Abstract
5-Enolpyruvylshikimate-3-phosphate synthase (EPSPS) and glyphosate N-acetyltransferase (GAT) can detoxify glyphosate by alleviating the suppression of shikimate pathway. In this study, we obtained transgenic tobacco plants overexpressing AM79 aroA, GAT, and both of them, respectively, to evaluate whether overexpression of both genes could confer transgenic plants with higher glyphosate resistance. The transgenic plants harboring GAT or AM79 aroA, respectively, showed good glyphosate resistance. As expected, the hybrid plants containing both GAT and AM79 aroA exhibited improved glyphosate resistance than the transgenic plants overexpressing only a single gene. When grown on media with high concentration of glyphosate, seedlings containing a single gene were severely inhibited, whereas plants expressing both genes were affected less. When transgenic plants grown in the greenhouse were sprayed with glyphosate, less damage was observed for the plants containing both genes. Metabolomics analysis showed that transgenic plants containing two genes could maintain the metabolism balance better than those containing one gene after glyphosate treatment. Glyphosate treatment did not lead to a huge increase of shikimate contents of tobacco leaves in transgenic plants overexpressing two genes, whereas significant increase of shikimate contents in transgenic plants containing only a single gene was observed. These results demonstrated that pyramiding both aroA and GAT in transgenic plants can enhance glyphosate resistance, and this strategy can be used for the development of transgenic glyphosate-resistant crops.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Glyphosate (N-phosphonomethyl glycine), an important and potent herbicide, is widely used to control weeds in agricultural fields. Glyphosate plays an important role in weed control because of its efficacy against all plant species, low mammalian toxicity, minimal environmental impact and low cost in the production and application process. Glyphosate inhibits the enzyme 5-enolpyruvylshikimate-3-phosphate synthase (EPSPS;EC2.5.1.19), which converts phosphoenolpyruvate (PEP) and shikimate-3-phosphate (S3P) to 5-enolpyruvylshikimate-3-phosphate (EPSP), shuts down the shikimate pathway and leads to plant death (Herrmann and Weaver 1999).
Two major types of EPSPSs have been classified. Type I EPSPSs have been identified mainly in plants and bacteria, and type II EPSPSs have been identified in some forms of bacteria. In E. coli, EPSPS is encoded by aroA. Type I EPSPSs are naturally sensitive to glyphosate, whereas type II EPSPSs are resistant to glyphosate (Funke et al. 2006). Apart from these, a novel EPSPS gene isolated from Pseudomonas putida 4G-1 was classified as a new type III EPSPS (Sun et al. 2005). After the primary target of glyphosate was identified as EPSPS in the 1980s, EPSPS became the top choice for the development of transgenic glyphosate-resistant crops. The transgenic glyphosate-resistant crops overexpressing CP4 EPSPS gene, a member of type II EPSPS, have been commercially used for many years.
The increased use of glyphosate over multiple years has led to glyphosate resistance of some weed species (Gaines et al. 2010). At present, researches on glyphosate focused on the target of glyphosate or other mechanisms involved in the resistance, which were also defined as target-site or non-target-site glyphosate resistance (Yu et al. 2009; Yuan et al. 2007). The mutation of EPSPS, especially the mutation at Pro106, is a common resistance mechanism which has been reported in several weedy species (Baerson et al. 2002; Wakelin and Preston 2006). Reduced glyphosate translocation is also a resistance mechanism which can lead to a higher level of resistance than EPSPS mutations (Preston and Wakelin 2008). It was also reported that Amaranthus palmeri developed glyphosate resistance by gene amplification to increase the EPSPS expression (Gaines et al. 2010).
In spite of resistance problems, it is still important to obtain glyphosate-resistant EPSPS genes for the development of transgenic glyphosate-resistant crops. Numerous EPSPSs were identified or screened from plants, bacteria or fungi. AM79 EPSPS is a type I EPSPS which was isolated from a glyphosate resistant bacteria strain through the metagenomic method. The transgenic experiment and the analysis of EPSPS enzyme kinetic parameters indicated that AM79 EPSPS could be a good candidate for the development of herbicide-resistant crops (Cao et al. 2012). Glyphosate N-acetyltransferase (GAT) can detoxify the herbicide glyphosate by N-acetylation of glyphosate, alleviating the suppression of shikimate pathway. The first reported GAT gene was isolated from Bacillus licheniformis and optimized by 11 cycles of DNA shuffling (Castle et al. 2004). The mutation collected from the eleventh cycle presented an extremely high glyphosate resistance when it was integrated into E. coli, Arabidopsis, tobacco and maize (Castle et al. 2004; Delaney et al. 2008). Structural analysis and kinetic data suggest that high glyphosate resistance is owing to the cooperative effects of some additional substitutions relocated to the active site (Siehl et al. 2007). The gene was widely used in transgenic maize, soybean and other plants by Pioneer Hi-bred, International (DuPont) company and could be a novel gene for developing transgenic glyphosate-resistant plants. A new GAT gene was identified from glyphosate polluted soil using the conservative primers refer to B. licheniformis, followed be optimized by DNA shuffling (Jin et al. 2007). This gene showed a high glyphosate resistance in recombinant Escherichia coli, and possessed a lower identity with the GAT acquired by Castle et al. (2004) and Jin et al. (2007).
