Abstract
Tomato is a key vegetable crop cultivated in nearly all agricultural regions of the world. It is a protective food very rich in vitamin A and C. Worldwide two major biotic stresses namely root-knot nematode (Meloidogyne incognita) and tomato leaf curl virus (ToLCV) pose a serious threat to its production. Serious environmental concerns associated with the chemical pesticides used for management of these pests compel researchers to look for alternate management strategies. Many recent publications indicate engineering resistance through host delivered RNAi to be an important, sustainable and efficient tool for nematode and virus management. In this study, we have attempted to engineer resistance against M. incognita and ToLCV by stacking dsRNA constructs of the Integrase gene of M. incognita and AC4 genes of ToLCV, using co-transformation protocol in tomato. Transgenic events, so produced, were confirmed at the molecular level by PCR and southern blot. Such T2 events showed absolute absence of leaf curl virus disease symptoms and 62–63% reduction in the number of galls, 51–70% reduction in the number of eggs, 31–38% reduction in number of eggs per egg mass and 66–81% reduction in nematode multiplication factor when compared to the untransformed control plants.The study provides an evidence for generating resistance through RNAi against multiple biotic stresses.
Key message
This study provides an insight into generating resistance against multiple biotic stresses in tomato through host generated RNA
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Plant parasitic nematodes (PPNs) are accredited to cause an annual economic loss of about$173 billion to agricultural crops worldwide (Elling 2013). Amongst the PPNs, the sedentary endo-parasite, Root-knot nematodes (Meloidogyne spp.) are economically the most detrimental as they parasitize nearly every agri-horticultural crop of the world. They feed by modifying the living cells of host roots and induce small/large galls or knots as a symptom of disease on them. The RKN parasitism affects water and nutrient uptake and also their upward translocation in the root system (Moens et al. 2009). More than 90 species of RKNs have been documented, but the most predominant are M. incognita, M. javanica, M. arenaria and M. hapla (Jones et al. 2013). Among these major RKN species, M. incognita is most widespread and regarded as the most favored parasite of tomato, Solanum lycopersicum L. (Dropkin 1980; Khan et al. 2000).M. incognita does not produce any particular above ground symptoms in tomato but affected plants show an unthrifty appearance. In India, about 27.21% losses in tomato production are attributed to M. incognita alone (Jain et al. 2007). Apart from M. incognita, tomato leaf curl virus (ToLCV) is another foremost, destructive pathogen of tomato causing the leaf curl disease. ToLCV belongs to the genus Begomovirus and is vectored by whitefly; Bemisia tabaci. To date, more than 15 species of Begomovirus have been found associated with tomato leaf curl disease in India (Yadava and Mukherjee 2012). Symptoms of ToLCV in tomato include curling, cupping, yellowing of leaves and stunting of plant growth eventually affecting the crop productivity (Rataul and Brar 1989).
Chemical pesticides are the most effective means for managing both these pathogens but are highly detrimental to the environment and as a result alternative strategies need to be explored for their management. Engineering resistance through host delivered RNAi has proved very effective for pest management (Banerjee et al. 2017). The first such successful demonstration of RNAi against a plant parasitic nematode was demonstrated in tobacco by silencing the housekeeping genes i.e.Integrase and splicing factor (dsRNA) of M. incognita (Yadav et al. 2006). Silencing of the pathogenecity factor,AC4 gene (Krake et al. 1998; Chellappan et al. 2005), of ToLCV by RNAi resulted in complete virus resistant tomato (Ramesh et al. 2007).
Boulter et al. (1990) proposed stacking or pyramiding of transgenes for enhancing and improving the effectiveness and longevity of resistance against multiple pathogens. Gene pyramiding/ gene stacking require identifying and introducing multiple genes, wherein each gene expresses and imparts resistance to an independent pathogen/insect pest/plant parasitic nematodes/weed etc. With this background, we proceeded to introduce two genes; M. incognita-Integrase and ToLCV-AC4via Agrobacterium mediated co-transformation in tomato and evaluated the level of resistance against the each of the pathogens.
Materials and methods
Pure culture of root-knot nematode, M. incognita
Tomato cultivar, Pusa Ruby was used to propagate and multiply M. incognita, pure culture in a glasshouse. Tomato plants were uprooted 30 days after inoculation of the 2nd stage RKN juveniles and roots washed using double distilled water (DDW). Egg masses were handpicked in a cavity block using a sterile forceps. These egg masses were surface sterilized using 0.1% HgCl2 for 1 min and then rinsed 3–4 times with DDW so as to wash away the effect of chemical. The collected egg masses were placed on a double-layered tissue paper supported on wire mesh in a petri dish containing DDW at 28 °C for hatching (Hooper 1986). Perineal pattern morphology was used to confirm RKN species (Fig. 1). For subsequent experimentation the freshly hatched second stage juveniles (J2s) were used.
Plant material, dsRNA constructs & bacterial culture
Tomato (S. lycopersicumcv. PusaRuby) seeds were washed withTween-20 for 15 min, sterilized by dipping them in70% ethanol for 1 min and post this treated with 4% sodium hypochlorite (NaOCl) for 15–20 min. The seeds were thoroughly washed with sterile DDW three to four times after each treatment. They were then placed in square magenta boxes containing half strength MS agar medium (Murashige and Skoog 1962) and grown aseptically under controlled conditions at 26 °C and 16/8 h (light/dark). Cotyledonary leaves of 12 to 14 days-old plants were served as explants for Agrobacterium co-transformation.
M. incognita RNAi construct pBC-142 harboring Integrase gene (NCBI Accession number: AW871671 & Fig. 1 Supplementary) was obtained from Dr. Subramaniam, IIT, Kanpur (now at IIT Madras) and transformed into Agrobacterium tumefaciens strain GV-3101.A. tumefaciens strain LB4404 harboring a binary vector pCAMBIA2301-AC4(Accession number: U15015.2 & Fig. 2 Supplementary) and the target gene of Tomato leaf curl New Delhi virus (ToLCNDV) was obtained from the Dr. Shelly Praveen’s laboratory, ICAR-IARI, New Delhi, India.
