Abstract
A triazene derivative and its transition metal complexes were prepared and characterized using molar conductance, magnetic susceptibility measurements, IR, UV–visible, NMR spectral studies wherever possible and applicable. The structure of the ligand and metal complexes was further confirmed using DFT calculations with the help of B97d method with 6-311++G(d,p) basis set. The antidiabetic and antioxidant activities of the ligand and the metal complexes were studied. The ligand showed potential biological activities which increased on chelation with metal ion. Apart from this, the molecular docking studies were carried out in order to understand the binding interaction of the ligand and its metal complexes with active sites of the target proteins.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Triazene derivatives with characteristic diazoamino group have attracted much attention because of their variable structural diversities, bonding interaction and promising biological activities. These compounds possess three nitrogen atoms which are presumably responsible for their interesting chemical properties and promising biological activities including their usage as oestrogen receptor modulators, antiviral, antimalarial agents and so on [1,2,3]. These compounds are capable of exhibiting versatile coordination modes with the transition metals and thereby achieve intermolecular, metal–ligand and ligand–ligand interactions and can also assemble as supramolecular structures [4]. The heterocyclic systems especially those containing sulphur have been reported to exhibit diverse biological and pharmacological activities due to the similarities with naturally occurring and synthetic molecules of known potential [5]. The uracil-based compounds with the pyrimidine nucleus also exhibit a wide range of biological activities such as diuretic, antitumour, anti-HIV activities. [6, 7]. In this communication, a triazene derivative obtained by coupling a diazotised benzothiophene derivative with 5-amino uracil has been used as a prospective ligand for the drug design. Diabetes mellitus is now considered as a common chronic disease characterized by elevated blood glucose levels [8]. Although a large number of anti-diabetes drugs are available in market, the need for effective anti-diabetes drugs still persists. In the treatment of type 2 diabetes, oral hypoglycaemic agents are used either individually or in a combination therapy, depending upon the individual. The antioxidant compounds prevent the oxidation of cellular substances and thereby reduce the risk of chronic diseases. Certain benzothiophene derivatives are reportedly used as novel pharmacophore for the treatment of type(II) diabetes and oxidative stress [9]. Moreover, the antidiabetic activity and antioxidant activity studies of triazene derivatives have received only sporadic attention so far. In view of the potential biological application of the parent compounds, the new ligand formed is expected to exhibit enhanced activities and effective in bond formation with the metal ions. This expectation was borne true as evidenced from the biological studies. Theoretical calculations with the basis set of B97d/6-311++G(d,p) were performed to determine the structural properties and to understand the orientation of the highest occupied molecular orbitals (HOMOs) and lowest unoccupied molecular orbitals (LUMOs). The DFT computational studies were also used to investigate the reactive sites, and the MEP surfaces were predicted at the same level. Furthermore, molecular docking studies were performed to understand the interactions of the metal complexes with active sites of the protein targets which are responsible for antidiabetic and antioxidant activities. Herein, we report the synthesis, characterization, antidiabetic, antioxidant activities and molecular docking studies of some 3d-metal complexes derived from a triazene derivative.
Materials and methods
The analar grade chemicals purchased from Sigma-Aldrich were used for synthesis. Solvents used for physico-chemical measurements were purified by standard methods. Elemental analysis for C, H and N was carried out by Elementar Vario EL III CHNS analyser. Molar conductance measurements were taken using 10–3 M solutions of the complexes in suitable solvent at room temperature using a Systronic direct reading conductivity meter Type 305. Infrared spectra were recorded using Agilent Cary 630 FT-IR spectrometer. The Far IR spectra were recorded using Polytec FIR 30 Fourier Far IR spectrophotometer. The electronic spectra were recorded on a Hitachi 320 UV–Vis spectrophotometer. Proton NMR spectra were recorded using on a JEOL GSX 400 NB 400 MHz spectrometer in DMSO-d6. X-ray diffraction experiments were taken on a Siemens D 5005 model spectrometer. The fluorescence property was measured using JASCO 8300 spectrofluorometer. The antidiabetic property was studied using alpha amylase inhibitory assay. The antioxidant activity was studied using DPPH assay, reducing power method and nitric oxide scavenging assay.
Computational details
The geometry optimization of the structure of ligand and metal complexes was performed using the B97d hybrid density functional method with 6-311++G(d,p) basis set. The Natural Bond Orbital analysis was also done at B97d/6-311++G(d,p) level. All the calculations were performed using Gaussian 09 programme package with the help of the GaussView visualization program. During optimization, no symmetry constraints were applied. No imaginary frequency was obtained after frequency calculations.
