models are currently in development to determine the precise mechanisms and extent to which frictional shear affects cell mechanics and regulation [7, 8]. Efforts to recapitulate the physical interactions between biomedical implants and biological surfaces in vitro may use sliding pairs of soft materials against living mammalian cells grown in monolayer or multilayer under cell culture conditions that maintain homeostasis. Tribological examination of hypotheses aimed at elucidating fundamental mechanisms of action may involve simplified systems of one or two cell types under laboratory-controlled conditions of material pairings, applied load, contact pressure, sliding speed, and duration [9,10,11,12,13,14]. The low elastic modulus of cells and the relative thinness and fragility of cell monolayers present challenges to multi-cell contact testing [15]. Fluorescence microscopy is one of the most useful tools among cell biologists, and it is almost essential for biotribology measurements to permit examination of the cell layers at a minimum before-and-after testing but ideally in situ during testing.
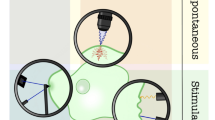
The in situ biotribometer (Fig. 1a) is an instrument that can directly measure dynamic interactions of biological interfaces during contact and sliding. As an experimental apparatus, the biotribometer should be capable of in situ optical capabilities while performing indentation [16, 17] and friction experiments [8, 14] on living biological materials and allow for statistically significant gene expression analysis. Ultimately, in situ biotribology instrumentation can help to build strong connections across the fields of tribology, rheology, and biology.
Illustration of the biotribometer. a The biotribometer performs sliding experiments against cell monolayers within an incubator that maintains physiologically relevant conditions. b The glass-bottom culture dish has two stadium-shaped wells within which two identical cell populations grow under the same conditions separated by a thin partition. Prior to testing, some growth media is removed to isolate both cell populations
2 Flexure-Based Design Strategy and Measurement Uncertainties
The biotribometer is based on earlier designs of the microtribometer [11, 18,19,20], which measures normal and friction forces through measurements of cantilever deflections. The cantilevers are fabricated in a double-leaf configuration in an effort to maintain rectilinear motion with a minimal amount of parasitic motions (rotations). Capacitance probes (5 µm/V sensitivity, Lion Precision) are mounted in the normal and tangential directions. Experience with soft aqueous gels, cells, and tissues shows that the friction coefficients are generally below µ < 0.1, and therefore one of the design approaches is to design the cantilevers with greater compliance in the tangential direction (e.g., normal and tangential stiffness: Kn = 160 µN/µm and Kt = 75 µN/µm, respectively [14, 21,22,23]). Applied normal loads have been maintained at Fn ~ 25 µN and friction forces as low as Ff = 2 µN have been measured; the experimental uncertainties are lower than the noise floor of the measurement, which is on the order of ± 1 µN (Fig. 1a) and is used in all of the propagation of uncertainty calculations.
The uncertainty in friction coefficient follows methods derived in Schmitz et al. [24] and Burris et al. [25], and is given by Eq. 1. Applying two substitutions in this expression unique to biotribology and the biotribometer design of mixed compliance cantilevers (µ < 0.1 and u(Ff) ~ 0.1u(Fn)) gives an approximate solution for friction coefficient uncertainty, Eq. 2.
Eqn. 1 should be used to guide instrument design for tribometers and is discussed in greater detail in [24, 25]. To illustrate the advantage of the mixed compliance cantilever in biotribology and a noise floor on the order of 1 µN, the uncertainty in friction coefficient, u(µ), for a 100 µN experiment is u(µ) ~ 1.4 × 10− 3 (which enabled measurements of superlubricity [23] in gels at applied loads of 500 µN, u(µ) < 3 × 10− 4). In biotribology measurements of cells and tissues, there are often spatial heterogeneities that lead to fluctuations in the measurements far in excess of the experimental uncertainties and it is common to report the standard deviation of the measurement instead of the experimental uncertainty.