Generation and selection of tomato transgenic events
Tomato transgenic events harboring dsRNAi construct of Integrase and AC4 genes were generated. One cm2 cotyledonary leaves were cut from 14 days aged tomato seedlings and plated in petri plate containing pre-cultivation medium (MS salts + 0.5 mg/l IAA and 1 mg/l Zeatin Riboside). Agrobacterium-mediated co-transformation of two days old pre-cultivated leaves by mixture method was performed (Walawage et al. 2013). A.tumefaciens strains GV-3101 and LB-4404 harboring binary vectors pBC-142 and pCAMBIA-2301 carrying respective transgenes were grown to 0.6–0.8 OD600 overnight in YEP liquid medium. This medium contained antibiotics kanamycin, (50 mg/l), rifampicin (25 mg/l) and gentamycin (50 mg/l) for A. tumefaciens GV-3101 and kanamycin (50 mg/l) and streptomycin (100 mg/l) for A. tumefaciens LB-4404 strain. Centrifugation at 5,000 g, 4 °C for 5 min pelleted down the bacterial culture. This pellet was re-suspended in 1:1 MS liquid medium and used for co-transformation of the pre-cultivated leaves by keeping them in it for 15 min. with gentle shaking. The Agrobacterium infected leaves were blot dried and placed on the co-cultivation medium (CCM = MS salts + 0.5 mg/l IAA and 1 mg/l ZR). After 48 h, cotyledonary leaf discs were treated with 250 mg/l cefotaxime for 15–20 min. and then transferred onto regeneration medium (RM = MS Salts + 1 mg/l Zeatin Riboside, 0.5 mg/l IAA + 50 mg/l kanamycin and 250 mg/l cefotaxime). The plates were kept in the tissue culture lab at 25 °C under 16 h light and 8 h dark cycle. After every 15 days, the healthy plants were subcultured into fresh shooting medium for shoot induction and elongation. The regenerated shoots were excised from the callus and transferred on to the rooting medium (MS + 0.5 mg/l IAA + 50 mg/l kanamycin + 250 mg/l cefotaxim). After 20–25 dyas, tomato plantlets with well developed shoots and roots were transferred to 10 cm diameter pots containing 50% soil rite mixed with autoclaved soil for hardening. After hardening, the plants were shifted to growthe chambers and maintained under the controlled condition at 26 °C with photoperiod of 16/8 h (light/dark) at National Phytotron Facility,ICAR-IARI, New Delhi (Koulagi and Sirohi 2015). A set of untransformed plants were grown to serve as control.
Molecular analysis of putative transgenic events
DNA isolation and PCR confirmation
The genomic DNA from leaves of T0 events was isolated using CTAB method (Murray and Thomson 1980). DNA integrity was evaluated on 0.8% agarose gel and PCR confirmations of putative transgenic events was done by using nptII, Integrase and AC4 gene specific primers (Table 1) with BIO-RAD C1000™ thermal cycler. The amplified products were analyzed on1% agarose gel.
Southern blot of T1 tomato events
Seeds of T0 generations were grown on MS medium containing 100 mg/l kanamycin and fifteen days old healthy plants were shifted to pots filled with soil sand mixture and grown at 25 °C, 70% RH, 16 h light and 8 h dark for 10–15 days in the phytotron chamber. DNA was extracted from the leaves and was analyzed by PCR using respective gene specific primers as listed in Table 1.
DNA from the PCR confirmed transgenic plants was used for southern blot analysis for ensuring integration of T-DNA. 20 µg of DNA from each sample was digested with 50U BamHI and HindIII(New England Biolabs, UK) separately, for 16 h at 37 °C. Digested DNA was electrophoresed on 0.8% agarose gel and transferred onto a nitro-cellulose membrane (PALL-Life Science) by capillary action in 10X saline sodium citrate (SSC) buffer. Integrase (624 bp) and AC4 (250 bp) gene fragments were labeled with α [32P]-dCTP using mega prime DNA labeling kit (Amersham Pharmacia Biotech) to as probes. Hybridization was carried out at 65 °C for 18 h and thereafter the membranes were washed with 3XSSC and 0.1% SDS, followed by 0.5X SSC and 0.1% SDS buffers, for 30 min each at 65 °C. The last wash of the membranes was done with 0.1 × SSC and 0.1% SDS for 30 min. (Southern 1975) and after this they were exposed to Fujifilm (Kodak) for 3 days at − 80 °C and developed.
Quantification of Integrase and AC4gene expression in T1 events
Quantitative gene expression analysis was studied in the southern hybrid positive plants by real time PCR (qRT-PCR). Total plant RNA was isolated from freshly harvested leaves by using the Trizol method and cDNAwas synthesized using Verso cDNA kit (Thermo scientific) following manufacturer’s protocol. Master mixes for qRT PCR were prepared using 2X KAPA SYBR FAST qPCR master mix (Sigma Aldrich) following manufacturer’s protocol and the amplification reactions were carried out 95 °C for 3 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 min. using the real time PCR machine [Applied Biosystems]. Specificity of amplification was assessed by disassociation or melt curve analysis at 60–90 °C after 40 cycles. Tomato actin gene served as the internal reference gene and was used to normalize the expression. The reactions were conducted for each of the analyzed samples with two biological replicates and three technical replicates. The average Ct values were documented to calculate the fold changeusing2−ΔΔCT (Livak and Schmittgen 2001).
Validation of dual resistance in T1 events
Surface sterilized T1 seeds were placed on MS media containing 100 mg/l kanamycin and healthy grown plants were transferred to pots filled with autoclaved soil mixture. These pots were maintained at 26 °C and 70% RH with a photoperiod of 16:8 (light: dark). DNA was extracted from the fresh leaves of these transgenic plants as explained earlier and was analyzed by PCR using respective gene specific primers.