Molecular docking
The X-ray crystal structures of human pancreatic alpha amylase in complex with montbretin A (PDB ID: 4W93) at 1.35 Å resolution and human myeloperoxidase in complex with thiocyanate (PDB ID: 1DNU) at 1.85 Å resolution were retrieved from the RCSB protein databank. The 3D structures of ligand and metal complexes were drawn using ChemSketch 2017.2.1 version. The in silico docking studies were carried out by using the C Docker of receptor–ligand interaction protocol section of BIOVIA Discovery Studio 2018. Initially, there was a pre-treatment process for both the proteins and the ligands. Protein preparation was done using the Prepare Protein module of receptor–ligand interaction tool. The protein structures were prepared by removing native ligands and active site water. This was followed by the addition of missed hydrogen atoms. Ligand preparation was done using the Prepare Ligand module in receptor–ligand interaction tool. AUBT and its complexes were prepared for molecular docking by adding hydrogen atoms and, the conformational search was applied to all the compounds and the best conformers underwent energy decrease using the CHARMM force field. The C Dock score estimated the binding energy of the compounds.
Synthesis of ethyl 2-[N-(5-(aminouracil)azo]-4,5,6,7-tetrahydro [b] thiophene -3-carboxylate (AUBT)
The ligand, ethyl 2-[N-(5-(aminouracil)azo]-4,5,6,7-tetrahydro [b] thiophene -3-carboxylate (AUBT) were prepared by a following procedure as shown in Scheme 1. The starting material, 2-amino-3-carboxyethyl-4,5,6,7-tetrahydrobenzo [b] thiophene was prepared by Gewald synthesis [10]. A quantity (0.01 mol) of 2-amino-3-carboxyethyl-4,5,6,7-tetrahydrobenzo [b] thiophene was dissolved in 2 M HCl and kept below 5 °C in an ice–salt bath. This solution was added to an aqueous solution of sodium nitrite (0.01 mol) and stirred for another 1 h keeping the temperature at 0 °C throughout the preparation. The excess nitrous acid was destroyed by adding urea and the resulting solution was filtered. The diazotized product was coupled with an aqueous solution of 5-aminouracil (0.01 mol) keeping the pH between 7.0 and 7.5 using 40% NaOH solution and allowed to keep for 1 h in an ice bath. The solid product separated was filtered off and further purified by recrystallization from ethanol.
Yield 80%; orange colour; m. p = 90 °C. FT-IR (cm−1): 1486 ν(N=N), 3292 ν(N1H), 3149 ν(N2H), 1532 δ(NH), 3406 ν(N–H), 1698 ν(C=O).1H NMR (DMSO-d6, δ, ppm): 8.70 (s, 1H, NH), 11.70(s, 1H, NH), 11.80(s, 1H, NH) 7.05–7.69 (m,Ar), 1.35–1.78 (m,CH3), 2.51–2.83 (m,CH2 of benzothiophene ring), 4.41–4.46 (CH2 of-OCH2CH3).13C NMR (DMSO-d6, δ,ppm): 115.10-131.11(Ar), 163.23(C2=O), 164.02(C4=O), 166.11(ester carbonyl), 59.08(–O–CH2), 14.85–26.97(CH3). Anal. Calcd (found) for C15H17O4N5S (%): C,49.61(49.10); H, 4.56(4.58); N,19.60(19.30); S, 8.42 (8.82).
Synthesis of metal complexes
Synthesis of copper (II) complex
Copper complex was prepared by using the following procedure:
A methanolic solution of the copper(II) chloride (0.01 M) was added in small portions to a magnetically stirred and hot methanolic solution of ligand (0.01 M). The mixture was subjected to magnetic stirring for 1 h and then refluxed for 4–6 h. The pH of the solution was adjusted to 7.0–7.5 using alcoholic ammonia solution. The resulting solution was concentrated and allowed to cool. The metal complex formed was filtered, washed successively with methanol and diethyl ether and finally dried in vacuum over P4O10. The purity of the complexes was examined using TLC (silica gel).
Yield 73%; black colour; FT-IR (cm−1):1471 ν(N=N), 1672 ν(C=O). Anal. Calcd (found) for CuC15H17O4N5SCl2 (%): C, 36.10 (36.18); H, 3.38(3.42); N, 14.01(14.07); S, 6.20 (6.43).
Synthesis of cobalt (II) complex
A solution of cobalt(II) chloride (0.01 M) dissolved in methanol(10 ml) was added gradually to a methanolic solution (30 ml) of the ligand (0.01). The mixture was refluxed for 6 h after magnetic stirring for 1 h. The pH of the solution was adjusted to 7.0–7.5 using alcoholic ammonia solution. The resulting solution was concentrated and allowed to cool. The metal complex formed was filtered, washed several times and dried in vacuum over P4O10.
Yield 70%; dark brown colour; FT-IR (cm−1):1468 ν(N=N), 1667 ν(C = O). Anal. Calcd (found) for CoC15H17O4N5SCl2 (%): C, 36.10 (36.18); H, 3.38(3.42); N, 14.01(14.07); S, 6.20 (6.43).
