Abstract
Indole derivatives are reported in the literature for their excellent kinase inhibition activity, so understanding their structural requirement is important. For their further development, ligand-based pharmacophore, atom and field-based 3D-QSAR, and ADME studies of the 3, 5-disubstituted indole derivatives were carried out. Ligand-based pharmacophore, atom and field-based 3D-QSAR models were developed using the Phase module of Schrodinger suite. In silico ADME and drug-likeness properties were studied using the Quikprop module of Schrodinger suite. Five-point pharmacophore model (DHRRR _1) with one hydrogen bond donor (D), one hydrophobic site (H), and three aromatic rings (R) was developed. 3D-QSAR models yielded with good statistical results as the models were characterized by PLS factors 4 and validated by parameters like R2, R2 CV, Stability, F-value, P value, RMSE, Q2, and Pearson-r.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Proto-oncogene serine/threonine-protein kinase (Pim-1) is an enzyme that is encoded by the Pim-1 gene [1, 2]. Pim proteins are of three forms Pim-1, Pim-2, and Pim-3, which regulate various signaling pathways in the development of cancer. [3] Pim kinases are often activated in various downstream substrates that are thought to contribute to tumor growth and survival in both hematologic and solid cancers [4]. The Pim-1 oncogene is identified as being the locus most frequently triggered by the Moloney murine leukemia virus in relation to murine T cell lymphomas [5].
Pim isoforms have different levels of expression and distinct roles. All three Pim genes in cancer, there is compelling evidence of a compensatory mechanism [6,7,8,9].
Adenosine triphosphate-competitive Pim inhibitors, SGI-1776, AZD1208, TP-36544, and LGH447 are in clinical trials for the treatment of acute prostate cancer, myelogenous leukemia, and lymphoma (Fig. 1) [10].
Indole derivatives are widely studied for their kinase inhibitory activity as reported in literature. [11,12,13,14] The novel series of meridianin C derivatives reported in the literature [15] and have shown strong inhibitory activities against Pim-1 and Pim-3 kinases that encouraged us to study 3D QSAR of these compounds.
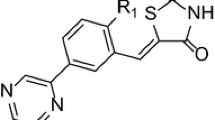
3, 5-disubstituted indole derivatives as Pim kinase inhibitors were selected for 3D QSAR study because authors noted that the electronic properties of the substituents on the phenyl ring did not have any significant effect on the inhibitory function of compounds [15] and the 3D QSAR model can reveal features influencing the activity. It was also stated by the author that the inhibitory activity, in particular for Pim-2 kinase, depended on the position of the substituent [15].
Indole derivatives, specifically 3,5 substituted and meridianin C derivatives have been in focus as Pim kinase inhibitors [11, 16,17,18]. The development of the QSAR model can direct further lead optimization in Pim Kinase inhibitor’s discovery by studying their features viz. steric, electrostatic, hydrophobic, H-bond Acceptor, and H-bond Donor in three-dimensional space. The aim of the study was to disclose the minimum structural requirements for Pim kinase inhibitors based on the indole nucleus that may guide the rational design of potent compounds.
In the present work, the pharmacophore and 3D-QSAR models of 3, 5-disubstituted indole derivatives were generated and validated using parameters like R2, Q2, RMSE, Pearson R, and F value. Also ‘Drug likenesses and ADME properties of molecules were evaluated.
Materials and method
All the molecular modeling work was carried out using the Schrodinger Suite [19, 20].
Selection of ligand library for developing 3D-QSAR studies
All the selected 3,5-disubstituted indole derivatives were taken from the literature [15] (Table 1). The structures of ligands were drawn using 2D sketcher of Schrödinger maestro 2019–3. Structures were optimized for using LigPrep modulea where the structures converted to 3D and energy-optimized using OPLS2005. Least energy conformation was generated for ligands at pH 7.2 using Epik, in the LigPrep module with all other default options.
Pim kinase inhibitors in clinical trials [10]
Data set for modeling the atom-based and field based 3D QSAR studies
For developing an atom and field-based QSAR model, the data set comprised of 40 compounds was selected. All the models were developed through a random selection of training and test set into ratio, 70:30%, respectively, by PHASE moduleb.
Pharmacophore modeling
A minimum of four to a maximum of five pharmacophore features were selected for the development of the pharmacophore hypothesis. A hypothesis difference criterion was kept at 0.50 defaults. There were 8 actives and 32 inactive. Among the hypotheses developed, the top-ranked hypothesis was selected on the basis of survival score.