Agroinoculation of ToLCNDV
Tomato seedlings were inoculated with A. tumefaciens strain EHA 105 harboring DNA-A and DNA-B of Tomato leaf curl New Delhi virus (ToLCNDV), through syringe infiltration of Agrobacterium by stem pricking method (Jyothsna et al. 2013). Observations on the development of typical leaf curl disease symptoms were documented after 20 and 50 days post inoculation (DPI). Leaf samples were collected and DNA isolated for PCR based validation of presence or absence of the gene in the viral genome by using AC4 gene specific primers.
Resistance validation in T1 events against M. incognita
T1 tomato plants were first inoculated with ToLCNDV and allowed to grow for 20 days. Thereafter these plants along with untransformed control plants were transferred to 10 cm diameter pots containing autoclaved soil medium and inoculated with 1000 freshly hatched M. incognita J2s. Plants were uprooted after 30 days post inoculation and roots were washed and observations of the total number of galls, egg masses and eggs per egg mass were recorded for each transgenic event. Multiplication factor of RKN [(Number of egg masses × number of eggs peregg mass) ÷ initial nematode inoculum] was calculated to assess the effect of targeted gene on development and multiplication of M. incognita. The recorded observations were compared with the untransformed, RKN inoculated control plants grown under similar environmental conditions. Experiment was conducted with four replicates of each event. The root galls of transformed and untransformed plants were stained (Byrd et al. 1983) and dissected to detect the morphological variation in the development of M. incognita.
Statistical analysis
The research data documented for all the experiments was statistically evaluated using the analysis of variance (ANOVA). Observations were testified as significant or non-significant using the CRD test, means were separated by using Duncan’s multiple range test at P < 0.01 significance level using software, SAS 9.3.
Phenotypic validation of dual resistance in T2 events
T2 seeds were surface sterilized and placed on MS media containing 100 mg/l kanamycin and healthy grown plants were transferred to pots filled with autoclaved soil mixture. These pots were maintained at 26 °C and 70% RH with a photoperiod of 16:8 (light: dark). DNA was extracted from the fresh leaves of these transgenic plants as explained earlier and was analyzed by PCR using respective gene specific primers. The gene expression of Integrase and AC4 gene was studied from the PCR confirmed T2 transgenic plants using the earlier described protocol for qRT-PCR. These tomato seedlings were inoculated with A. tumefaciens strain EHA 105 harboring DNA-A and DNA-B of Tomato leaf curl New Delhi virus (ToLCNDV) as per the protocol described for T1 generation. Visual observations were recorded for symptom development after 20 and 50 days post inoculation (DPI). To know the presence or absence of the AC4 gene in the viral genome, a PCR based validation was done by using AC4 gene specific primers.
Absolute quantification of ToLCNDV titer T2 events
The absolute copy number of ToLCNDV in transgenic events was calculated using the linear equation obtained from the standard curve. Total RNA was isolated from all virus inoculated transgenic and non-transgenic plants using Trizol method. cDNA was synthesized from 500 ng RNA usingVerso cDNA kit (Thermo scientific) following manufacturer’s protocol. qRT-PCR was performed, as explained earlier, by using AC4 primers. The mean Ct values were used for calculating the absolute copy number of the virus.
Preparation of standard curve
Standard curve was prepared by serial dilution (10) of a plasmid containing ToLCNDV-AC4 gene. qRT-PCR was performed at different serial concentrations (9 ng/μl, 0.9 ng/μl, 0.09 ng/μl, and 0.009 ng/μl) of plasmid DNA by using AC4gene specific primers (Table 1). Log copy number was calculated for each concentration and graph was plotted between mean Ct values vs log copy number (Fig. 2). The linear equation obtained from this graph was used for calculation of absolute copy number of ToLCNDV in transgenic and non-transgenic plants. Three technical replicates were used for each concentration.
Meloidogyne incognita infection analysis of T2 tomato events
T2 tomato plants were first inoculated with ToLCNDV and allowed to grow for 20 days. Thereafter these plants along with untransformed control plants were transferred to 10 cm diameter pots containing autoclaved soil medium and inoculated with 1000 freshly hatched M. incognita J2s. 30 DPI of M. incognita, the plants were uprooted and observed for nematode infection assay parameters including number of galls, egg masses and eggs per egg mass. Multiplication factor of RKN was considered to assess the effect of Integrase gene on M. incognita development. The data recorded was used for statistical analysis.
Transcript analysis of Meloidogyne incognita-Integrase gene
The RNA was mined from adult females of M. incognita, isolated from the roots of transgenic and non-transgenic plants. The cDNA was synthesized using 500 ng RNA by Verso cDNA kit (Thermo-scientific) following manufacturer’s protocol. Integrase transcript analysis was done by using qRT-PCR and it’s relative change in expression was calculated by using 2−ΔΔCT. The qRT-PCR analyses were replicated six times biologically and thrice technically.
Results
Molecular confirmation of putative transgenic events through PCR
Tomato (S. lycopersicumcv. Pusa Ruby) plants were co-transformed simultaneously using gene constructs containing binary vectors pBC-142 and pCAMBIA-2301, harboring M. incognita-Integrase and ToLCNDV-AC4 genes by the Agrobacterium mediated method and T0 plants generated. PCR analysis of the co-transformed tomato plants was carried out to confirm the integration of the transgenes by using gene specific primers (Table 1). Gene amplification results of 624 & 250 bp indicated presence of (Integrase &AC4 genes respectively in the transgenic events (Fig. 3). Seeds of the PCR confirmed plants were collected and used for growing the next generation of transgenic events.