Synthesis of nickel(II) complex
Nickel(II) chloride (0.01 M) solution in methanol (20 ml) was added to a magnetically stirred solution of ligand (0.01 M) in methanol. The mixture was refluxed for 4–5 h. The pH of the solution was adjusted to 7.5-8.0 using alcoholic ammonia solution. The metal complex obtained was filtered, washed with diethyl ether and finally dried in vacuum over P4O10.
Yield 73%; light brown colour; FT-IR (cm−1): 1476 ν(N=N), 1676 ν(C=O). Anal. Calcd (found) for NiC15H17O4N5SCl2(%): C, 36.12 (36.53); H, 3.42(3.42); N, 14.11(14.20); S, 6.38(6.49).
Synthesis of zinc(II) complex
The zinc(II) complex was prepared according to the same procedure adopted for preparing the other complexes. About 0.01 M zinc(II) chloride in methanol was added to the 30 ml methanolic solution of ligand (0.01 M). The mixture was magnetically stirred for 1 h and then refluxed for 6 h. The pH of the solution was adjusted to 7.0–7.5 using alcoholic ammonia solution. A brown coloured precipitate was formed which was filtered, washed successively with methanol and diethyl ether and finally dried in vacuum over P4O10.
Yield 76%; light orange colour; FT-IR (cm−1):1469 ν(N=N), 1679 ν(C=O). 1H NMR (DMSO-d6, δ, ppm): 8.52 (s, 1H, NH), 7.04–7.59 (m, Ar). 13C NMR (DMSO-d6, δ, ppm): 103.09-130.76 (Ar), 165.88 (ester carbonyl). Anal. Calcd (found) for ZnC15H17O4N5SCl2(%): C, 36.01(36.04); H, 3.39(3.40); N, 14.12(14.02); S, 6.40 (6.42).
Antidiabetic activity
The in vitro antidiabetic activity of the ligand and the metal complexes was evaluated by α-amylase inhibition assay. The α-amylase inhibition assay was carried out by the method of Apostolidis [11]. The different concentrations of 10 mg/ml sample were taken and made up to 100 μl using 25 mM phosphate buffer containing 25 μl of porcine α-amylase at a concentration of 0.5 mg/ml were incubated at 25 °C for 10 min. After preincubation, 25 μl of 0.5% starch solution in 25 mM phosphate buffer (pH = 6.9) was added. The reaction mixtures were then incubated at 25 °C for 10 min. The reaction was stopped with 50 μl of 3, 5 dinitrosalicylic acid colour reagent. The microplate was then incubated in a boiling water bath for 5 min and cooled to room temperature. The solution thus prepared was taken as the test solution. Absorbance was measured at 540 nm using a microplate reader. A control was prepared using the same procedure replacing the sample with distilled water. All the tests were run in triplicate in this assay.
Antioxidant activity
DPPH assay
1,1-diphenyl-2-picryl hydrazyl is a stable free radical with red colour which turns yellow when scavenged. This property of DPPH is used in free radical scavenging activity. The radical scavenging activity of ligand and metal complexes was determined by using DPPH assay. The decrease in the absorption of the DPPH solution after the addition of an antioxidant was measured at 517 nm. Ascorbic acid (10 mg/ml DMSO) was used as reference. 1,1-diphenyl-2-picryl hydrazyl is a stable free radical with red colour which turns yellow when scavenged. This property of DPPH is used in free radical scavenging activity. The degree of discoloration indicates the scavenging potential of the antioxidant compounds or extracts in terms of hydrogen donating ability. Different volumes (2.5–40 μl) of samples were made up to a final volume of 40 μl with DMSO, and 2.96 ml DPPH (0.1 mM) solution was added. The reaction mixture incubated in dark condition at room temperature for 20 min. After 20 min, the absorbance of the mixture was read at 517 nm. The DPPH solution was taken as control.
Reducing power assay
The ferric-reducing antioxidant power (FRAP) assay is carried out by the method of Oyaizu which measures the ability of antioxidants to reduce ferric iron [12]. Different concentrations of sample such as 12.5-200 µg/mL from a stock concentration of 1 mg/mL were mixed with 2.5 ml of phosphate buffer (200 mM), and 2.5 ml of 1% potassium ferricyanide was added and boiled for 20 min at 50 °C. A control without the test compound, but an equivalent amount of distilled water was taken. After incubation, 2.5 ml of 10% TCA was added to the mixtures followed by centrifugation for 10 min. The upper layer (5 ml) was mixed with 5 ml of distilled water, and 1 ml of 0.1% ferric chloride was added and the absorbance was read at 700 nm.
