Abstract
Erythrinus erythrinus presents extensive karyotypic diversity, with four karyomorphs (A–D) differing in the number of chromosomes, karyotype structure or sex chromosomes systems. Karyomorph A has 2n = 54 chromosomes in males and females without heteromorphic sex chromosomes, while karyomorph C has 2n = 52 chromosomes in females and 2n = 51 chromosomes in males, due a X1X1X2X2/X1X2Y sex chromosome system. Three allopatric populations of the karyomorph A and one population of the karyomorph C were now in deep investigated by molecular cytogenetic analyses, using repetitive DNAs as probes. The results reinforced the relatedness among populations of the karyomorph A, despite their large geographic distribution. Karyomorph C, however, showed a remarkably difference in the genomic constitution, especially concerning the amount and distribution of the 5S rDNA and Rex3 sequences on chromosomes. In addition, although karyomorphs C and D share several features, exclusive chromosomal markers show the derivative evolutionary pathway between them. Thus, besides the classical chromosomal rearrangements, the repetitive DNAs were useful tools to reveal the biodiversity, relatedness and differentiation of this fish group. The chromosomal set strongly corroborates that E. erythrinus corresponds to a species complex instead of a single biological entity.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Erythrinidae, a small Neotropical fish family, is characterized by great karyotype diversity (Bertollo 2007). Particularly, in the genus Erythrinus four karyomorphs (A–D) have been identified in the nominal species E. erythrinus, which differ in chromosome number and morphology, and sex chromosomes (Bertollo et al. 2004; Cioffi et al. 2010). Karyomorph A has 2n = 54 chromosomes (6m + 2st + 46a), without heteromorphic sex chromosomes. Karyomorph B has 2n = 54 chromosomes in females and 2n = 53 chromosomes in males (♀ 6m + 2st + 42a + X1X1X2X2; ♂ 6m + 2st + 42a + X1X2Y. Karyomorph C has 2n = 52 chromosomes in females and 2n = 51 chromosomes in males (♀ 6m + 2sm + 6st + 34a + X1X1X2X2; ♂ 6m + 2sm + 6st + 34a + X1X2Y). Karyomorph D has 2n = 52 chromosomes in females and 2n = 51 chromosomes in males (♀ 4m + 2sm + 2st + 40a + X1X1X2X2; ♂ 4m + 2sm + 2st + 42a + X1X2Y. In all the cases, the Y chromosome is a large metacentric, the biggest chromosome of the set. Bertollo et al. (2004) proposed that the Y chromosome was originated by a centric fusion between two non-homologous acrocentric chromosomes. Consequently, the unpaired acrocentrics in the male karyotype correspond to the X1 and X2 chromosomes.
It is well now known that a substantial fraction of the eukaryotic genomes consists of repetitive DNA sequences, including satellites, minisatellites, microsatellites and transposable elements (Jurka et al. 2005; López-Flores and Garrido-Ramos 2012). The chromosomal mapping of these repeated sequences by means of the fluorescence in situ hybridization (FISH) provide a better picture of the genome organization, and the characterization of the biodiversity (Cioffi et al. 2012a).
Some classes of repetitive DNAs were already previously used providing relevant information about the evolutionary diversification of the karyomorphs in E. erythrinus. In fact, the use of telomeric probes clearly evidenced that the large Y chromosome of karyomorph D harbors a characteristic interstitial telomeric site (ITS) on its centromeric region (Cioffi et al. 2010), thus confirming the previous proposed origin for this sex chromosome. A notable spreading of the retrotransposon Rex3 co-localized with the 5S rDNA sites was also found to occur on the centromeric region up to 22 chromosomes in karyomorph D. It was hypothesized that the retrotransposable element has been inserted into the 5S rDNA sequence and that the 5S rDNA/Rex3 complex dispersed in the karyotype (Cioffi et al. 2010). In addition, chromosome painting with probes derived from microdissection of the Y chromosome suggested that the multiple X1X2Y sex system was derived from a simple XY sex system, still morphologically undifferentiated, as present in karyomorph A (Cioffi et al. 2011).