PCR confirmation of T0 primary tomato (cv. Pusa Ruby) events using respective gene specific primers. a With nptII gene specific primers, M-100 bp DNA ladder, 12-PR-Int+AC4-12(T0), 55-PR-Int+AC4-55(T0), 56-PR-Int+AC4-56(T0), 58-PR-Int+AC4-58(T0), 61-PR-Int+AC4-61(T0), 68-PR-Int-68(T0),74-PR-Int+AC4-74(T0) and 80-PR-Int+AC4-80(T0) transgenic lines, W-untransformed tomato plant, -ve-Negetive control, + v Positive control (Plasmid DNA). b With M. incognita-Integrase gene specific primers. c With ToLCNDV-AC4 gene specific primers
Southern blot analysis of T1 events
PCR+veT1events of Integrase &AC4 genes were put to southern blot analysis to confirm the integration of T-DNA. Southern blot analysis identified six events harboring Integrase gene [PR-Int-12(T1), PR-Int-55(T1), PR-Int-58(T1), PR-Int-68(T1), PR-Int-74(T1) and PR-Int-80(T1)] four events harboring AC4 gene [PR-AC4-61(T1), PR-AC4-68(T1), PR-AC4-74(T1) and PR-AC4-80(T1)] and three events [PR-Int+AC4-68(T1), PR-Int+AC4-74(T1) and PR-Int+AC4-80(T1) exhibited integration of both Integrase and AC4 genes. No gel band corresponding to the genes under study was detected in untransformed plants and in undigested sample used as negative control (Fig. 4a and b).
Southern blot analysis of the T1 tomato (cv. Pusa Ruby) events harboring dsRNA of M. incognita-Integrase and ToLCNDV-AC4 genes. a Southern analysis to confirm the integration of M. incognita-Integrase gene. Lanes—M: Lambda HindIII digest, WT-Digested untransformed tomato plant DNA: 12-PR-Int+AC4-12(T1), 55-PR-Int+AC4-55(T1), 56-PR-Int+AC4-56(T1), 57-PR-Int+AC4-57(T1), 58-PR-Int+AC4-58(T1), 61-PR-Int+AC4-61(T1), 68-PR-Int+AC4-68(T1), 74-PR-Int+AC4-74(T1), 76-PR-Int+AC4-76(T1), 80-PR-Int+AC4-80(T1), and 82-PR-Int+AC4-82(T1) -DNA samples from T1 tomato events digested with BamHI restriction enzyme. b Southern analysis to confirm the integration of ToLCNDV-AC4gene. Lanes—M: Lambda HindIII digest, WT-Digested untransformed tomato plant DNA: DNA samples from T1 tomato events digested with HindIII restriction enzyme
Expression analysis of Integrase and AC4 genes in T1 events.
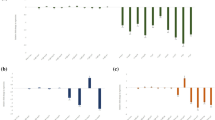
The qRT-PCR analysis was performed to observe the abundance of the targeted gene transcript and expression pattern in the southern blot confirmed T1 events. All the transgenic events showed increased transcript abundance in comparison to untransformed plants with respect to both the genes (Fig. 5). The expression pattern was observed to be variable in all the transgenic events, however, AC4 gene was found to have more abundant expression in all the events than Integrase gene. The maximum expression of Integrase and AC4 transcripts was recorded in PR-Int-12(T1) and 74PR-Int+AC4-74(T1) events, respectively.
Quantification and expression of T1 tomato (cv. Pusa Ruby) events through real time PCR; a M. incognita-Integrase gene expression in different events; 12-PR-Int-12(T1), 55-PR-Int-55(T1), 58-PR-Int-58(T1), 68-PR-Int+AC4-68(T1), 74-PR-Int+AC4-74(T1), and 80-PR-Int+AC4-80(T1). b ToLCNDV-AC4 gene expression in various events; 61- PR-AC4-61(T1), 68- PR-Int+AC4-68(T1), 74-PR-Int+AC4-74(T1) and 80-PR-Int+AC4-80(T1); fold change was calculated by using 2−ΔΔCT method. Each bar represents the mean ± SE of n = 3, at P < 0.05
Validation of dual resistance T1events
Assessment of ToLCNDV resistance inT1 events
In order to assess the effect of AC4dsRNA construct on ToLCNDV, three transgenic events {68 [PR-Int+AC4-68(T2)],74 [PR-Int+AC4-74(T2)] and 80 [PR-Int+AC4-80(T2)]} expressing Integrase + AC4 dsRNA and one transgenic event {61[PR-AC4-61(T2)} expressing only AC4 in dsRNA were tested with ToLCNDV through Agrobacterium infiltration assay. Symptoms of infection were recorded at 20 and 50 DPI. None of the infection symptoms of such as curling, cupping, yellowing of leaves and stunting of plants were observed in any of the four events (Table 2, Fig. 6).To prove this at molecular level, DNA from the virus inoculated transgenic events and non transformed tomato plants was extracted and PCR analysis was performed using AC4 gene specific primers. Suppression of the viral disease was confirmed by non-amplification of specific gene fragment in the transgenic plants vis a vis untransformed plants. The untransformed plants showed typical tomato leaf curl virus symptoms and PCR confirmed amplification of AC4 gene (Fig. 7).