Nitric oxide scavenging activity
The procedure is based on the principle that sodium nitroprusside in aqueous solution at physiological pH spontaneously generates nitric oxide which interacts with oxygen to produce nitrite ions that can be estimated using Griess reagent. Sodium nitroprusside in phosphate buffered saline pH 7.4 was mixed with different concentrations of the sample such as 12.5–200 μg/mL from a stock concentration of 1 mg/mL and incubated at 25 °C for 30 min. A control without the test compound, but an equivalent amount of distilled water was taken. After 30 min, 1.5 mL of the incubated solution was removed and diluted with 1.5 mL of Griess reagent (1% sulphanilamide, 2% phosphoric acid and 0.1% N-1-naphthyl ethylene diaminedihydrochloride). Absorbance of the chromophore formed during diazotization of the nitrate with sulphanilamide, and subsequent coupling with N-1 naphthylethylene diaminedihydrochloride was measured at 546 nm, and the percentage scavenging activity was measured with reference to the standard (Gallic acid 1 mg/mL) [13].
Results and discussion
Analytical data indicated that the coupling reaction of 5-aminouracil and 2-amino-3-carboxyethyl-4,5,6,7-tetrahydrobenzo [b] thiophene occurred in 1:1 molar ratio. The ligand is soluble in common organic solvents like methanol, ethanol, benzene, toluene, etc. Analytical data of the complexes are also in good agreement with their formulation as in Table 1. All the complexes are stable and non-hygroscopic solids soluble in DMSO and DMF. The molar conductance values of the complexes in DMSO are in the range 6.3–11.1 O−1cm2 mol−1 which confirms its non-electrolytic nature [14].
IR spectra
Infrared spectra of the ligand and the metal complexes were recorded in order to study their coordination properties. Infrared spectra of the ligand and the copper(II) complex are given in Figs. 1 and 2, respectively. The infrared spectra of the ligand show a medium intensity band at 1486 cm−1, characteristic of ν(N=N) stretching vibration. This band is shifted to a lower frequency by 15–20 cm−1 indicating the involvement of the azo nitrogen in coordination to the metal ion [15]. The IR spectrum of the ligand showed a broad band centred at 3292 and 3149 cm−1 indicating νN(1)H and νN(2)H vibrations of uracil moiety [16]. A band was observed at 3406 cm−1 which represents (νNH) vibration of triazene group. The ester carbonyl groups appear at 1698 cm−1 in the ligand which showed considerable shift to lower frequency in the case of complexes indicating the participation of ester carbonyl group in complexation [17]. The complexes showed non-ligand bands in the region of 529–550, 424–430 and 340–358 cm−1 which can be assigned to ν(M–O), ν(M–N) and ν(M–Cl) vibrations, respectively [18].
NMR spectra
The 1H NMR spectra of both ligand and zinc(II) complex were recorded in DMSO-d6 (Fig. 3a, b). The 1H NMR spectra of the ligand showed a multiplet in the region 7.05–7.69 ppm due to aromatic protons. The complex showed a marginal downfield shift of aromatic protons in the range of 7.04–7.59 ppm which confirmed the bond formation. The 1H NMR spectrum showed characteristic singlets at δ = 11.7 and 11.8 ppm for two NH groups which remain almost unchanged in the complexes [19]. The ligand showed a singlet at δ = 8.72 ppm which was downshifted to 0.2 ppm in zinc(II) complex. This can be assigned to the NH proton of triazene moiety. The spectrum shows peaks in the region 1.35–1.78 ppm due to methyl protons, 2.51–2.83 ppm due to protons of benzothiophene ring, 4.41-4.46 ppm due to methylene protons of the ester carbonyl group.
13C NMR spectra of the ligand and zinc (II) complex were recorded in DMSO-d6 using TMS as internal standard (Fig. 4a, b). In the case of 13C NMR spectra, the aromatic carbons in the ligand showed signal in the range of 115–131 ppm which was shifted downfield in the zinc(II) complex. The signal due to C2=O and C4=O carbon of the ligand showed a signal at 163.33 and 164.02 ppm, respectively, which remained unchanged in the complexes showing the non-participation of carbonyl groups of uracil moiety. The ester carbonyl carbon appears at 166.52 ppm which was downshifted to 0.64 ppm in the complex. The –O–CH2– carbon appears at 59.08 ppm and the methyl carbons appear at the range 14.85–26.97 ppm which was shifted slightly in the metal complexes.