GAT and EPSPS participate in different glyphosate resistance mechanisms. EPSPS blocks the suppression of the shikimate pathway by producing more EPSPS or slowing down the binding capacity of glyphosate, whereas GAT detoxifies the glyphosate by N-acetylation. Either AM79 aroA or GAT has been shown to confer transgenic plants with high glyphosate resistance (Castle et al. 2004; Cao et al. 2012). Pyramiding both genes might be able to make transgenic plants more tolerant to glyphosate, that make it possible that farmers could use higher concentration of glyphosate to kill the tolerant weeds. Due to that GAT and EPSPS has different function mechanisms, combining both genes could also delay the development of new glyphosate-tolerant weeds. The effects of co-expression of glyphosate-resistant genes with different resistance mechanisms on transgenic plants have not previously been reported.
In this study, we transformed AM79 aroA and GAT gene into tobacco plants and assessed the glyphosate resistance of the transgenic plants containing two genes or only one gene. Transgenic plants containing two genes could acquire a higher glyphosate resistance by maintaining the stability and balance of key metabolic pathways.
Materials and methods
Plasmid construction
The GAT gene (Lin et al. 2006) was amplified using PCR method with BamHI and SacI restriction site added to the upstream and downstream end of the gene. The sequence of the forward primer was 5′-CTGGATCCATGATTGACGTGAAC-3′, with the restriction enzyme BamHI at the 5′ end, and the reverse primer was 5′-CAGAGCTCTTATGCGATCCTCTTG-3′, with SacI at the 5′ end. Then the PCR product was ligated to pGEM-T Easy vector (Promega Company) for sequencing. The plasmid p3301-121spAM79 (Cao et al. 2012) was used as the donor vector. The AM79 aroA gene was replaced by GAT fragment digested with BamHI and SacI to construct the vector p3301-121spGAT (renamed as pSpGAT). For constructing non signal peptide GAT plant transformation vector p3301-121GAT (renamed as pGAT), we used the GAT gene to replace the G2 fragment in the plasmid p3301-121G2 with the added BamH I and Sac I sites (Cao et al. 2012).
Plant transformation
Plant expression plasmids pSpGAT and pGAT were transferred into competent cells of the Agrobacterium tumefaciens strain EHA105 through freeze–thaw treatment. The transformed A. tumefaciens colonies were selected on YEB-agar plates containing 100 µg/mL of kanamycin and 50 µg/mL of rifampicin. The positive colonies were identified by PCR amplification of the GAT gene and used for the tobacco (Nicotiana tabacum var. Samsum) transformation as previously described (Horsch et al. 1985) using 10 mg/L phosphinothricin (PPT) as the selected reagent. The transgenic plants were confirmed by PCR amplification of the GAT gene and transferred to the greenhouse for harvesting.
Production of hybrid plants
The transgenic lines with a single GAT copy and the transgenic plants containing one copy of AM79 aroA (Cao et al. 2012) were used for the cross-pollination experiments. To prepare maternal acceptor plants, the stamens were removed and the blooms were covered to prevent cross contamination with undesired pollen. At the flowering stage, we hand-pollinated the plants with tweezers, and reciprocal crosses were carried out. F1 generation seeds were harvested and further confirmed to have both genes by PCR method.
Glyphosate resistance analysis of transgenic tobacco
To evaluate the glyphosate resistance of tobacco seedlings in MS medium, heterozygous tobacco seeds of the T1 generation were germinated on MS medium containing 10 mg/L PPT and grown for 7 days at 100 μmol m−2 s−1 with a 16 h light/8 h dark period. The living seedlings with similar size were transferred onto MS medium in plates containing different amounts of glyphosate and grown vertically. 2 weeks later, injury was analyzed according to the percent of chlorotic leaves. Besides this, we used the homozygous T2 generation seeds for glyphosate resistance identification directly. Tobacco seeds were sterilized and germinated on MS medium containing 2 mM glyphosate and grown vertically for 14 days at 100 µmol m−2 s−1 with a 16 h light/8 h dark period at 25 °C. Injury was observed according to the percent of chlorotic leaves, and the fresh weight and root length was measured.
To evaluate the glyphosate resistance of tobacco plants grown in greenhouse, one-week-old seedlings germinated on MS medium were transferred to soil and grown in a greenhouse for about 1 month. Six-to-eight leaf stage transgenic plants were sprayed with 4 times (9 L/ha Roundup®, for GC/MS analysis) or 8 times (18 L/ha Roundup®, for glyphosate resistance evaluation) of the recommended field application concentration of glyphosate. Injury was observed and fresh weight was measured 2 weeks after treatment.