PCR based validation of resistance against ToLCNDV in T2 tomato plants Transgenic events - 61-PR-AC4-61(T2), 68-PR-Int+AC4-68(T2), 74-PR-Int+AC4-74(T2) and 80-PR-Int+AC4- 80(T2); W-untransformed tomato plant: -ve-Water control: +ve-Plasmid DNA: M-100bp DNA marker:−̶ sign indicates absence of ToLCNDV and + sign indicates presence of ToLCNDV
Meloidogyne incognita bioassay in T1 events
RKN resistance bioassay of the transgenic events {12 [PR-Int-12(T1)], 55 [PR-Int-55(T1)], 58 [PR-Int-58(T1)], 68 [PR-Int+AC4-68(T1)], 74 [PR-Int+AC4-74(T1)] and 80 [PR-Int+AC4-80(T1)]}having Integrase dsRNA, was done by inoculating freshly hatched M. incognita J2s and observations recorded after 30 days. Mean number of galls per plant reduced significantly (P < 0.01) by 60–93% (Table 3) in transgenic events as compared to untransformed plants (Fig. 8).Fecundity of RKN in the RNAi transgenic events was highly affected. Average number of egg masses per plant reduced between 83 to 97% and the number of eggs per egg mass reduced between 24 to 41% (P < 0.01). The nematode multiplication factor, which is indicative of nematode reproductive potential and parasitic success got reduced significantly (P < 0.01) by 88–98% in transgenic events in comparison to untransformed tomato plants. Dissection of stained galls revealed detrimental effect on the development of M. incognita female. In untransformed plants, saccate shaped adult females with normal development were observed, where as in transgenic root galls they were elongate in shape, had deformed neck and transparent body (Fig. 9).
Quantification and expression of Integrase and AC4 genes in T2 events
An increase in the transcript level of M. incognita-Integrase and ToLCNDV-AC4genes was observed with respect to the untransformed plants in all the T2transgenic tomato events (Fig. 10). However, the expression of both these genes was less in T2events as compared to their respective T1 events.
Quantification and expression of T2 tomato (cv. Pusa Ruby) events through real time PCR; a M. incognita-Integrase gene expression in different events; 12-PR-Int-12(T2), 55-PR-Int-55(T2), 58-PR-Int-58(T2), 68-PR-Int+AC4-68(T2), 74-PR-Int+AC4-74(T2) and 80-PR-Int+AC4-80(T2). b ToLCNDV-AC4 gene expression in various events; 61- PR-AC4-61(T2), 68- PR-Int+AC4-68(T2), 74-PR-Int+AC4-74(T2) and 80-PR-Int+AC4- 80(T2); fold change was calculated by using 2-ΔΔCT method. Each bar represents the mean ±SE of n = 3, at P < 0.05
Assessment of ToLCNDVresistanceinT2 events
The same T1 transgenic events were used to assess the effect of AC4 dsRNA construct on ToLCNDV, through Agrobacterium infiltration assay. The similar results were obtained as of T1 events (Table 2). These were further confirmed by calculating the absolute copy number of ToLCNDV by using standard curve. Significant reduction in the viral copy number was recorded in all the transgenic events as compared to untransformed tomato plant (Fig. 11). Event 74 [PR-Int+AC4-74(T2)] showed least copy number (0.115 × 104) of virus in comparison to untransformed plant. These results prove that dsRNA suppressed the leaf curl disease in the in transgenic events of tomato.
Assessment of Meloidogyne incognita resistance in tomato transgenic events
Freshly hatched, M. incognita J2s (2J2s/cc soil) were used to inoculate the tomato events having single{12 [PR-Int-12(T2)], 55 [PR-Int-55(T2)], 58 [PR-Int-58(T2)]} and dual genes {68 [PR-Int+AC4-68(T2)], 74[PR-Int+AC4-74(T2)], 80 [PR-Int+AC4-80(T2)]}. The inoculated transgenic plants were uprooted after 30 DPI and observations on the reproduction and development of RKN (Table 4) recorded. Transgenic plants with single gene Integrase, exhibited reduced growth and reproduction of M. incognita. Average number of root galls reduced by 70–77%. Reduction in egg mass and eggs per egg mass was in the range of 72–94% and 29–52%, respectively. Nematode multiplication factor of the transgenic tomato plants reduced by 80–97% in comparison to the control plants. One transgenic event- 68 [PR-Int+AC4-68(T2)] expressing Integrase dsRNA construct didn’t show significant reduction in the number of galls in relation to the untransformed plants, but reduced number of egg masses and eggs per egg mass lead to reduction in nematode multiplication factor by 66%. In co-transformed plants (Integrase + AC4), the average number of galls per plant was reduced significantly (P < 0.01) by 62 to 63% as compared to untransformed plants and also they were of smaller size. Significant reduction in egg mass and eggs per egg mass (P < 0.01) in the range of 51 to 70% and 31 to 38% respectively was noticed in transformed plants w.r.t. untransformed plants. Nematode multiplication factor, an indicator of its reproductive ability was also significantly reduced in the range of 66 to 81%. All these results establish that transgenic events carrying dual gene confer the resistance to both ToLCNDV and the RKN..
Meloidogyne incognita-Integrase gene transcript analysis of adult females
To analyze the abundance of expressed transcript level of Integrase dsRNA in M. incognita, qRT-PCR was performed using cDNA of M. incognita adult females isolated from T2 transgenic events 12[PR-Int-12(T2)], 55[PR-Int-55(T2)], 58[PR-Int-58(T2)], 68[PR-Int+AC4-68(T2)], 74[PR-Int+AC4-74(T2)] and 80[PR-Int+AC4-80(T2)] at 30DPI. The expression level of Integrase transcript in the RKN females isolated from transgenic events was significantly lower in the range of 20–90% when compared with untransformed plants (Fig. 12).
Discussion
Tomato (S. lycopersicum L.) cultivation worldwide is greatly threatened by the root-knot nematode, M. incognita and ToLCV. Despite of high economic damage caused to tomato cultivation worldwide by these two disease causing agents, we don’t have any effective and environmentally safe method of their management. Host resistance is possibly the best natural attribute to combat disease, but none of the plant species are reported to carry resistance against both, the RKN and ToLCV together. Effectiveness of host delivered RNAi was first successfully demonstrated by Yadav et al. (2006) in tobacco by incorporating dsRNA constructs of the housekeeping genes target i.e. Integrase and Splicing factor of M. incognita. The study showed more than 90% reduction in nematode population in transgenic tobacco and paved way to engineer resistance in crop plants against these ubiquitous parasites.