UV–Visible spectra
The electronic absorption spectra of the ligands and its complexes were recorded at room temperature using DMSO as the solvent with the concentration of 1 × 10−4 M. The absorption spectra of the ligand show bands at 312 nm and 391 nm due to П–Π* and n-П* transitions, respectively. The electronic spectral value of ligand is slightly red shifted in the complexes which clearly indicate the complex formation [20]. This is assignable to spin–orbit coupling from the heavy atom effect of the transition metal ions as well as MLCT transition of ligand on binding to the transition metal ions. The copper(II) complex exhibited a broad band at 730 nm. The magnetic moment value was found to be 1.84 BM which confers distorted square planar geometry for the complex. The cobalt(II) complex exhibited bands at 645 nm and 520 nm which corresponds to 4A2 → 4 T1(F) (υ2) and 4A2 → 4 T1(P) (υ3), supporting an tetrahedral geometry for the cobalt(II) complex. It possesses a magnetic moment value of 4.30 BM, which is in agreement with tetrahedral environment. The Ni(II) complex showed bands at 544 nm and 730 nm which gave rise to 1A1g → 1B1g and 1A1g → 1A2g transitions, respectively, indicating a square planar geometry. The magnetic moment measurements clearly indicate the diamagnetic nature of nickel(II) complex. The zinc(II) complex owing to their completely filled d10 electronic configuration preferred a tetrahedral geometry.
Powder XRD study
The X-ray diffractograms of ligand and cobalt (II) complex were recorded with the scan angle in the range of 5° to 80°. The diffractograms of the ligand indicated its crystalline nature (Fig. 5). The diffractogram of the ligand showed 37 reflections with maxima at 2θ = 11.3399°. The interplanar distance was found to be d = 3.62952 Å (Online Resource 1). The ligand was indexed to orthorhombic crystal system with lattice constants a = 12.6442 Å, b = 9.9309 Å, c = 7.0488 Å and cell volume = 885.0988 \(A^{^\circ 3}\). The XRD pattern of the cobalt(II) complex indicated its amorphous nature.
The average crystalline size of the ligand was calculated using the Scherrer formula, dxrd = \(\frac{0.9\lambda }{\beta \cos \theta }\).
The crystallite size of ligand was found to be 27 nm.
From all these spectral studies, the structure of the ligand and the metal complexes is assigned as shown in Fig. 6.
Computational studies
The DFT studies have gained much attention as this method can reproduce the experimental data and also can predict various properties of compounds. In the present study, the geometry of ligand and its metal complexes was optimized using B97D density functional method with 6-311++G(d, p) basis sets as incorporated in the Gaussian 09 programme in gas phase. The optimized structures of the ligand and the metal complexes along with the numbering are given in Fig. 7. The bond angles and bond lengths of these compounds are given in Table 2. The spectral studies clearly indicated that the ligand coordinated to the metal ion through the O15 atom of the ester group and N24 atom of the triazene moiety which is further confirmed by the considerable shift in their bond lengths observed in the theoretical studies. The M–Cl bond lengths were also found to be shifted to a considerable extent confirming their participation in the complex formation. The bond angles obtained were found to be in good agreement with the geometry obtained experimentally.
The Frontier molecular contour surfaces of ligand AUBT and metal complexes are displayed in Fig. 8. The HOMO of AUBT is spread over the whole ligand moiety, and LUMO is mainly spread over the azo unit and uracil moiety. The HOMO of the metal(II) complexes was spread over the azo unit and the metal atom and the LUMO encompasses the pyrimidine moiety, azo group and metal atom. The observations indicate that the complexes are highly reactive due to the extensive overlapping of orbitals which in turn enhance its interaction with the biological molecules. The atomic charge distribution is determined by NBO analysis and is given in Table 3. It was observed that the negative charge of the coordinating nitrogen atom of azo group decreased considerably on complexation which might be presumably due to the back donation of the electrons from metal to ligand. The electron density of oxygen atom of the ester group was also reduced on coordination with the metal ion. The charge on the metal atom was found to be lower than their formal charge and this clearly indicate the possibility of L-M charge transfer. Thus, the NBO analysis provides the adequate support for the spectral studies which further confirm that the coordination occurs through the azo nitrogen and the ester carbonyl group.
Global reactivity descriptors
The HOMO–LUMO energy gap of ligand and the metal complexes was used for the determination of the global reactivity descriptors. The energy gap of the ligand and the metal complexes follows the order Ni(II) > Co(II) > Zn(II) > Cu(II) > HNAP (Table 4). The smaller energy gap indicates that the metal complexes are softer molecules with higher reactivity. The compounds with higher HOMO energy are electron donors and hence good antioxidants. The parameters global hardness (η), global softness (S) and the absolute softness (ϭ) were used to determine the hardness and softness of ligand and the metal complexes. The Ni(II) complex showed lowest hardness which can contribute to its higher biological activity. The mathematical expression of η, S, ϭ can be written as: [21]
The Ni(II) complex showed lowest hardness which may lead to its enhanced biological activity. The evaluation of chemical hardness would be helpful for the determination of influential biological efficiency. The global electrophilicity index (ω) [22].
The electrophilicity may define the biological activity of proposed drugs. The increase in the electrophilicity index values of metal complexes than the ligand indicates the higher biological activity of the complexes in a biological system. The order of electrophilicity index is as follows: Ni(II) > Zn(II) > Cu(II) > Co(II) > HNAP.