RNA isolation and RT-PCR
Total RNA was isolated from tobacco leaves using Trizol reagent (Invitrogen, Carlsbad, CA, USA) and first-strand cDNA synthesis was performed with M-MLV reverse transcriptase (Promega) using oligo-dT primer. Specific primer for AM79, GAT and internal actin gene were shown in Supplemental Table 1. For RT-PCR, 1 μL tenfold diluted cDNA were used for template and 22–30 amplification cycles were selected for PCR project based on the amplification efficiency of each primer. The amounts of the PCR products were screened on 1 % agarose.
Metabolic profiling
Six-to-eight leaf stage transgenic tobacco harboring GAT, AM79 aroA and dual genes were sprayed with four times of the recommended field application concentration of glyphosate. 5 days later, each leaf sample was collected from three independent plants and total six samples were prepared for each treatment. The leaf tissue of each category were frozen in liquid nitrogen and then stored in a low temperature refrigerator of −80 °C for subsequent processing. The leaves of transgenic plants which have not been treated by glyphosate were also prepared as control samples.
For GC/MS analysis, each grated powder sample of 20 mg was transferred to a glass centrifuge tube followed by 1 mL 100 % methanol. 10 μL phenylalanine (Sigma Aldrich) as internal standard was added to each sample and the mixture was ultrasonicated at ambient temperature (approximately 60 °C) for 15 min and then vortex-mixed. The samples were subsequently centrifuged at 2200 rpm for 15 min and 0.4 mL of the supernatant was collected separately from each sample into a test tube. Then 0.2 mL precooled 100 % acetonitrile and 0.4 mL ultra-pure water were added and vortex-mixed. Samples were centrifuged at 2200 rpm for 15 min and 0.2 mL selected supernatant was transferred to a screw vial (2 mL) which could be used for derivatization and evaporated to dryness under a stream of nitrogen gas. 30 μL 20 mg/mL methoxyamine pyridine hydrochloride solution was added to each tube and shaken to mix, then maintained at a temperature 37 °C for 90 min. Then, we added 30 μL BSTFA with 1 % TMCS to each vial, and left the mixture to be incubated for 60 min at a temperature of 70 °C. After incubation preservation for 30 min on room temperature, the samples were selected and prepared for metabolic profiling based on GC/MS principle (Wu et al. 2010).
Analysis was performed on an Agilent 7890A/5975C GC/MS system equipped with a 30 m × 0.25 mm i.d. fused-silica capillary column with 0.25-μm HP-5MS stationary phase (Agilent J&W Scientific, Shanghai, China). The injection temperature was set at 280 °C. High purity helium (>99.999 %) was used as the carrier gas at a constant flow rate of 1 mL/min. 1 μL aliquot of derivatized sample was injected splitless into the injection port and the column temperature was initially kept at 80 °C for 2 min and then increased to 320 °C at a rate of 10 °C/min, where it was held for 6 min. The ion source temperature was set at 230 °C and the MS quadrupole temperature at 150 °C. Using the full scan mode for mass spectrometric detection, the masses were acquired from 50 to 550 m/z. Random sampling for analyzing consecutive samples was selected to avoid the impact of possible fluctuations of the machine’s signal.
The chromatograms were collected and subjected to noise reduction using MassHunter workstations. After correction of the retention time and analysis of the mass fragments, the data were converted to MZ format, followed by converting to Computable Document Format (CDF). The Automated Mass Spectral Deconvolution and Identification System (AMDIS) were used to make the data for deconvolution. Each compound had a unique fragmentation pattern composed of a number of split molecular ions. The mass abundance and charge ratios could be compared with a standard mass chromatogram in the National Institute of Standards and Technology (NIST) mass spectra library and a self-built library by Shanghai Sensichip Infotech Co., Ltd. by the ChemStation Software. The intensity information of each peak was expressed and the comparison between different categories was performed.
Statistical analysis
Comparisons of the values for significant differences were performed using Student’s T test in Excel (Microsoft 2010) at P < 0.05 or P < 0.01 level. All the photographs were treated using Adobe Illustrator CS5 or Origin 8.0 software.
Results
Overexpression of GAT conferred transgenic tobacco plants with high glyphosate resistance
A GAT gene was cloned using metagenomic screening method from a bacterium grown in glyphosate contaminated soil (Jin et al. 2007). To investigate whether this GAT gene is suitable for developing glyphosate-resistant transgenic crops, the glyphosate resistance conferred by GAT gene in transgenic tobacco plants was assessed. Vector pSpGAT where GAT gene was fused with the signal peptide sequence of the pea rib-1,5-bisphospate carboxylase (rbcS) small subunit and vector pGAT where GAT gene was not fused with the signal peptide sequence were constructed, respectively (Supplemental Fig. 1). The vectors were transformed into tobacco plants and transgenic plants were identified using PCR method (data not shown).