The present study attempted to engineer resistance against RKN and ToLCV in tomato cv. Pusa Ruby via co-transformation of RNAi genes and assessed the level of resistance against M. incognita in T1& T2 generations and of ToLCV in T2 generation. The attained results indicate that stacking of transgenes in tomato is an achievable strategy to develop resistance against these two disease-causing agents. Seven transgenic tomato events were generated using binary vectors (pBC142-Intgrase and pCAMBIA2301-AC4) and Agrobacterium based transformation. Molecular validation revealed that events 12[PR-Int-12]; 55 [PR-Int-55] and 58[PR-Int-58] incorporated Integrase dsRNA and events 68[PR-Int+AC4-68]; 74[PR-Int+AC4-74] and 80[PR-Int+AC4-80] had both Integase + AC4 dsRNAs and event 61[PR-AC4-61] had only AC4 dsRNA construct. Transgenic events containing dual dsRNA{68;74 and 80}exhibited complete suppression of ToLCV and a mean reduction of 75% in the multiplication factor of M. incognita. Transgenic events with only Integrase dsRNA {12; 55 and 58} showed 88% control of M. incognita. Our results are in tandem with the results of Ramesh et al. (2007) and Yadav et al. (2006) who independently demonstrated the impact of host delivered RNAi induced resistance against ToLCV and M. incognita in tomato and tobacco respectively, using the same genes. Many researchers have successfully demonstrated the effectiveness of host delivered RNAi against M. incognita by targeting different genes in various crops and with varying degree of success (Niu et al. 2012; Choudhary et al. 2012; Xue et al. 2013; Papoluet al. 2013; Lourenço-Tessutti et al. 2015; Dutta et al. 2015; Niu et al. 2016; Kumar et al. 2017; Banerjee et al. 2018). Kumar et al. (2017) Host delivered RNAi approach was used to target the M. incognita- Integrase gene in Arabidopsis. The results showed significant mean reduction in the number of galls, number of females and number of egg masses to the tune of 59.5%, 66.8% and 63.4%, respectively. RKN bioassay studies conducted on the transgenic tomato events (T1) incorporating dsRNA targeting RKN Integrase gene showed significant reduction in the number of galls, number of egg mass and number of eggs per egg masses in the range of 60–93%, 83–97% and 24–41%, respectively. The RKN multiplication factor also got reduced by 88 to 98% in comparison to the RKN inoculated untransformed tomato plants. On perusal of the results of RNAi silencing of RKN integrase gene in tobacco (Yadav et al. 2006), Arabidopsis (Kumar et al. 2017) and tomato (present study), it is observed that the resistance level offered by the same gene (Integrase) against M. incognita is higher in tomato and tobacco than Arabidopsis. This may be due to host preferability of the nematode and variation in host response to the parasite, as tobacco and tomato belong to Solanaceae family and Arabidopsis belongs to Brasssicaceae family. Literature review suggests that plants belonging to Solanaceae family are good hosts to RKN where as plants classified under Brassicaceae family are not preffered hosts. The nematode adult females isolated from the roots of the transgenic plants were not normal pear shaped looking. They had a distorted transparent body with deformed neck and looked similar to those found in transgenic tobacco and Arabidopsis (Yadav et al. 2006; Kumar et al. 2017). The nematode deformation can be directly correlated with the RNAi based functional obstruction or silencing of the house keeping gene Integrase. This gene of M. incognita (Acc. No. AW871671) is involved in pre-mRNA splicing (Joining of exons after removal of introns).
In order to judge the resistance against the ToLCNDV, the T2 generation transgenic events harbouring ToLCNDV-AC4 + Integrase or alone ToLCND-AC4 were challenged with ToLCNDV using the Agrobacterium infiltration method. None of the events showed any virus symptom throughout their growth stage where as the untransformed plants, showed typical leaf curl symptoms.AC4-gene specific PCR assay showed a complete absence of the particular gene fragment in transgenic events. However, results of qRT-PCR analysis showed presence of viral titer in all the transgenic events but significantly less as compared to untransformed plants. Our results were consistent with many studies on transgenic resistance to Geminiviruses through RNAi approach (Arago et al. 1998; Ramesh et al. 2007; Praveen et al. 2010; Nahid et al. 2011; Lin et al. 2011; Ammara et al. 2015). AC4 gene codes for a pathogenicity determination factor (Moriones et al. 2017) and as the factor got silenced, no viral disease symptom was observed.
After 20 DPI of viral infection, the same transgenic events were challenged with M. Incognita and the observations revealed after 30 DPI of M. incognita, significant reduction in the number of galls (62–63%), number of egg mass per plant (51–70%), number of eggs per plant (31–38%) and multiplication factor (66–81%). However transgenic event 68[PR-Int+AC4-68] was less effective in reducing the number of galls as compared to transgenic events 74[PR-Int+AC4-74] and 80[PR-Int+AC4-80].This kind of result may be due to the low expression of Integrase in this event as correlated by the result obtained from qRT analysis. Similar variation in gene expression result was reported by Papolu et al. (2013) in tobacco against M. incognita in which one of the events (A 39.1) expressing flp-18 was ineffective in reducing M. incognita due to very low gene expression. These results indicated that a minimum threshold level of dsRNA expression in the host is essential for an effective silencing of a particular gene/s.
There are very few studies similar to ours where RNAi has been used to engineer resistance against a nematode and another pathogen within the same plant. Walawage et al. (2013) stacked two RNAi genes in a single walnut (Juglans regia) rootstock genotype for lesion nematode and crown gall disease resistance. A. tumefaciens, carrying self-complimentary iaaM and ipt transgenes, and Agrobacterium rhizogenes, having a self-complimentary Pv010 gene from P. vulnus, served as co-transformation vectors. Complete suppression of the crown gall disease was observed in transgenic event having both the genes but only 32% reduction in nematode number in the roots of this event was recorded where as transgenic events incorporating only the nematode gene Pv010 showed 79 to 100% reduction in nematode population.