The molecular electrostatic surface potential
The molecular electrostatic potential (MEP) surface provides valuable information regarding the polarity of the compounds Fig. 9 [23, 24]. The electrostatic potential (ESP) map of ligand and the metal complexes illustrates the three-dimensional charge distributions of the molecule computed at the 0.0004 a. u. isodensity surface. They are also used to study the molecular interactions. The red and yellow colour regions in ESP map indicate the negative potential region which is related to electrophilic reactivity and the blue colour region represents the positive potential region which is the favourable site for nucleophilic attack. The green colour region depicts the neutral electrostatic region. These regions give informations regarding the molecular interactions which would further help in the investigations related to its bioactivity. The most negative regions are associated with the oxygen atom of ester group and nitrogen atom of triazene group. In particular, the oxygen atom O15 in ester group and N24 with electrostatic potentials ranging from −6.102 × 10−2 a. u to 7.459 × 10−2 a. u are found to be the most suitable sites for binding of metal ions.
Fluorescence studies
The fluorescence properties of the triazene ligand and the metal complexes were studied at room temperature in DMSO (Fig. 10). The free ligand exhibits fluorescence emission maximum at 407 nm upon excitation at 339 nm. The zinc(II) complex showed an intense blue fluorescent emission with a broad band at 463 nm under 337 nm light excitation. The copper(II) complex showed a green emission with two emission peaks at 485 nm and 536 nm on excitation at 360 nm. The irradiation of nickel(II) complex at 340 nm results in a slightly red shifted emission band at 421 nm. The cobalt(II) complex also showed a blue emission band at 425 nm on the excitation at 340 nm. The large red-shift emission observed in the complexes could be attributed to the metal-perturbed intraligand charge transfer [25].
Upon chelation, the intensities of emissions in the metal complexes were found varying depending on the nature of the metal and was observed in the order Zn(II) > Ni(II) > Co(II) > Cu(II) complex. The fluorescence enhancement in the zinc(II) complex may be due to the lack of vacant d-orbital which would prevent ligand-to-metal charge transfer and also the absence of absorption bands prevents energy transfer. The fluorescence quenching observed in the case of Co(II) and Cu(II) complex could be attributed to the loss of rigidity on chelation which may lead to fluorescence attenuation [26]. The fluorescence studies provide the scope for the utilization of these metal complexes as fluorescent probes to study the molecules and its in vivo interaction.
Antidiabetic activity
The α-amylase inhibitory activity studies were carried out to study antidiabetic property of the ligand and the metal complexes and the results are given in Fig. 11. The ligand and the complexes inhibit α-amylase in a dose-dependent manner. The IC50 value of ligand and metal complexes is given in Table 5. The IC50 values for ligand were found to be 5.38 µg/ml. Among complexes nickel (II) showed the IC50 value of 2.17 µg/ml close to that of the standard acarbose. It has been observed that the alpha amylase inhibitory activity of ligand enhanced on complexation with metal ions. The nickel (II) complex has higher percentage of -amylase inhibition even at low concentrations. The enhanced activity might be attributed to the enzyme deactivation through chelation. In addition, the metal complexes formed might lower the blood glucose level by activating the glucose transport into cell of the peripheral tissues. The higher activity of nickel(II) complex might be due to its square planar structure which facilitates better interaction between the active sites of the enzyme and the metal complex.
Antioxidant studies
The reactive oxygen species which is essential for various physiological processes such as cell proliferation, intracellular signalling, phagocytosis are products of normal metabolism. The abnormal production of ROS can lead to oxidative stress. The ligand and the metal complexes were subjected to different assays in order to evaluate their antioxidant potential. The triplicate measurements of each experiment were taken.
Free radical scavenging activity
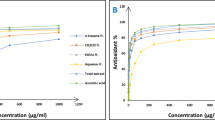
The antioxidant potential of the ligand and the metal complexes were determined by DPPH assay. This method is widely used to evaluate the radical scavenging ability of antioxidants. These radicals absorb strongly in the visible region and absorption decreases proportionally upon interaction with the antioxidants. Thus, the radical scavenging capacity of the antioxidants can be obtained based on the absorption change. The interaction of ligand and the metal complexes with the DPPH was found to be concentration dependent. The free radical scavenging activities were calculated as the percentage of DPPH decolorization in the presence of the synthesised ligand and the metal complexes. The metal complexes showed enhanced antioxidant potential when compared to that of the ligand. The nickel(II) complex showed IC50 value at 2.12 µg/ml which is close to that of the standard. The percentage inhibition of the nickel(II) complex was found to be 65% at IC50 value (Fig. 12). The presence of thiophene moiety and the NH groups might be the reason for the antioxidant activity of the ligand. The enhanced activity of the metal complexes can be attributed to the chelation of the metal ion to the bioactive ligand moiety.