3 Contact Pressures
Physiologically relevant experiments in biotribology should match contact pressure at a minimum, and most experiments involve scaled testing where the contact patch is much smaller in vitro than in vivo. This results in in vitro loads that are many orders of magnitude lower than the physiological loads. Hertzian contact mechanics [26,27,28,29] and Winkler foundation models [18, 28, 29] have been widely used to determine pressures for biotribology applications, and the selection is generally based on considerations of the contact width to sample thickness. Because of the relative stiffness of most gels and silicones, as compared to cells and tissues, matching contact pressures over contact widths on the order of 100 µm yields predictions of unrealistically large radius probes. Recently, the design and use of membrane probes [30], which are constant thickness curved spherical caps, has revealed that these probes have a constant contact pressure (i.e., area scales linearly with applied load). These probes have been fabricated by molding and 3D printing techniques [31,32,33,34], and the design of a targeted contact pressure depends on the thickness, radius, and elastic modulus. These probes allow for small radius of curvature, finite contact width, and a contact pressure that can be tuned by altering the membrane thickness rather than the material properties.
4 Sliding Speeds
Sliding speeds in biological applications can range from 10 nm/s (e.g., motor protein motility [35]) to over 100 mm/s (e.g., blink speeds [36] and joint motions [37]). Rather than matching the sliding speeds, it is suggested that matching lubrication conditions (boundary, mixed, soft EHL, and fluid film lubrication) is the more important parameter. Soft EHL equations by Hamrock and Dowson have been widely used to predict minimum film thickness and transitions into full fluid film lubrication [18, 38]. For full fluid film lubrication, Kapitza’s classical equations for a sphere-on-flat configuration can be used to predict film thickness [39]. Under fluid film lubrication, the shear stress is determined by the shear rate and fluid viscosities, and studies of mechanotransduction with cells and tissues should aim to match shear stress.
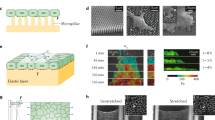
Many biotribology applications are outside the hydrodynamic regime and involve contact between cells, extracellular matrix, mucins, gels, or implant materials (and in some cases all of these materials). At low loads and physiological contact pressures, the resulting sliding speeds are on the order of 1 mm/s or below, which introduces challenges in stage motion control. Linear piezo stages have extremely smooth motion profiles across all speed ranges but have limited displacement ranges (often below 1 mm). Servo-motors often “jitter” unless they are carefully tuned to specific masses, speeds, and damping ratios and are often best at higher speeds. Multiphase-micro-stepper-motor stages have been shown to produce smooth motion profiles through a combination of fine-pitched drive screws and multiple phase excitation of the drive voltages to allow for intermediate positioning between steps to reduce vibrations. The injection of any frequencies of motion pulses, jitter, or steps should be an order of magnitude larger than the natural frequency of the flexures. For the design of this biotribometer, a two-phase micro-stepping linear x-stage (Physik Instrumente, PLS-85 #623493112-0001) capable of up to 52 mm of travel and sliding speeds up to 20 mm/s was used in reciprocated sliding at 1 mm/s; the highest natural frequency of the cantilevers was approximately 35 Hz, and no motor-induced vibrations could be detected using spectral analysis. Representative indentation and friction data are shown in Fig. 2, and analyzed following the methods described in [18, 21, 24,25,26, 29, 40], respectively.
Biotribometers should be designed to address physiologically relevant sliding speeds, contact pressures, and shear stresses during in vitro testing of cells, cell layers, and tissues. The design described herein enables microscale force measurements during indentation and friction studies of biological materials over macroscale contact areas and sliding distances. These data are from non-destructive a indentation and b, c tribological studies of mucinated cell monolayers using high water content aqueous gel probes. Arrows indicate loading and sliding directions and typical experimental conditions are provided. (b) The friction coefficient of each cycle is calculated from the free sliding region of the friction force trace, and (c) average friction coefficients are calculated following the methods described in [18, 24, 25, 40]. See Supplementary Material for in situ imaging during cell friction studies
5 In Situ Imaging and Microscopy
Mounting the biotribometer onto an inverted microscope (optical [41], interferometric [19], epifluorescent [14, 16, 17], or confocal [26, 42]) allows for direct imaging of the contact in situ (see Supplementary Material). Monitoring the responses of cells with fluorescence microscopy [43] allows for a wide variety of fluorescent stains, dyes, and reporters [44] to be quantitatively imaged in situ, and dynamically enables mechanistic studies of cellular responses to direct contact shear. These cellular responses can span a wide range of timescales from milliseconds (e.g., calcium fluctuations [45]) to hours (e.g., cytokine production [46, 47]), and it is important to maintain a growth environment that is conducive to homeostasis for the cells and tissues being studied (Table 1), which frequently requires the development of an incubation chamber around the entire tribometer [7] but at a minimum the samples [14] (Fig. 1a).