The glyphosate resistance of T1 generation of transgenic tobacco plants overexpressing GAT or AM79 aroA (Cao et al. 2012) gene were investigated. One-month-old transgenic tobacco plants grown in the greenhouse were sprayed with Roundup® at an equal dose of four times of the recommended field application concentration (approximately 9 L/ha). 2 weeks after the treatment, all WT plants were killed, and the transgenic plants transformed with pSpGAT were also injured severely (Fig. 1a). On the contrary, the transgenic plants containing only GAT gene showed good glyphosate resistance similarly to the plants overexpressing AM79 aroA (Fig. 1). Tobacco tissues of aerial parts from ten plants were 5.00 g and 7.02 g for WT plants and transgenic plants transformed with vector pSpGAT, respectively. The fresh weight of transgenic plants overexpressing GAT or AM79 aroA was 23.08 and 22.80 g, respectively (Fig. 1c). Transgenic plants transformed with vector pGAT had significantly higher fresh weight than plants transformed with vector pSpGAT (Fig. 1c), indicating that the location of GAT in the chloroplast decreased the efficiency of GAT to detoxify glyphosate.
Glyphosate resistance of transgenic tobacco plants in a greenhouse. T1 tobacco seeds were germinated on the MS medium containing 10 mg/L PPT and grown for 7 days at 100 μmol s−1m−2 with a 16 h light/8 h dark period. The live seedlings were transferred to soil in pots and grown for another month. Six-to-eight leaf stage transgenic plants were sprayed with 9 L/ha Roundup®. Two weeks after treatment, injury was observed, and fresh weight was measured. AM79, transgenic plants overexpressing AM79 aroA; GAT, transgenic plants overexpressing GAT; SP + GAT, transgenic plants overexpressing GAT which is fused with signal peptide; a photograph of tobacco plants harboring AM79 aroA and SP + GAT 2 weeks after glyphosate treatment. b Photograph of tobacco plants harboring AM79 aroA and GAT 2 weeks after glyphosate treatment. c Fresh weight of tobacco plants. Data are shown as the average ± SD of ten independent transgenic lines. Asterisk indicates significant difference at P < 0.01 level
On the medium without glyphosate, transgenic tobacco seedlings grew as well as the WT plants (Supplemental Fig. 2). The transgenic plants harboring only GAT gene showed as good performance (a heavier fresh weight and uninhibited root elongation) as the AM79 aroA transformants which we previously reported (Supplemental Fig. 2; Cao et al. 2012).
Transgenic tobacco plants harboring both GAT gene and AM79 aroA showed higher glyphosate resistance
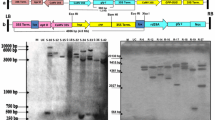
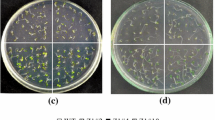
GAT and AM79 EPSPS participate in different glyphosate resistance mechanisms. We further investigate whether pyramiding of both genes could confer plants with higher glyphosate resistance than those containing one gene. GAT transgenic line GAT-15 was determined to have a single transgene insertion in chromosome. The T2 generation of homozygous transgenic tobacco plant lines GAT-15 and AM79-4 which has one copy of AM79 aroA (Cao et al. 2012) were selected for the crossing to obtain F1 generation seeds. Reciprocal cross was performed between AM79 aroA and GAT transformants, and the glyphosate resistance was not different between the hybrid plants. The F1 generation plants were confirmed to harbor both two genes using PCR method (Fig. 2a). We also used RT-PCR method to confirm the transcriptional level of transgenes, and the results showed that both genes were expressed normally and not affected by each other in the hybrid plants (Fig. 2b).The glyphosate resistance of the hybrid plants containing both genes and the transgenic plants harboring only one gene was compared. On the MS medium without glyphosate,all the seedlings grew well and displayed a similar root length and fresh weight (data not shown), indicating that the seeds selected from WT, hybrids and single gene transformants retained similar initial seed quality. 2 mM glyphosate inhibited the growth of WT plants which turned yellow completely (Fig. 3a). Hybrid plants harboring two genes grew well under the glyphosate stress and did not show any signs of glyphosate injury. The root length was measured and the results showed that hybrid plants containing both genes had significantly longer root length than the transgenic plants containing only one gene (Fig. 3b). These results clearly showed that pyramiding of both genes could enhance the glyphosate resistance of transgenic plants.