Reducing power method
Some reducing substances in the sample could provide electrons to reduce Fe3+ to Fe2+. The Fe2+ can be monitored by measuring the formation of Perl’s Prussian blue at 700 nm. The study showed that on complexation relatively stronger FRAP activity was observed (Fig. 13). The higher absorbance indicates the higher reducing power of the compounds. The reducing ability of a compound ensures good antioxidant activity. The compounds that possess reducing power can reduce the reactive radicals into stable unreactive radicals [27]. The nickel(II) and cobalt(II) complexes showed greater reducing power as the concentration was increased from 12.5 to 200 µg/ml. The zinc(II) and copper(II) complexes also showed moderate activity when compared to the ligand. The higher reducing power of nickel(II) complex might be attributed to its higher electron donor ability which is evident from the theoretical studies. The reductant reacts with the free radical chain and terminates the reaction responsible for the oxidative stress.
Nitric oxide scavenging activity
Nitric oxide is a small biologically active signalling molecule which can diffuse and react rapidly with molecular oxygen (Kusum 2013). It serves as a neurotransmitter in vascular dilation and mediation of cellular defence. The chronic exposure to NO can alter the structure and function of cellular components leading to chronic diseases such as various carcinomas and inflammatory conditions [28]. The nitric oxide radical is more toxic when it reacts with superoxide leading to the formation of peroxynitrite anion which can decompose into. OH and NO2. The metal complexes showed promising activity when compared to the ligand (Fig. 14). The nickel(II) and zinc(II) complex showed potent activity with IC50 value of 17.38 µg/ml and 22.77 µg/ml, respectively. These compounds showed much better activity than the standard gallic acid with IC50 value of 51.5 µg/ml as shown in Table 6. The mechanism of action of the nickel(II) complex involves a redox process centred on nickel(II) complex [29].
Molecular docking studies
The molecular docking technique has played a vital role in drug design and discovery by providing insight into the drug–enzyme interactions. In our study, the molecular docking studies were carried out in order to describe the antidiabetic and antioxidant potential and also to predict the binding mode of chemical moieties into the enzyme pocket [30]. It provides information regarding the binding affinity and binding site along with the sterically acceptable conformations. The lower the binding free energy, the more potent the binding affinity between the receptor protein and ligand molecules and a stable ligand-enzyme complex will be formed. The appropriate orientation of compounds with respect to the selected protein target and its geometric and energetic conformations were obtained. The conformations of the docked complex were also analysed in terms hydrogen bonding interactions between the compounds and target protein. The docking analysis of the selected targets was conducted by the computational simulation of protein targets with the designed ligands. The binding affinities of the compounds were evaluated by C Docker energy score.
In order to understand the antidiabetic potential of the synthesised compounds, a human pancreatic alpha amylase in complex with montbretin A with PDB ID: 4W93 was used as the protein target [31]. The protein consists of a single polypeptide chain with sequence length of 496 amino acids. The binding site of protein interaction with its ligand montbretin is Glu 233, Arg 195, Asp 197, His 101, His 201, Ile 235, Glu 240 and Lys 200. The target protein and ligands were prepared prior to docking. The ligand generated 40 conformers and the remaining metal complexes generated 10 conformers during ligand preparation. The negative values of the binding free energy of docked complexes suggested that these complexes reasonably bind to the human pancreatic alpha amylase. The ligand and the metal complexes interact with protein by hydrogen bonding interactions. The nickel(II) complex showed the lowest C Docker energy value of − 41.1605 kcal mol−1 which suggested its strong interaction to the human pancreatic alpha amylase. The Ni(II) complex exhibited four hydrogen bonding interactions with aminoacid residues Arg195, Ile235, His299, Glu233 and Val234 (Fig. 15a, b). The results were in good agreement with the wet lab experiments (Table 7). The antioxidant potential of the compounds was evaluated by docking with the protein target of PDB ID: 1DNU which is human myeloperoxidase in complex with thiocyanate. The protein consists of two polypeptide chain with sequence length of 104 amino acids. AUBT generates 40 conformers and the remaining metal complexes generated 10 conformers during ligand preparation. The nickel(II) complex showed hydrogen interactions with the aminoacid residues His336, Gln91, His95, Arg239, Arg424 which resulted in strong binding interactions with the protein target than the other metal complexes (Table 8). The strong hydrogen bond interaction might be the reason for the enhanced biological efficiency of nickel(II) complex than other compounds (Fig. 16a, b).