6 Strategies for Differential Measurements of Biological Responses
Unlike tribological testing with reference materials, biological samples have significant variabilities from batch-to-batch, patient factors, and history of handling and preparation. An approach that has been recently adopted is to make culture dishes that allow for differential measurements between two populations that were cultured as a single population and then physically isolated prior to testing (Fig. 1b).
Established gene expression analyses (e.g., quantitative reverse-transcription polymerase chain reaction (RT-qPCR) for quantifying RNA; enzyme-linked immunosorbent assay (ELISA) for detecting and quantifying secreted analytes: proteins, peptides, antibodies, or hormones) recommend using roughly 1 million cells to harvest RNA and collect secreted analytes. Table 2 details “rules of thumb” for a suite of molecular biology assays, input material, and corresponding minimum detection thresholds. Direct contact stimulation and shear of 0.1–1.0 million cells require a significant contact area (from 100 mm2 to 1,000 mm2). Spreading out contact with membrane probes (Fig. 3a, b) and sliding over extended track lengths can provide hundreds of thousands of cells for analysis (Fig. 3c). In comparison to the membrane probe, which may have a diameter on the scale of millimeters, mammalian cells range from about 5–20 µm in diameter, or 500–4,000 µm3 in volume [48]. Although cell type, developmental stage, and physiological state greatly influence the genetic content and makeup of a cell, each cell is estimated to contain 10–30 pg of total ribonucleic acid (RNA) [49], 200,000 messenger RNA (mRNA) molecules [50], and upwards of 10 billion total proteins [51]. The majority of RNA molecules are transfer RNA (tRNA) and ribosomal RNA (rRNA), but approximately 1–5% of total RNA is messenger RNA (mRNA) [48]. Following transcription of primary transcript mRNA (pre-mRNA), mature mRNA is translated into a protein. In our experience with corneal epithelial cells (hTCEpi), a ~ 100 mm2 well contains a confluent monolayer of ~ 200,000 cells, which yield a total RNA concentration of about 100 ng/µL, only a small percent of which contains the target sequences of mRNA capable of producing specific proteins (pg/mL). This amount of input RNA is amenable to gene expression analyses using RT-qPCR or cDNA-microarray-based analysis of transcription.
a The in situ biotribometer enables direct observations of contact and sliding of a spherical shell hydrogel probe against cell monolayers in the custom glass-bottom culture dish. b The spherical shell hydrogel probe enables large contact areas and low contact pressures against living cell monolayers. c Contact pressures and shear stresses from tribological testing may induce changes in cellular responses compared to the reference cell population (e.g., cytokine secretion, presentation of cell surface receptors, integrin reorganization, ion flow through gap junctions, cytoskeletal remodeling, gene expression). Changes in cellular responses may be measured in situ with microscopy or with molecular biology techniques (e.g., RT-qPCR, ELISA)
7 Closing Remarks
The widespread use of soft implant materials in biomedicine and the need to examine biocompatibility through tribology have introduced opportunities to collaborate and develop specialized instrumentation for tribological studies on model systems. Cells respond to shear through complex signaling pathways that can be activated in vitro through direct contact shear under physiological pressures and lubrication conditions. The biotribometer is designed with increased sensitivity in the friction direction to reduce uncertainties associated with measurements of low-friction aqueous gels, cells, and tissues. Matching physiological contact conditions from an application to a small population of cells in vitro requires the scaling of the applied load and contact geometry to match contact pressures across the interface. Although it is often intuitive to match sliding speeds between the application and the biotribology testing, the more important scaling considerations are to match lubrication regimes and to maintain smooth, low-vibration stage motions. The inclusion of in situ fluorescence optical microscopy provides a quantitative method of examining mechanotransduction during biotribology studies. Real-time observations of cell signaling and function during biotribology testing provide further opportunities to explore detailed pathways and mechanisms of cellular responses to direct contact shear. A method of performing differential analysis in biotribology testing between two match populations of cells is enabled through an integral separation wall within the sample plate that isolates the populations only at the onset of testing. Roughly, 0.1 to 1 million cells are needed for gene expression analysis such as RT-qPCR and ELISA, which give high-fidelity quantitative analysis of RNA expression and protein secretion and signaling from the cells, respectively. The biotribometer platform and design considerations outlined here provide an opportunity to develop experimental instrumentation designed for in vitro studies in biotribology, mechanotransduction, and biocompatibility, with quantitative in situ characterization of cellular responses that ultimately can assist in the informed design of biomedical implants and devices.