Molecular identification of transgenic plants harboring GAT and AM79 aroA. a Multiplex-PCR detection of the hybrid tobacco plants, M, DL2000 plus DNA ladder; 1–8, different hybrid lines; 9, H2O; 10, WT; 11, plasmid pGAT; 12, plasmid p3301-121spAM79; b the transcriptional level of AM79 aroA and GAT in transgenic tobacco plants
Glyphosate resistance of transgenic tobacco seedlings grown on medium containing 2 mM glyphosate. T2 homozygous tobacco seeds were germinated on the MS medium containing 2 mM glyphosate and grown vertically for 14 days at 100 μmol s−1m−2 with a 16 h light/8 h dark period. Photographs were taken 2 weeks later, and the root length was measured at the same time. a Photograph of WT plants and transgenic tobacco harboring GAT, AM79 or both gene. b Root length of the tobacco plants. Data are shown as the average ± SD of seven to ten independent transgenic lines. Asterisk means significant difference at P < 0.01 level
The glyphosate resistance was also assessed using plants grown in the greenhouse. Plants with six-to-eight leaves were sprayed with high concentration of glyphosate at a dose of eight times of the recommended field application concentration (approximately 18 L/ha). 2 weeks after treatment, injury was observed and fresh weight was measured (Fig. 4). As expected, all the WT plants were killed by the herbicide and possessed a minimum weight. The hybrid plants harboring GAT and AM79 aroA gene displayed a powerful capacity for coping with the impact of high concentration glyphosate. The leaves of the hybrid plants harboring dual genes had no visible damage, whereas the leaves of the transgenic lines containing GAT or AM79 aroA showed necrosis of leaf margin (Fig. 4a). The fresh weight of ten plants of hybrid plants, GAT plants and AM79 aroA plants was 64.41, 23.08 and 22.80 g, respectively, and the difference between the hybrid plants harboring two genes and the one gene transformants was significant (Fig. 4b). These results further confirmed that pyramiding of both genes could enhance the glyphosate resistance of transgenic plants.
Glyphosate resistance of transgenic tobacco plants in a greenhouse when sprayed with 18 L/ha Roundup®. T2 homozygous tobacco seeds and F1 hybrids tobacco seeds were germinated on the MS medium containing 10 mg/L PPT and grown for 7 days at 100 μmol s−1m−2 with a 16 h light/8 h dark period. The live seedlings were transferred to soil in pots and grown for another month. Six-to-eight leaf stage transgenic plants were sprayed with 18 L/ha Roundup®. Two weeks after treatment, injury was observed, and fresh weight of ten plants was measured. a Photograph of tobacco plants harboring AM79, GAT, or both genes. b Fresh weight of tobacco plants. Data are shown as the average ± SD of ten independent transgenic lines. Asterisk means significant difference at P < 0.01 level
Overexpression of both GAT and AM79 aroA could maintain better metabolic balance in glyphosate-treated hybrid plants
To investigate the mechanism of the higher glyphosate resistance of hybrid plants, metabolomics analysis was performed. Sixty metabolites were identified in the tobacco plants (Supplemental Table 2). Most of these compounds are secondary metabolites, and a portion of the compounds are primary metabolites that play important roles in specific metabolic pathways. In transgenic tobacco plants overexpressing AM79 aroA, 24 metabolites were up-regulated and 2 metabolites were down-regulated by treatment with glyphosate, respectively (Fig. 5a). Seven metabolites were up-regulated and two metabolites were down-regulated in transgenic plants overexpressing GAT (Fig. 5b). In hybrid plants overexpressing both AM79 aroA and GAT, only two metabolites were up-regulated and one metabolite was down-regulated by glyphosate treatment, respectively (Fig. 5c).
Metabolic changes in the leaves of transgenic tobacco overexpressing AM79 aroA (a), GAT (b) or dual genes (c). Six-to-eight leaf stage transgenic tobacco plants were sprayed with four times of the recommended field application concentration of glyphosate. Five days later, leaf samples were collected for metabolic profiling. Data were presented with average ± SE of six independent replicates. Experimental data was tested by Students’ t test analysis at P < 0.05 or P < 0.01 level
High accumulation of shikimate is an indicator of the response of plants to glyphosate stress. Before the application of glyphosate, there were no significant differences in metabolites between the tobacco plants (data not shown). Under glyphosate stress, transgenic tobacco plants expressing AM79 aroA exhibited 2.19 fold increase of shikimate (Supplemental Table 2; Supplemental Fig. 3) because of the enhancement of catalysis reaction given by AM79 EPSPS. GAT could confer transgenic tobacco with high glyphosate resistance as similarly to AM79 EPSPS, but transgenic tobacco harboring GAT showed a 101 fold increase of shikimate (Supplemental Table 2; Supplemental Fig. 3). Nevertheless, glyphosate treatment did not cause an accumulation of shikimate in hybrid tobacco plants, indicating that the shikimate pathway in hybrid plants was not affected (Supplemental Table 2; Supplemental Fig. 3). A three folds increase of acetic acid was observed in transgenic plants overexpressing AM79 aroA or GAT, whereas there was not observable change in hybrid plants expressing both genes (Supplemental Table 2). Tetradecanoic acid accumulations were also up-regulated in transgenic plants expressing AM79 aroA or GAT, and no change was observed in hybrid plants containing both genes. Up-regulation of glycoside was detected in all the tobacco plants treated with glyphosate, possibly because glycoside has multiple functions in plants suffering abiotic stress. Comprehensive analysis of metabolite change in single gene transformants showed that some compounds had opposite change patterns. Both nicotine and d-Glycero-d-gulo-Heptose were up-regulated in AM79 aroA transformants, whereas they were down-regulated in GAT transformants. In transgenic tobacco plants overexpressing one or two genes, glyphosate treatment led to the increased accumulation of pyroglutamic acid.