3D QSAR
All ligands were superimposed upon each other by utilizing the Flexible ligand alignment from the maestro. The assignment of training and test set molecules was totally performed by random splitting into a ratio of 70:30% by software, i.e., 28 training set and 12 test set molecules. The QSAR models were developed using PLS factor of 4 for atom-based and field-based QSAR models development. Grids spacing were kept as 1 Å. Gaussian based field style was utilized for field-based models which include five features like Gaussian Steric, Gaussian Electrostatic, Gaussian Hydrophobic, Gaussian H-bond Acceptor, and Gaussian H-bond Donor. In the building of field-based models, the steric force field, and electrostatic force fields were kept at 30.0 kcal/mol.
Absorption, distribution, metabolism, and excretion (ADME)
ADME predictions were carried out using the QikProp module.c The pharmacologically relevant characteristics of many organic molecules were predicted by the QikProp module.c ADME properties like CYP2C19 inhibition, CaCo2, CYP2C9 inhibition, CYP3A4 inhibition, CYP3A4 substrate, and percent Plasma protein binding were investigated in PreADMET online server (https://preadmet.bmdrc.kr/) (Scheme 1).
Result and discussion
Determination of pharmacophore
The pharmacophore models were developed on energy optimized and aligned structures of molecules. A common pharmacophore models were generated for Pim-1 and Pim-3 kinase inhibitors by selecting top 10 most active molecules based Pim-1 and Pim-3 kinase inhibitory potential and out of those molecules, 8 compounds (3, 12, 17, 31, 32, 35, 38, and 40) were found common in both the selected sets. These 8 molecules were set as active in the generation of pharmacophore models. Pharmacophore models were developed and hypotheses were studied. The selected top-ranked five-point hypothesis DHRRR_1 based on a survival score of 5.6678, site score of 0.7699 (Fig. 2). All the active molecules were aligned on selected pharmacophore DHRRR_1 (Fig. 3).
Structures and their drug-likeness properties
All the molecules were subjected to in silico ‘drug-likeness’ analysis as suggested by CA Lipinski [21]. Almost all molecules were found within acceptable ‘drug-likeness’ parameters or ‘Rule of five’ except few molecules which are failed to satisfy drug-likeness because of their predicted logP values (Table 1).
3D QSAR
Predictive 3D QSAR models were developed and validated by various software-generated statistical parameters. Standard Deviation (SD) should not be more than the standard deviation of actual activities. The model to considered predictive when R2 for the regression (the coefficient of determination) is above 0.6 and Q2 (Q-squared Value is directly analogous to R-squared, but based on the test set predictions) is above 0.5. The difference between R2 and Q2 is accepted when it is not more than 0.3. F that is the ratio of the model variance to the observed activity variance value should be large. P (probability of error) should be as low as possible. RMSE (Root-mean-square error in the test set predictions) should be close to zero for a model to be predictive. Pearson R (value for the correlation between the predicted and observed activity for the test set) with a value close to 1 indicates less scattered predicted activities in the graph of actual vs. predicted activities [22,23,24].
The significance of each influencing factor was studied by evaluating their contribution as mentioned in contribution charts. Plots of actual vs. Predicted activities reflect the predictivity of the models (Fig. 4).
Atom-based QSAR for Pim-1 kinase inhibitors
The atom-based 3D QSAR model was developed in order to study structural requirements for Pim-1 kinase inhibition (Fig. 5). The atom-based QSAR, the maps of occlusion, are either blue or red or both in the form of color cubes. The increases in activity were well associated with blue occlusion cubes in the atom-based models; while the red cubes associated with decrease in activtty. It was clearly observed that the region associated with phenyl ring directly attached with indole shows the blue colored cubes indicating that electron-withdrawing functionalities favor the activities where the region of phenyl ring attached to pyrazine shows both red and blue maps indicating that the further electron-withdrawing substituents may favor/disfavor the activity (1A and 1B). The region around the substituents of phenyl ring directly attached with indole shows the blue color for the hydrogen bond donor region which indicates favorable for activity (2A and 2B). The hydrophobic substituents were well associated with the decrement in activity as seen in the red contours around the phenyl ring directly attached to indole and incremental biological activity with longer alkyl chains on phenyl ring attached to pyrazine (3A and 3B) (Table 2, 3, and 4).