References
Reeve, L., Baldrick, P.: Biocompatibility assessments for medical devices – evolving regulatory considerations. Exp. Rev. Med. Devices. 14, 161–167 (2017). https://doi.org/10.1080/17434440.2017.1280392
Center for Devices and Radiological Health U.S. Food and Drug Administration. Anaplastic Large Cell Lymphoma (ALCL) In Women with Breast Implants: Preliminary FDA Findings and Analyses, http://wayback.archive-it.org/7993/20171115053750/https:/www.fda.gov/MedicalDevices/ProductsandMedicalProcedures/ImplantsandProsthetics/BreastImplants/ucm239996.htm. Accessed Nov 9 2017
U.S. Food and Drug Administration. Medical Device Reports of Breast Implant-Associated Anaplastic Large Cell Lymphoma, https://www.fda.gov/MedicalDevices/ProductsandMedicalProcedures/ImplantsandProsthetics/BreastImplants/ucm481899.htm. Accessed April 15 2018
Nichols, J.J., Willcox, M.D.P., Bron, A.J., Belmonte, C., Ciolino, J.B., Craig, J.P., Dogru, M., Foulks, G.N., Jones, L., Nelson, J.D., Nichols, K.K., Purslow, C., Schaumberg, D.A., Stapleton, F., Sullivan, D.A.: The TFOS international workshop on contact lens discomfort: executive summary. Investig. Ophthalmol. Vis. Sci. 54, 7–13: (2013). https://doi.org/10.1167/iovs.13-13212
Efron, N.: Contact lens wear is intrinsically inflammatory. Clin. Exp. Optom. 100, 3–19 (2017). https://doi.org/10.1111/cxo.12487
Efron, N.: Rethinking contact lens discomfort. Clin. Exp. Optom. 101, 1–3 (2018). https://doi.org/10.1111/cxo.12629
Qin, G., Baidouri, H., Glasser, A., Raghunathan, V., Morris, C., Maltseva, I., McDermott, A.M.: Development of an in vitro model to study the biological effects of blinking. Ocul. Surf. (2018). https://doi.org/10.1016/j.jtos.2017.12.002
Pitenis, A.A., Urueña, J.M., Hart, S.M., O’Bryan, C.S., Marshall, S.L., Levings, P.P., Angelini, T.E., Sawyer, W.G.: Friction-induced inflammation. Tribol. Lett. 66, 81 (2018). https://doi.org/10.1007/s11249-018-1029-7
Cobb, J.A., Dunn, A.C., Kwon, J., Sarntinoranont, M., Sawyer, W.G., Tran-Son-Tay, R.: A novel method for low load friction testing on living cells. Biotechnol. Lett. 30, 801–806 (2008). https://doi.org/10.1007/s10529-007-9623-z
Dunn, A.C., Zaveri, T.D., Keselowsky, B.G., Sawyer, W.G.: Macroscopic friction coefficient measurements on living endothelial cells. Tribol. Lett. 27, 233–238 (2007). https://doi.org/10.1007/s11249-007-9230-0
Dunn, A.C., Cobb, J.A., Kantzios, A.N., Lee, S.J., Sarntinoranont, M., Tran-Son-Tay, R., Sawyer, W.G.: Friction coefficient measurement of hydrogel materials on living epithelial cells. Tribol. Lett. 30, 13–19 (2008). https://doi.org/10.1007/s11249-008-9306-5
Straehla, J.P., Limpoco, F.T., Dolgova, N.V., Keselowsky, B.G., Sawyer, W.G., Perry, S.S.: Nanomechanical probes of single corneal epithelial cells: Shear stress and elastic modulus. Tribol. Lett. 38, 107–113 (2010). https://doi.org/10.1007/s11249-010-9579-3