Conclusion
A novel triazene-based ligand and its metal complexes were prepared and characterized by elemental analysis, IR,UV, 1 H and 13C NMR spectral studies. The ligand coordinated to the metal ion through azo nitrogen and the ester carbonyl oxygen of benzothiophene moiety. The fluorescence studies revealed that the ligand and metal complexes were fluorescent and provide further scope for its utilization as fluorescent probe in biological applications. The metal complexes showed enhanced biological activities than the corresponding ligand. The nickel(II) complex exhibited promising antidiabetic and antioxidant potential. The relevant antidiabetic and antioxidant activity of the triazene-based metal complexes proved the potential of metal-based enzyme inhibitors which can effectively interact with the active sites of enzyme through its three-dimensional architecture. Molecular docking studies were carried out to substantiate the wet lab experiments and also to find out the active binding sites of the synthesised compounds with that of the target protein.
References
D.B. Kimball, M.M. Haley, Angew. Chem. Int. Ed. 41, 3338 (2002)
B.R. Henke, T.G. Consler, N. Go, J. Med. Chem. 45, 5492 (2002)
V.K. Pandey, S. Tusi, Z. Tusi, M. Joshi, S. Bajpai, Acta Pharm. 54, 1 (2004)
D.F. Back, M. Horner, F. Broch, G.M. de Oliveira, Polyhedron 31, 558 (2012)
L. D. Quin, J. Tyrell, Fundamentals of heterocyclic chemistry: Importance in nature and in the synthesis of pharmaceuticals, Wiley-Interscience, 2010
C.O. Kappe, Tetrahedron 49, 6937 (1993)
G. Meng, Y. Liu, A. Zheng, F. Chen, W. Chen, Eur. J. Med. Chem. 82, 600 (2014)
S. Wild, G. Roglic, A. Green, R. Sicree, H. King, Diabetes Care 27, 1047 (2004)
R.M. Poole, R.T. Dungo, Drugs 74, 611 (2014)
K. Gewald, E. Chinke, H. Bottcher, Chem. Ber. 99, 94 (1966)
E. Apostolidis, Y.I. Kwon, K. Shetty, Innov. Food. Sci. Emerg. Technol. 8, 46 (2007)
M. Oyaizu, J. Nutr. 44, 307 (1986)
D.C Garrat, The Quantitative analysis of Drugs. Chapman and Hall Ltd., Japan, 3, 456 (1964)
W.J. Geary, Coord. Chem. Rev. 7, 81 (1971)
J. Wang, H. Niino, A. Yabe, Appl. Surf. Sci. 154, 571 (2000)
S.I. Mostafa, M.A. Kabil, E.M. Saad, A.A. El-Asmy, J. Coord. Chem. 59, 279 (2006)
M. Thankamony, S.B. Kumari, G. Rijulal, K. Mohanan, J. Therm. Anal. Cal. 95, 259 (2009)
K.V. Sharma, V. Sharma, R.K. Dubey, U.N. Tripathi, J. Coord. Chem. 62, 493 (2009)
R. M. Shaker, M. A Elrady, K.U Sadek, Mol. Divers. 2016, 20,153 (2016)
H.M. Wen, Y.H. Wu, Y. Fan, L.Y. Zhang, C.N. Chen, Z.N. Chen, Inorg. Chem. 249, 2210 (2010)
A. Arab, M. Habibzadeh, Comput. Theor. Chem. 1068, 52 (2015)
R.G. Pearson, Proc. Natl. Acad. Sci. 83, 8440 (1986)
P. Politzer, J.S. Murray, Theor. Chem. Acc. 108, 134 (2002)
S. Murray, P. Politzer, Comput. Mol. Sci. 2, 153 (2011)
S.Q. Zhang, F.L. Jiang, M.Y. Wu, L. Chen, J.H. Luo, M.C. Hong, Cryst. Eng. Comm. 15, 3992 (2013)
W. Zhang, W. Dou, W.S. Liu, X.L. Tang, W.W. Qin, Eur. J. Inorg. Chem. 5, 748 (2011)
A.L. Dawidowicz, D. Wianowska, M. Olszowy, Food Chem. 131, 1037 (2012)
R.E. Huie, S. Padmaja, Free Radical Res. Commun. 18, 1951 (1993)
M.N. Patel, D.S. Gandhi, P.A. Parmar, Inorg. Chem. Commun. 13, 618 (2010)
H.Q. Qurrat-ul-Ain, A. Nadhman, M. Sirajuddin, Inorg. Chim. Acta 423, 220 (2014)
M. Taha, M.S. Baharudin, N.H. Ismail, S. Imran, M.N. Khan, F. Rahim, M. Selvaraj, S. Chigurupati, M. Nawaz, F. Qureshii, S. Vijayabalan, Bioorg. Chem. 80, 36 (2018)
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Philip, S., Jayasree, E.G. & Mohanan, K. Antidiabetic, antioxidant, DFT and molecular docking studies of a triazene derivative and its transition metal complexes. Res Chem Intermed 46, 75–99 (2020). https://doi.org/10.1007/s11164-019-03936-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11164-019-03936-8