Atom-based QSAR for PIM 3 kinase inhibitors
The atom-based 3D QSAR model was developed in order to study structural requirements for Pim-1 kinase inhibition (Figs. 6 and 7). It was clearly observed that the region associated with phenyl ring directly attached with indole shows the blue colored cubes indicating that electron-withdrawing functionalities favor the activities where the region of phenyl ring attached to pyrazine shows both red and blue maps indicating that the further electron-withdrawing substituents may favor/disfavor the activity and the electron-withdrawing group on alkyl chain favors the activity (4A and 4B). The region around the substituents of phenyl ring attached with pyrazine shows mixed region blue/red color for the hydrogen bond donor region which indicates favorable/disfavourable for activity (5A and 5B) (Fig. 8). The hydrophobic substituents were well associated with the decrement in biological activity as seen in the red contours around the phenyl ring directly attached to indole and mixed biological activity for substituents with phenyl ring attached to pyrazine (6A and 6C) (Table 5, 6 and 7).
Field-based QSAR of Pim-1 kinase inhibitors
The field-based 3D QSAR model was developed in order to study structural requirements for Pim-1 kinase inhibition (Fig. 9). The electrostatic contours (1P) associated with Gaussian electrostatic fields around shows mixed blue/red colored region around the substituents of attached to pyrazine ring may favors/disfavours the activity. The substituent region shown by red color around phenyl ring attached to indole moiety disfavours the activity with electrostatic groups. H-bond acceptors around phenyl ring attached to indole ring shown with red color suggest favored electropositive features and the magenta region around substituents close to pyrazine indicated disfavoured region (1Q). The contours for hydrogen bond donor shows cyan region around the phenyl ring attached to indole rings indicated disfavourable for activity and substituents close to pyrazine ring with violet-colored polyhedron indicated favorable for activity, also it should be taken into consideration that H-bond donor contribution in this model is minor (1R). The green contour for steric groups indicated favorable at the terminus substituents attached to pyrazine ring but also show disfavouable around either side of substituents on phenyl ring attached to the indole structure (1 T). The yellow contour for hydrophobic groups indicated favorable at the terminus and close to pyrazine ring but also show disfavouable for substituents on phenyl ring attached to indole structure (1S). The contribution chart indicated the high impact of steric and hydrophobic regions. The statistical parameters and field fractions for the developed field-based models were tabulated and also revealed good results for Q2 and R2 (Table 8, 9, and 10).
Field-based for Pim-3 kinase inhibitors
The field-based 3D QSAR model was developed in order to study structural requirements for Pim-3 kinase inhibition (Figs. 10 and 11). The contours associated with Gaussian electrostatic fields around shown with blue/red region around the phenyl ring attached to indole suggests favorable/disfavourable for the activity. Terminus and close proximity substituent at nitrogen attached to pyrazine ring showed by blue color contour suggest favorable region for the activity with electrostatic groups where red contour suggests disfavourable for activity (2P). For Hydrogen bond acceptors magenta colored contour around close proximity of pyrazine ring disfavors the activity where the red colored polyhedrons around terminus substituents attached to pyrazine and phenyl ring attached to indole are suggesting favored region (2Q). The contours for hydrogen bond donor shown violet colored region around the phenyl ring attached to indole ring suggests favorable for activity. The contours for hydrogen bond donor shown cyan/violet colored counters around the pyrazine ring suggests favorable/disfavourable for activity (2R). Contours for steric groups suggest disfavourable for activity around phenyl ring attached to indole and substituents close to pyrazine ring. The contour for steric groups found favorable around terminus substituents of pyrazine ring (2 T).The yellow contour of hydrophobic groups close to pyrazine ring favors activity. The white colored contour of hydrophobic groups around phenyl ring attached to indole and terminus region of pyrazine substituents suggest disfavor to activity (2S). The contribution chart suggests the high impact of steric regions also considerable impact on the hydrophobic and electrostatic region (Table 13). The statistical parameters and field fractions for the developed field-based models were tabulated and also revealed good results for Q2 and R2 (Table 11, 12, and 13).
Statistical quality of models
The QSAR models are considered predictive when R2 for the regression (the coefficient of determination) is above 0.6 and RMSE should be close to zero or less than 0.5. All the models have shown RMSE value much below 0.5 and almost all the models have shown r2 values above 0.6 that indicates the models are highly predictive (Table 14).