Sterner, O., Aeschlimann, R., Zürcher, S., Scales, C., Riederer, D., Spencer, N.D., Tosatti, S.G.P.: Tribological classification of contact lenses: from coefficient of friction to sliding work. Tribol. Lett. 63, 9 (2016). https://doi.org/10.1007/s11249-016-0696-5
Pitenis, A.A., Urueña, J.M., Hormel, T.T., Bhattacharjee, T., Niemi, S.R., Marshall, S.L., Hart, S.M., Schulze, K.D., Angelini, T.E., Sawyer, W.G.G.: Corneal cell friction: survival, lubricity, tear films, and mucin production over extended duration in vitro studies. Biotribology. 11, 77–83 (2017). https://doi.org/10.1016/j.biotri.2017.04.003
Pitenis, A.A., Urueña, J.M., McGhee, E.O., Hart, S.M., Reale, E.R., Kim, J., Schulze, K.D., Marshall, S.L., Bennett, A.I., Niemi, S.R., Angelini, T.E., Sawyer, W.G., Dunn, A.C.: Challenges and opportunities in soft tribology. Tribol. Mater. Surfaces Interfaces. 11, 180–186 (2017). https://doi.org/10.1080/17515831.2017.1400779
Zehnder, S.M., Wiatt, M.K., Uruena, J.M., Dunn, A.C., Sawyer, W.G., Angelini, T.E.: Multicellular density fluctuations in epithelial monolayers. Phys. Rev. E. 92, 032729 (2015). https://doi.org/10.1103/PhysRevE.92.032729
Schulze, K.D., Zehnder, S.M., Urueña, J.M., Bhattacharjee, T., Sawyer, W.G., Angelini, T.E.: Elastic modulus and hydraulic permeability of MDCK monolayers. J. Biomech. 53, 210–213 (2017). https://doi.org/10.1016/j.jbiomech.2017.01.016
Rennie, A.C., Dickrell, P.L., Sawyer, W.G.: Friction coefficient of soft contact lenses: measurements and modeling. Tribol. Lett. 18, 499–504 (2005). https://doi.org/10.1007/s11249-005-3610-0
Krick, B.A., Vail, J.R., Persson, B.N.J., Sawyer, W.G.: Optical in situ micro tribometer for analysis of real contact area for contact mechanics, adhesion, and sliding experiments. Tribol. Lett. 45, 185–194 (2012). https://doi.org/10.1007/s11249-011-9870-y
Urueña, J.M., Pitenis, A.A., Harris, K.L., Sawyer, W.G.: Evolution and wear of fluoropolymer transfer films. Tribol. Lett. 57, 9 (2015). https://doi.org/10.1007/s11249-014-0453-6
Pitenis, A.A., Urueña, J.M., Schulze, K.D., Nixon, R.M., Dunn, A.C., Krick, B.A., Sawyer, W.G., Angelini, T.E., Sawyer, G., Angelini, T.E.: Polymer fluctuation lubrication in hydrogel gemini interfaces. Soft Matter. 10, 8955–8962 (2014). https://doi.org/10.1039/C4SM01728E
Urueña, J.M., Pitenis, A.A., Nixon, R.M., Schulze, K.D., Angelini, T.E., Sawyer, W.G.: Mesh Size control of polymer fluctuation lubrication in gemini hydrogels. Biotribology. 1–2, 24–29 (2015). https://doi.org/10.1016/j.biotri.2015.03.001
Pitenis, A.A., Manuel Urueña, J., Cooper, A.C., Angelini, T.E., Sawyer, W.G.: Superlubricity in gemini hydrogels. J. Tribol. 138, 042103 (2016). https://doi.org/10.1115/1.4032890
Schmitz, T.L., Action, J.E., Ziegert, J.C., Sawyer, W.G.: The difficulty of measuring low friction: Uncertainty analysis for friction coefficient measurements. J. Tribol. 127, 673 (2005). https://doi.org/10.1115/1.1843853