Discussion
In this study, we investigated the function of a novel N-acetyltransferase gene (GAT) which was isolated from glyphosate polluted soil using metagenomics method (Jin et al. 2007; Lin et al. 2006). It has been reported that glyphosate treatment reduced the shoot and root dry matter production and micronutrient status in WT plants and glyphosate-resistant plant (Bott et al. 2008; Ozturk et al. 2008). In greenhouse, fresh weight of plants was used for glyphosate resistance evaluation. Transgenic plants transformed with vector pGAT had significantly higher fresh weight than WT plants and transgenic plants transformed with vector pSpGAT (Fig. 1), confirming that GAT could detoxify glyphosate in transgenic plants and indicating that the location of GAT in the chloroplast decreased the efficiency of GAT to detoxify glyphosate. However, we did not know whether the GAT gene used in this study has similar glyphosate-resistance efficiency as the reported GAT gene by Castle et al. (2004).
Due to the fact that GAT and EPSPS detoxify glyphosate with different mechanisms, transgenic plants pyramiding both of them should have better glyphosate resistance performance than those plants containing only one gene. For glyphosate resistance evaluation of transgenic plants which were grown on MS medium, root length was selected as an indicator. As expected, transgenic tobacco harboring both GAT and AM79 aroA showed longer root length (Fig. 3) and higher fresh weight than plants containing one gene and WT plants (Fig. 4). The higher glyphosate resistance of transgenic plants expressing both AM79 aroA and GAT, confirmed that the synergistic effect could detoxify glyphosate more efficiently.
In this study, reciprocal crosses were carried out to obtain hybrid plants containing both AM79 aroA and GAT. The glyphosate resistance of AM79 × GAT and GAT × AM79 hybrid plants were assayed, and no significant differences appeared in the evaluation index of survival rate, root length and fresh weight. The hybrids born from reciprocal cross should possess unanimous glyphosate resistance capacity and the cytoplasm effect could be omitted. The transgenic plant lines used for the reciprocal cross have only one copy of the transgene, avoiding the gene dosage imbalance caused by differences in the copy numbers of AM79 aroA or GAT genes.
To further explain the better performance of transgenic plants containing both genes, metabolic analysis was performed. Our previous work had showed higher accumulation of shikimate and other metabolites in WT tobacco and maize plants than transgenic plants expressing AM79 aroA or G2 aroA (Cao et al. 2012; Liu et al. 2014). In this study, we focused on the metabolite change among the transgenic plants expressing different genes. Transgenic tobacco expressing AM79 aroA displayed a maximum amount of metabolite changes. Our previous work showed that transgenic plants expressing AM79 aroA or other aroA genes displayed a higher shikimate level 5 days after glyphosate treatment before the shikimate level returning to normal, possibly might due to the maintenance of the overexpressed EPSPS proteins (Cao et al. 2012). Under glyphosate treatment conditions, there was a two-fold increase of shikimate accumulation in transgenic plants expressing AM79 aroA (Supplemental Table 2; Supplemental Fig. 3), indicating that the shikimate pathway was not severely affected by glyphosate. However, around 100-fold increase of shikimate was observed in glyphosate-treated transgenic plants overexpressing GAT (Supplemental Table 2; Supplemental Fig. 3). Glyphosate could occupy the active sites of EPSPS and rapidly inhibit the shikimate pathway, leading to the high accumulation of shikimate, while the process that GAT uses to detoxify glyphosate is a relatively slow reaction because of the necessary step for searching for the active site to acetylate glyphosate (Siehl et al. 2007). This might be the reason why there was high shikimate accumulation in GAT transformants. Shikimate accumulation was not affected by glyphosate treatment in transgenic plants overexpressing AM79 aroA and GAT (Supplemental Fig. 3), confirming that EPSPS and GAT can function together to detoxify glyphosate.
Plant metabolism regulation is an extremely important strategy to maintain normal growth and development of plants (Prins et al. 2011; Tzin and Galili 2010; Vivancos et al. 2011). Changes in metabolites could be a positive signal for plants in response to abiotic stress (Zhu 2003). A number of other metabolites were found to be changed in transgenic plants treated with glyphosate, and some metabolites remain unchanged during the glyphosate’s application (Fig. 5). Metabolite features associated with glyphosate resistance could be used as biomarkers to facilitate genetic improvement of plants. A huge number of non-target site glyphosate resistance genes participated in the herbicide transportation through Cytochrome P450 s, ABC transporters, and so on (Vila-Aiub et al. 2012, 2013; Yuan et al. 2007). Compared with a limited number of site-target glyphosate resistance genes (EPSPS or GAT), there are a mass of genes which may play specific roles in non-target site glyphosate resistance (Ge et al. 2010; Wakelin et al. 2004). It is valuable to identify the pivotal pathway and excavate novel genes involved in glyphosate adaptive regulation.