Biological activity distribution in test and training sets
The minimum Pim-1 and Pim-3 inhibition (Biological activity) of the test set were greater than or equal to the minimum activity of the training set, and the maximum activity of the test set was less than the maximum activity of the training set for all the developed models. This indicates that the test sets are within the activity domain of the training set. A higher mean value of the test set than of the training set indicates the presence of relatively more potent compounds, as compared to inactive ones (Table 15) [23].
In silico ADMET
CaCo2 (gut-blood barrier) and Madin-Darby Canine Kidney (MDCK) cell permeability is considered Low if the value was less than 4, average permeability if the value was within 4~70 and High permeability if the value was more than 70 and almost all the compounds found Highly permeable. BBB is a blood-brain barrier permeability for drugs and acceptable Compounds considered CNS active if the value of BBB is more than 1. All Compounds were found CNS inactive as per In Silico predictions. Also predicted Percent Human Oral Absorption found 65–100% for all the compounds. Percent of human intestinal absorption (% HIA) also found excellent which was around 88–94% (Table 16).
Conclusion
Developed field-based and atom-based QSAR models for 3, 5-disubstituted indole derivatives were found predictive for their Pim kinase inhibitory activities. The developed models were validated by using software-generated parameter like R2, Q2, RMSE and Pearson R. The results of the ligand-based pharmacophore hypothesis and atom/field-based 3D-QSAR revealed detailed structural insights of the novel of 3,5-disubstituted indole derivatives as Pim kinase inhibitors which could be guidance for the rational design of novel potent indole derivatives for PIM kinase inhibitory activity.
Notes
aLigPrep, Schrödinger, LLC, New York, NY, 2017.
bPHASE, Schrödinger, LLC, New York, NY, 2017.
cQikProp, Schrödinger, LLC, New York, NY, 2017.
References
B.A. Domen J, Von Lindern M, Hermans A, Breuer M, Grosveld G (1987) Comparison of the human and mouse PIM-1 cDNAs: nucleotide sequence and immunological identification of the in vitro synthesized PIM-1 protein. Oncogene Research 1:103–112
C.C. Meeker TC, Nagarajan L, Ar-Rushdi A, Rovera G, Huebner K (1987) Characterization of the human PIM-1 gene: a putative proto-oncogene coding for a tissue specific member of the protein kinase family. Oncogene Research 1:87–101
Nawijn MC, Alendar A, Berns A (2011) For better or for worse: the role of Pim oncogenes in tumorigenesis. Nat Rev Cancer 11:23–34. https://doi.org/10.1038/nrc2986
Brault L, Gasser C, Bracher F, Huber K, Knapp S, Schwaller J (2010) Pim serine/threonine kinases in the pathogenesis and therapy of hematologic malignancies and solid cancers. Haematologica. 95:1004–1015. https://doi.org/10.3324/haematol.2009.017079
Bachmann M, Möröy T (2005) The serine/threonine kinase Pim-1. Int J Biochem Cell Biol 37:726–730. https://doi.org/10.1016/j.biocel.2004.11.005
Qian KC, Wang L, Hickey ER, Studts J, Barringer K, Peng C, Kronkaitis A, Li J, White A, Mische S, Farmer B (2005) Structural basis of constitutive activity and a unique nucleotide binding mode of human Pim-1 kinase. J Biol Chem 280:6130–6137. https://doi.org/10.1074/jbc.M409123200
van der Lugt NM, Domen J, Verhoeven E, Linders K, van der Gulden H, Allen J, Berns A (1995) Proviral tagging in E mu-myc transgenic mice lacking the Pim-1 proto-oncogene leads to compensatory activation of Pim-2. EMBO J 14:2536–2544. https://doi.org/10.1002/j.1460-2075.1995.tb07251.x
Davuluri RV, Grosse I, Zhang MQ (2001) Computational identification of promoters and first exons in the human genome. Nat Genet 29:412–417. https://doi.org/10.1038/ng780