Burris, D.L., Sawyer, W.G.: Addressing practical challenges of low friction coefficient measurements. Tribol. Lett. 35, 17–23 (2009). https://doi.org/10.1007/s11249-009-9438-2
Schulze, K.D., Hart, S.M., Marshall, S.L., O’Bryan, C.S., Urueña, J.M., Pitenis, A.A., Sawyer, W.G., Angelini, T.E.: Polymer osmotic pressure in hydrogel contact mechanics. Biotribology. 11, 3–7 (2017). https://doi.org/10.1016/j.biotri.2017.03.004
Urueña, J.M., McGhee, E.O., Angelini, T.E., Dowson, D., Sawyer, W.G., Pitenis, A.A.: Normal load scaling of friction in gemini hydrogels. Biotribology. 13, 30–35 (2018). https://doi.org/10.1016/j.biotri.2018.01.002
Johnson, K.L.: Contact Mechanics. Cambridge University Press, Cambridge (1985)
Garcia, M., Schulze, K.D., O’Bryan, C.S., Bhattacharjee, T., Sawyer, W.G., Angelini, T.E.: Eliminating the surface location from soft matter contact mechanics measurements. Tribol. Mater. Surfaces Interfaces. 11, 187–192 (2017). https://doi.org/10.1080/17515831.2017.1397908
Marshall, S.L., Schulze, K.D., Hart, S.M., Urueña, J.M., McGhee, E.O., Bennett, A.I., Pitenis, A.A., O’Bryan, C.S., Angelini, T.E., Sawyer, W.G.: Spherically capped membrane probes for low contact pressure tribology. Biotribology. 11, 69–72 (2017). https://doi.org/10.1016/j.biotri.2017.03.008
Bhattacharjee, T., Zehnder, S.M., Rowe, K.G., Jain, S., Nixon, R.M., Sawyer, W.G., Angelini, T.E.: Writing in the granular gel medium. Sci. Adv. 1, e1500655–e1500655 (2015). https://doi.org/10.1126/sciadv.1500655
Bhattacharjee, T., Gil, C.J., Marshall, S.L., Urueña, J.M., O’Bryan, C.S., Carstens, M., Keselowsky, B., Palmer, G.D., Ghivizzani, S., Gibbs, C.P., Sawyer, W.G., Angelini, T.E.: Liquid-like solids support Cells in 3D. ACS biomater. Sci. Eng. 2, 1787–1795 (2016). https://doi.org/10.1021/acsbiomaterials.6b00218
O’Bryan, C.S., Bhattacharjee, T., Hart, S., Kabb, C.P., Schulze, K.D., Chilakala, I., Sumerlin, B.S., Sawyer, W.G., Angelini, T.E.: Self-assembled micro-organogels for 3D printing silicone structures. Sci. Adv. 3, e1602800 (2017). https://doi.org/10.1126/sciadv.1602800
O’Bryan, C.S., Bhattacharjee, T., Niemi, S.R., Balachandar, S., Baldwin, N., Ellison, S.T., Taylor, C.R., Sawyer, W.G., Angelini, T.E.: Three-dimensional printing with sacrificial materials for soft matter manufacturing. MRS Bull. 42, 571–577 (2017). https://doi.org/10.1557/mrs.2017.167
Svoboda, K., Block, S.M.: Force and velocity measured for single kinesin molecules. Cell. 77, 773–784 (1994). https://doi.org/10.1016/0092-8674(94)90060-4
Dunn, A.C., Tichy, J.A., Uruenã, J.M., Sawyer, W.G.G., Urueña, J.M., Sawyer, W.G.G.: Lubrication regimes in contact lens wear during a blink. Tribol. Int. 63, 45–50 (2013). https://doi.org/10.1016/j.triboint.2013.01.008
Dowson, D.: Paper 12: Modes of Lubrication in Human Joints. Proc. Inst. Mech. Eng. Conf. Proc. 181, 45–54: (1966). https://doi.org/10.1243/PIME_CONF_1966_181_206_02