Numerous metabolites were found to be up-regulated by glyphosate treatment, whereas a small amount of down-regulated metabolites were detected. Specially, no consistent metabolite was discovered for down-regulation in the different transgenic plants and hybrids. Galactose was specially detected in hybrids which was down-regulated by glyphosate treatment. In E. coli, galactose-proton symport system mediated by galactose permease can affect the shikimate pathway product (Yi et al. 2003). As KEGG analysis shows that galactose possesses multiple functions in many biological metabolic pathways, we speculate that the moderate down-regulation in transgenic tobacco plants may be an adverse adaptation of glyphosate treatment. Previous research showed that glyphosate could decrease the regeneration capacity of maize callus by changing the primary and secondary metabolism beyond the EPSPS inhibition metabolism (Ulanov et al. 2009). In our metabolomics analysis, numerous primary and secondary metabolites (amino acids and sugars) were detected (Supplemental Table 2). Different and specially expressed metabolites in the hybrid and its parents may play similar roles in the glyphosate stress response and show different glyphosate resistance.
Taken together, we confirmed that overexpressing a GAT gene could confer transgenic tobacco plants with high glyphosate resistance, and co-expressing both AM79 aroA and GAT gene could confer the tobacco plants higher glyphosate resistance than those expressing only one gene. The results here indicate that integration of the two genes is a viable strategy for the development of herbicide resistant crops.
References
Baerson SR, Rodriguez DJ, Tran M, Feng Y, Biest NA, Dill GM (2002) Glyphosate-resistant goosegrass. Identification of a mutation in the target enzyme 5-enolpyruvylshikimate-3-phosphate synthase. Plant Physiol 129:1265–1275
Bott S, Tesfamariam T, Candan H, Cakmak I, Roemheld V, Neumann G (2008) Glyphosate-induced impairment of plant growth and micronutrient status in glyphosate-resistant soybean (Glycine max L.). Plant Soil 312:185–194
Cao G, Liu Y, Zhang S, Yang X, Chen R, Zhang Y, Lu W, Wang J, Lin M, Wang G (2012) A novel 5-enolpyruvylshikimate-3-phosphate synthase shows high glyphosate tolerance in Escherichia coli and tobacco plants. PLoS ONE. doi:10.1371/journal.pone.0038718
Castle LA, Siehl DL, Gorton R, Patten PA, Chen YH, Bertain S, Cho HJ, Duck N, Wong J, Liu D, Lassner MW (2004) Discovery and directed evolution of a glyphosate tolerance gene. Science 304:1151–1154
Delaney B, Zhang J, Carlson G, Schmidt J, Stagg B, Comstock B, Babb A, Finlay C, Cressman RF, Ladics G, Cogburn A, Siehl D, Bardina L, Sampson H, Han Y (2008) A gene-shuffled glyphosate acetyltransferase protein from Bacillus licheniformis (GAT4601) shows no evidence of allergenicity or toxicity. Toxicol Sci 102:425–432
Funke T, Han H, Healy-Fried ML, Fischer M, Schonbrunn E (2006) Molecular basis for the herbicide resistance of Roundup Ready crops. Proc Natl Acad Sci USA 103:13010–13015
Gaines TA, Zhang W, Wang D, Bukun B, Chisholm ST, Shaner DL, Nissen SJ, Patzoldt WL, Tranel PJ, Culpepper AS, Grey TL, Webster TM, Vencill WK, Sammons RD, Jiang J, Preston C, Leach JE, Westra P (2010) Gene amplification confers glyphosate resistance in Amaranthus palmeri. Proc Natl Acad Sci USA 107:1029–1034
Ge X, d’Avignon DA, Ackerman JJ, Sammons RD (2010) Rapid vacuolar sequestration: the horseweed glyphosate resistance mechanism. Pest Manag Sci 66:345–348
Herrmann KM, Weaver LM (1999) The shikimate pathway. Annu Rev Plant Physiol Plant Mol Biol 50:473–503
Horsch RB, Fry JE, Hoffmann NL, Eichholtz D, Rogers SG, Fraley RT (1985) A Simple and General Method for Transferring Genes into Plants. Science 227:1229–1231
Jin D, Chen M, Ma RQ, Yang ZR, Chen J (2007) To identify sites related to glyphosate-tolerance in glyphosate N-aceltransferase by DNA shuffling technology (In Chinese). J Agric Sci Technol 9:110–114
Lin M, Dun B, Lu W, Jin D. (2006) Glyphosate acetyl transferase gene and its application. China Patent CN1772908 A
Liu Y, Zhang Y, Liu Y, Lu W, Wang G (2014) Metabolic effects of glyphosate on transgenic maize expressing a G2-EPSPS gene from Pseudomonas fluorescens. J Plant Biochem Biotechnol. doi:10.1007/s13562-014-0263-9