The results of our study make us conclude that stacking or pyramiding of genes for RNAi based resistance engineering against different set of pathogens/parasites is a doable option. To our knowledge, this is the second such publication reporting stacking of root-knot nematode, M. incognita and ToLCNDV resistance in tomato by RNAi approach. The results of this study unlock prospects for stacking multiple pest/pathogen resistance in important crop plants. This model/strategy could also be useful to engineer crops resistant to biotic as well as abiotic stress.
References
Ammara U, Mansoor S, Saeed M, Amin I, Briddon R, Mohammed A (2015) RNA interference-based resistance in transgenic tomato plants against Tomato yellow leaf curl virus-Oman (TYLCV-OM) and its associated betasatellite. Virol J 12:38. https://doi.org/10.1186/s12985-015-0263-y
Aragao FJL, Ribeiro SG, Barros LMG, Brasileiro AM, Maxwell DP, Rech EL, Faria JC (1998) Transgenic beans (Phaseolus vulgaris L.) engineered to express viral antisense RNAs show delayed and attenuated symptoms to bean golden mosaic geminivirus. Mol Breeding 4:491–499
Banerjee S, Banerjee A, Gill SS, Gupta OP, Dahuja A, Jain PK, Sirohi A (2017) RNA interference: a novel source of resistance to combat plant parasitic nematodes. Front Plant Sci 8:834. https://doi.org/10.3389/fpls.2017.00834
Banerjee S, Gill SS, Gawade BH, Jain PK, Subramaniam K, Sirohi A (2018) Host delivered RNAi of two cuticle collagen genes, Mi-col-1 and Lemmi-5 hampers structure and fecundity in Meloidogyne incognita. Front Plant Sci 8:2266. https://doi.org/10.3389/fpls.2017.02266
Boulter D, Edwards GA, Gatehouse AMR, Gatehouse JA, Hilder VA (1990) Additive protective effects of different plant-derived insect resistance genes in transgenic tobacco plants. Crop Prot 9(5):351–354. https://doi.org/10.1016/0261-2194(90)90005-R
Byrd DW, Krickpatrick T, Barker KR (1983) An improved technique for cleaning and staining of plant tissue for detection of nematodes. J Nematol 15:142–143
Chellappan P, Vanitharani R, Fauquet CM (2005) Micro-RNA-binding viral protein interferes with Arabidopsis development. Proc Natl Acad Sci 102(29):10381–10386. https://doi.org/10.1073/pnas.0504439102
Choudhary D, Koulagi R, Rohatagi D, Kumar A, Jain P K, Sirohi A (2012) Engineering resistance against root-knot nematode, Meloidogyne incognita, by host delivered RNAi, in Abstracts of International Conference on Plant Biotechnology for food Security: New Frontiers (New Delhi: National Agricultural Science Centre) 21–24.
Dropkin VH (1980) Introduction to plant nematology. Wiley, New York
Dutta TK, Papolu PK, Banakar P, Choudhary D, Sirohi A, Rao U (2015) Tomato transgenic plants expressing hairpin construct of a nematode protease gene conferred enhanced resistance to root-knot nematodes. Front Microbiol 6:260. https://doi.org/10.3389/fmicb.2015.00260
Elling AA (2013) Major emerging problems with minor Meloidogyne species. Phytopathology 103(11):1092–1102. https://doi.org/10.1094/PHYTO-01-13-0019-RVW
Hooper DJ (1986) Extraction of free-living stages from soil. In: Southey JF (ed) Laboratory methods for work with plant and soil nematodes. Ministry of Agriculture, Fisheries and Food, London, pp 5–30
Jain RK, Mathur KN, Singh RV (2007) Estimation of losses due to plant parasitic nematodes on different crops in India. Indian J Nematol 37(2):219–221
Jones JT, Haegeman A, Danchin EGJ, Gaur HS, Helder J, Jones MGK, Kikuchi T, Manzanilla-Lopez R, Palomares-Rius JE, Wesemae WIMML, Perry RN (2013) Top 10 plant-parasitic nematodes in molecular plant pathology. Mol Plant Pathol. 14(9):946–961. https://doi.org/10.1111/mpp.1205
Jyothsna P, Haq QMI, Singh P, Sumiya KV, Praveen S, Rawat R, Briddon RW, Malathi VG (2013) Infection of Tomato leaf curl New Delhi virus (ToLCNDV), a bipartite begomovirus with betasatellites, results in enhanced level of helper virus components and antagonistic interaction between DNA B and betasatellites. Appl Microbiol Biotechnol 97:5457–5471
Khan H, Ahmad R, Akhtar AS, Arshad M, Tahir B, Tariq N (2000) Effect of inoculums density of Meloidogyne incognita and plant age on the severity of root knot disease in tomato. Int J Agric Biol 2(4):360–363
Koulagi R, Sirohi A (2015) Gene stacking through Agrobacterium mediated co-transformation in tomato to engineer resistance against root-knot nematode, Meloidogyne incognita and tomato leaf curl virus. Indian J Nematol 45(2):161–168
Krake L, Rezaian MA, Dry IB (1998) Expression of the tomato leaf curl geminivirus C4 gene produces virus like symptoms in transgenic plants. Mol Plant-Microbe Interact 11:413–417. https://doi.org/10.1094/MPMI.1998.11.5.413
Kumar A, Kakrana A, Sirohi A, Subramaniam K, Srinivasan R, Abdin MZ, Jain PK (2017) Host-delivered RNAi-mediated root-knot nematode resistance in Arabidopsis by targeting splicing factor and integrase genes. J Gen Plant Pathol. https://doi.org/10.1007/s10327-017-0701-3
Lin CY, Tsai WS, Ku HM, Jan FJ (2011) Evaluation of DNA fragments covering the entire genome of a monopartite begomovirus for induction of viral resistance in transgenic plants via gene silencing. Transgenic Res 21(2):1–11. https://doi.org/10.1007/s11248-011-9523-9