Hamrock, B.J., Dowson, D.: Elastohydrodynamic Lubrication of elliptical contacts for materials of low elastic modulus I—fully flooded conjunction. J. Lubr. Technol. 100, 236 (1978). https://doi.org/10.1115/1.3453152
Kapitza, P.L.: Hydrodynamic theory of lubrication during rolling. Zh. Tekh. Fiz. 25, 747–762 (1955)
Dunn, A.C., Urueña, J.M., Huo, Y., Perry, S.S., Angelini, T.E., Sawyer, W.G.: Lubricity of surface hydrogel layers. Tribol. Lett. 49, 371–378 (2013). https://doi.org/10.1007/s11249-012-0076-8
Schulze, K.D., Bennett, A.I., Marshall, S., Rowe, K.G., Dunn, A.C.: Real area of contact in a soft transparent interface by particle exclusion microscopy. J. Tribol. 138, 041404 (2016). https://doi.org/10.1115/1.4032822
McGhee, E.O., Pitenis, A.A., Urueña, J.M., Schulze, K.D., McGhee, A.J., O’Bryan, C.S., Bhattacharjee, T., Angelini, T.E., Sawyer, W.G.: In situ measurements of contact dynamics in speed-dependent hydrogel friction. Biotribology. 13, 23–29 (2018). https://doi.org/10.1016/j.biotri.2017.12.002
Lichtman, J.W., Conchello, J.-A.: Fluorescence microscopy. Nat. Methods. 2, 910–919 (2005). https://doi.org/10.1038/nmeth817
Shaner, N.C., Steinbach, P.A., Tsien, R.Y.: A guide to choosing fluorescent proteins. Nat. Methods. 2, 905–909 (2005). https://doi.org/10.1038/nmeth819
Berridge, M.J., Bootman, M.D., Roderick, H.L.: Calcium signalling: dynamics, homeostasis and remodelling. Nat. Rev. Mol. Cell Biol. 4, 517–529 (2003). https://doi.org/10.1038/nrm1155
de Oliveira, C.M.B., Sakata, R.K., Issy, A.M., Gerola, L.R., Salomão, R.: Cytokines and Pain. Rev. Bras. Anestesiol. 61, 255–265 (2011). https://doi.org/10.1016/S0034-7094(11)70029-0
Zhang, J.-M., An, J.: Cytokines, inflammation, and pain. Int. Anesthesiol. Clin. 45, 27–37 (2007). https://doi.org/10.1097/AIA.0b013e318034194e
Milo, R., Phillips, R.: Cell biology by the numbers. Garland Science, Taylor & Francis Group, New York (2016)
Ramsköld, D., Luo, S., Wang, Y.-C., Li, R., Deng, Q., Faridani, O.R., Daniels, G.A., Khrebtukova, I., Loring, J.F., Laurent, L.C., Schroth, G.P., Sandberg, R.: Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nat. Biotechnol. 30, 777–782 (2012). https://doi.org/10.1038/nbt.2282
Shapiro, E., Biezuner, T., Linnarsson, S.: Single-cell sequencing-based technologies will revolutionize whole-organism science. Nat. Rev. Genet. 14, 618–630 (2013). https://doi.org/10.1038/nrg3542
Milo, R.: What is the total number of protein molecules per cell volume? A call to rethink some published values. BioEssays. 35, 1050–1055 (2013). https://doi.org/10.1002/bies.201300066
Acknowledgements
This work was supported by Alcon Laboratories.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Urueña, J.M., Hart, S.M., Hood, D.L. et al. Considerations for Biotribometers: Cells, Gels, and Tissues. Tribol Lett 66, 141 (2018). https://doi.org/10.1007/s11249-018-1094-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11249-018-1094-y