Ozturk L, Yazici A, Eker S, Gokmen O, Romheld V, Cakmak I (2008) Glyphosate inhibition of ferric reductase activity in iron deficient sunflower roots. New Phytol 177:899–906
Preston C, Wakelin AM (2008) Resistance to glyphosate from altered herbicide translocation patterns. Pest Manag Sci 64:372–376
Prins A, Mukubi JM, Pellny TK, Verrier PJ, Beyene G, Lopes MS, Emami K, Treumann A, Lelarge-Trouverie C, Noctor G, Kunert KJ, Kerchev P, Foyer CH (2011) Acclimation to high CO2 in maize is related to water status and dependent on leaf rank. Plant, Cell Environ 34:314–331
Siehl DL, Castle LA, Gorton R, Keenan RJ (2007) The molecular basis of glyphosate resistance by an optimized microbial acetyltransferase. J Biol Chem 282:11446–11455
Sun YC, Chen YC, Tian ZX, Li FM, Wang XY, Zhang J, Xiao ZL, Lin M, Gilmartin N, Dowling DN, Wang YP (2005) Novel AroA with high tolerance to glyphosate, encoded by a gene of Pseudomonas putida 4G-1 isolated from an extremely polluted environment in China. Appl Environ Microbiol 71:4771–4776
Tzin V, Galili G (2010) New insights into the shikimate and aromatic amino acids biosynthesis pathways in plants. Mol Plant 3:956–972
Ulanov A, Lygin A, Duncan D, Widholm J, Lozovaya V (2009) Metabolic effects of glyphosate change the capacity of maize culture to regenerate plants. J Plant Physiol 166:978–987
Vila-Aiub MM, Balbi MC, Distefano AJ, Fernandez L, Hopp E, Yu Q, Powles SB (2012) Glyphosate resistance in perennial Sorghum halepense (Johnsongrass), endowed by reduced glyphosate translocation and leaf uptake. Pest Manag Sci 68:430–436
Vila-Aiub MM, Gundel PE, Yu Q, Powles SB (2013) Glyphosate resistance in Sorghum halepense and Lolium rigidum is reduced at suboptimal growing temperatures. Pest Manag Sci 69:228–232
Vivancos PD, Driscoll SP, Bulman CA, Ying L, Emami K, Treumann A, Mauve C, Noctor G, Foyer CH (2011) Perturbations of amino acid metabolism associated with glyphosate-dependent inhibition of shikimic acid metabolism affect cellular redox homeostasis and alter the abundance of proteins involved in photosynthesis and photorespiration. Plant Physiol 157:256–268
Wakelin AM, Preston C (2006) A target-site mutation is present in a glyphosate-resistant Lolium rigidum population. Weed Res 46:432–440
Wakelin AM, Lorraine-Colwill DF, Preston C (2004) Glyphosate resistance in four different populations of Lolium rigidum is associated with reduced translocation of glyphosate to meristematic zones. Weed Res 44:453–459
Wu H, Xue R, Tang Z, Deng C, Liu T, Zeng H, Sun Y, Shen X (2010) Metabolomic investigation of gastric cancer tissue using gas chromatography/mass spectrometry. Anal Bioanal Chem 396:1385–1395
Yi J, Draths KM, Li K, Frost JW (2003) Altered glucose transport and shikimate pathway product yields in E. coli. Biotechnol Prog 19:1450–1459
Yu Q, Abdallah I, Han H, Owen M, Powles S (2009) Distinct non-target site mechanisms endow resistance to glyphosate, ACCase and ALS-inhibiting herbicides in multiple herbicide-resistant Lolium rigidum. Planta 230:713–723
Yuan JS, Tranel PJ, Stewart CN Jr (2007) Non-target-site herbicide resistance: a family business. Trends Plant Sci 12:6–13
Zhu JK (2003) Regulation of ion homeostasis under salt stress. Curr Opin Plant Biol 6:441–445
Acknowledgments
This work was funded by the National Major Project for Transgenic Organism Breeding (2013ZX08003-001). We gratefully thank the anonymous reviewers for their valuable suggestions. We acknowledge the technical staffs from Shanghai Sensichip Infotech Co., Ltd for metabolic profiling. We also give our thanks to Dr. Liang Li from Biotechnology Research Institute of CAAS for insightful suggestion.
Author information
Authors and Affiliations
Corresponding author
Additional information
Yunjun Liu and Gaoyi Cao have contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Liu, Y., Cao, G., Chen, R. et al. Transgenic tobacco simultaneously overexpressing glyphosate N-acetyltransferase and 5-enolpyruvylshikimate-3-phosphate synthase are more resistant to glyphosate than those containing one gene. Transgenic Res 24, 753–763 (2015). https://doi.org/10.1007/s11248-015-9874-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11248-015-9874-8