Martín-Sánchez E, Odqvist L, Rodríguez-Pinilla SM, Sá M, Roncador G, Domínguez-González B, Blanco-Aparicio C, Collazo AMG, Cantalapiedra EGL, Ndez JPF, Del Olmo SC, Pisonero H, Madureira R, Almaraz C, Mollejo M, Alves FJ, Menárguez J, Gonzál F, Rodríguez-Peralto JL, Ortiz-Romero PL, Real FX, García JF, Bischoff JR, Piris MA (2014) PIM kinases as potential therapeutic targets in a subset of peripheral T cell lymphoma cases. PLoS ONE 9. https://doi.org/10.1371/journal.pone.0112148
Xinning Zhang Z-ZL, Song M, Kundu JK, Lee M-H (2018) PIM kinase as an executional target in Cancer. Journal of Cancer Prevention 23:109–116
Rathi AK, Syed R, Singh V, Shin H-S, Patel RV (2016) Kinase inhibitor Indole derivatives as anticancer agents: a patent review. Recent Patents on Anti-Cancer Drug Discovery 12:55–72. https://doi.org/10.2174/1574892811666161003112119
More KN, Jang HW, Hong VS, Lee J (2014) Pim kinase inhibitory and antiproliferative activity of a novel series of meridianin C derivatives. Bioorganic and Medicinal Chemistry Letters. 24:2424–2428. https://doi.org/10.1016/j.bmcl.2014.04.035
Lee J, More KN, Yang SA, Hong VS (2014) 3,5-bis(aminopyrimidinyl)indole derivatives: synthesis and evaluation of pim kinase inhibitory activities. Bull Kor Chem Soc 35:2123–2129. https://doi.org/10.5012/bkcs.2014.35.7.2123
Dadashpour S, Emami S (2018) Indole in the target-based design of anticancer agents: a versatile scaffold with diverse mechanisms. Eur J Med Chem 150:9–29. https://doi.org/10.1016/j.ejmech.2018.02.065
More KN, Hong VS, Lee A, Park J, Kim S, Lee J (2018) Discovery and evaluation of 3,5-disubstituted indole derivatives as Pim kinase inhibitors. Bioorganic and Medicinal Chemistry Letters 28:2513–2517. https://doi.org/10.1016/j.bmcl.2018.05.054
Asati V, Mahapatra DK, Bharti SK (2019) PIM kinase inhibitors: structural and pharmacological perspectives. Eur J Med Chem 172:95–108. https://doi.org/10.1016/j.ejmech.2019.03.050
Wan Y, Li Y, Yan C, Yan M, Tang Z (2019) Indole: a privileged scaffold for the design of anti-cancer agents. Eur J Med Chem 183:111691. https://doi.org/10.1016/j.ejmech.2019.111691
Barberis C, Pribish J, Tserlin E, Gross A, Czekaj M, Barragué M, Erdman P, Maniar S, Jiang J, Fire L, Patel V, Hebert A, Levit M, Wang A, Sun F, Huang SMA (2019) Discovery of N-substituted 7-azaindoles as pan-PIM kinases inhibitors – Lead optimization – part III. Bioorganic and Medicinal Chemistry Letters. 29:491–495. https://doi.org/10.1016/j.bmcl.2018.12.015
Dixon RA, Smondyrev SL, Knoll AM, Rao EH, Shaw SN, Friesner DE (2006) PHASE: A New Engine for Pharmacophore Perception, 3D QSAR Model Development, and 3D Database Screening. 1. Methodology and Preliminary Results. Journal of Computer-Aided Molecular Design - Springer 20:647–671
Dixon SN, Smondyrev SL, Rao AM (2006) PHASE: A Novel Approach to Pharmacophore Modeling and 3D Database Searching. Chemical Biology & Drug Design 67:370–372
Lipinski CA (2004) Lead- and drug-like compounds: the rule-of-five revolution. Drug Discov Today Technol 1:337–341. https://doi.org/10.1016/j.ddtec.2004.11.007
Veerasamy R, Rajak H, Jain A, Sivadasan S, Varghese CP, Agrawal RK (2011) Validation of QSAR models - strategies and importance. International Journal of Drug Design and Disocovery 2:511–519
Zambre VP, Hambarde VA, Petkar NN, Patel CN, Sawant SD (2015) Structural investigations by in silico modeling for designing NR2B subunit selective NMDA receptor antagonists. RSC Adv 5:23922–23940. https://doi.org/10.1039/C5RA01098E
Ganjoo A, Prabhakar C (2019) In silico structural anatomization of spleen tyrosine kinase inhibitors: Pharmacophore modeling, 3D QSAR analysis and molecular docking studies. J Mol Struct 1189:102–111. https://doi.org/10.1016/j.molstruc.2019.04.009
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Varpe, B.D., Jadhav, S.B., Chatale, B.C. et al. 3D-QSAR and Pharmacophore modeling of 3,5-disubstituted indole derivatives as Pim kinase inhibitors. Struct Chem 31, 1675–1690 (2020). https://doi.org/10.1007/s11224-020-01503-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11224-020-01503-1