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2-Delta C (T) Method. Methods 25:402–408. https://doi.org/10.1006/meth.2001.1262PubMed: 11846609
Lourenço-Tessutti T, Souza Junior JDA, Martins-de-Sa D, Viana AAB, Carneiro RM, Togawa RC et al (2015) Knockdown of heat-shock protein 90 and isocitrate lyase gene expression reduced root-knot nematode reproduction. Phytopathology 105(5):628–637. https://doi.org/10.1094/PHYTO-09-14-0237-R
Moens M, Perry RN, Starr JL (2009) Meloidogyne spp.—a diverse group of novel and important plant parasite. In: Perry RN, Moens M, Starr JL (eds) Root-knot nematodes. CAB International, Wallingford, pp 1–17
Moriones E, Praveen S, Chakraborty S (2017) Tomato Leaf Curl New Delhi Virus: an emerging virus complex threatening vegetable and fiber crops. Viruses 9(10):264. https://doi.org/10.3390/v9100264
Murray MG, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 8:4321–4325
Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Plant Physiol 15:473–497
Nahid N, Amin I, Briddon RW, Mansoor S (2011) RNA interference based resistance against a legume mastrevirus. Virol J 8:499. https://doi.org/10.1186/1743-422X-8-499
Niu JH, Jian H, Xu J, Chen C, Guo Q (2012) RNAi silencing of the Meloidogyne incognita Rpn7 gene reduces nematode parasitic success. Eur J Plant Pathol 134:131–144. https://doi.org/10.1007/s10658-012-9971-y
Niu J, Liu P, Liu Q, Chen C, Guo Q, Yin J et al (2016) Msp40 effector of root-knot nematode manipulates plant immunity to facilitate parasitism. Sci Rep 6:19443. https://doi.org/10.1038/srep19443
Papolu PK, Gantasala NP, Kamaraju D, Banakar P, SreevathsaR RU (2013) Utility of host delivered RNAi of two FMRF amide like peptides, flp-14 and flp-18, for the management of root-knot nematode, Meloidogyne incognita. PLoS ONE 8:e80603. https://doi.org/10.1371/journal.pone.0080603
Praveen S, Mishra AK, Ramesh SV, Koundal V, Palukaitis P (2010) Silencing potential of viral derived RNAi constructs in Tomato leaf curl virus-AC4 gene suppression in tomato. Transgenic Res 19(1):45–55. https://doi.org/10.1007/s11248-009-9291-y
Ramesh SV, Mishra AK, Praveen S (2007) Hairpin RNA mediated strategies for silencing of tomato leaf curl virus AC1 and AC4 genes for effective resistance in plants. Oligonucleotide 17:251–257. https://doi.org/10.1089/oli.2006.0063
Rataul HS, Brar JS (1989) Status of tomato leaf curl virus research in India. Trop Sci 29(2):111–118
Rojas MR, Hagen C, Lucas WJ, Gibertson RL (2005) Exploiting chinks in the plant’s armor: evolution and emergence of geminiviruses. Annu Rev Phytopathol 43:361–394. https://doi.org/10.1146/annurev.phyto.43.040204.135939
Seal SE, van den Bosch F, Jeger MJ (2006) Factors influencing begomovirus evolution and their increasing global significance: Implications for sustainable control. Crit Rev Plant Sci 25(1):23–46. https://doi.org/10.1080/07352680500365257
Southern EM (1975) Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol 98:503–517. https://doi.org/10.1016/S0022-2836(75)80083-0PubMed:1195397
Walawage SL, Britton MT, Leslie CA, Uratsu SL, Li YY, Dandekar AM (2013) Stacking resistance to crown gall and nematodes in walnut rootstocks. BMC Genomics 14:668. https://doi.org/10.1186/1471-2164-14-668
Xue B, Hamamouch N, Li C, Huang G, Hussey RS, Baum TJ et al (2013) The 8D05 parasitism gene of Meloidogyne incognita is required for successful infection of host roots. Phytopathology 103:175–181. https://doi.org/10.1094/PHYTO-07-12-0173-R
Yadav BC, Veluthambi K, Subramaniam K (2006) Host generated double stranded RNA induces RNAi in plant-parasitic nematodes and protects the host from infection. Mol Biochem Parasitol 148:219–222
Yadava P, Mukherjee SK (2012) Artificial microRNA and its applications. In: Bibekanand M, Zhumur G (eds) Regulatory RNAs: basics, methods and applications. Springer, Berlin, pp 505–521
Acknowledgement
First author acknowledges the Indian Council of Agricultural Research (ICAR), New Delhi for financial support (ICAR-Senior Research Fellowship) during the research work. Authors are thankful to Head, Division of Nematology, ICAR-IARI, New Delhi for providing necessary facilities and resources for conducting experiments.
Funding
First author acknowledges the Indian Council of Agricultural Research (ICAR), New Delhi for financial support (ICAR-Senior Research Fellowship) during the research work.
Author information
Authors and Affiliations
Contributions
AS, PKJ, SP and RK conceived and designed the experiments. RK performed the experiments. RK, BHG and AKS analyzed the data. RK and SB wrote the manuscript. AS, PKJ, SP and KS critically revised the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
All the authors declare that they do not have any conflict of interest.
Consent for publication
All the authours agreed for the publication of present research.
Additional information
Communicated by Jose M. Segui-Simarro.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Koulagi, R., Banerjee, S., Gawade, B.H. et al. Host-delivered RNA interference in tomato for mediating resistance against Meloidogyne incognita and Tomato leaf curl virus. Plant Cell Tiss Organ Cult 143, 345–361 (2020). https://doi.org/10.1007/s11240-020-01921-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11240-020-01921-3