In the present study, three allopatric populations of the karyomorph A and one population of the karyomorph C of E. erythrinus were in deep investigated. Molecular cytogenetic analyses were employed in order to find useful markers for comparative genomics at the chromosomal level. The data highlighted unique or shared features present among populations and/or karyomorphs, their interrelationships and differentiation, providing new insights into the karyoevolutionary pathways of this fish group.
Materials and methods
Specimens and mitotic chromosome preparation
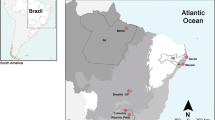
Samples of E. erythrinus from three populations of karyomorph A and one population of karyomorph C were obtained from distinct hydrographic basins (Fig. 1; Table 1), which are largely isolated by thousands of kilometers. Population from Barão de Melgaço (MT) is now being analyzed for the first time. The other populations were previously analyzed only by classical cytogenetic procedures.
Mitotic chromosomes were obtained from cell suspensions of the anterior kidney, using the conventional air-drying method (Bertollo et al. 1978). The experiments followed ethical conducts, and anesthesia was used prior to sacrificing the animals. Approximately 30 metaphase spreads were analyzed per specimen to confirm the diploid chromosome number and karyotype structure. Images were captured by the CoolSNAP system software, Image Pro Plus, 4.1 (Media Cybernetics, Silver Spring, MD, USA), coupled to an Olympus BX50 microscope (Olympus Corporation, Ishikawa, Japan). The chromosomes were classified as metacentric (m), submetacentric (sm), subtelocentric (st) or acrocentric (a) according to the arm ratios (Levan et al. 1964).
Chromosome probes and FISH experiments
Two tandemly-arrayed DNA sequences isolated from the genome of another Erythrinidae species, Hoplias malabaricus, were used. The first probe contained a 5S rDNA repeat copy and included 120 base pairs (bp) of the 5S rRNA transcribing gene and 200 bp of the non-transcribed spacer (NTS) (Martins et al. 2006). The second probe corresponded to a 1,400-bp segment of the 18S rRNA gene obtained via PCR from nuclear DNA (Cioffi et al. 2009). The 5S and 18S rDNA probes were cloned into plasmid vectors and propagated in DH5α Escherichia coli competent cells (Invitrogen, San Diego, CA, USA). The retrotransposable element Rex3 was obtained by PCR directly from the genome of E. erythrinus (Cioffi et al. 2010). The 5S rDNA probe was labeled with biotin-14-dATP by nick translation according to the manufacturer’s recommendations (BioNick™Labeling System; Invitrogen, San Diego, CA, USA). The 18S rDNA and Rex3 probes were labeled by nick translation with DIG-11-dUTP according to the manufacturer’s instructions (Roche, Mannheim, Germany).
Fluorescence in situ hybridization (FISH) was performed under high stringency conditions on mitotic chromosome spreads (Pinkel et al. 1986). The metaphase chromosome slides were incubated with RNAse (40 μg/ml) for 1.5 h at 37 °C. After denaturation of chromosomal DNA for 3 min in 70 % formamide/2× SSC (pH 7.0) at 70 °C, the spreads were incubated in 2× SSC for 4 min at 70 °C. The hybridization mixture (2.5 ng/μl probes, 2 μg/μl salmon sperm DNA, 50 % deionized formamide, 10 % dextran sulphate, pH 7.0) was dropped on the slides, and the hybridization was performed overnight at 37 °C in a moist chamber containing 2× SSC. Two posthybridization washes were carried out on a shaker (150 rpm) at 37 °C. The first wash was in 50 % formamide/2× SSC (pH 7.5) for 15 min, followed by a second wash in 2× SSC for 15 min. A final wash was performed at room temperature in 4× SSC for 15 min. Avidin-FITC (Sigma, St.Louis, MO, USA) was used for signal detection of the 5S rDNA probe and anti-digoxigenin-rhodamine (Roche, Mannheim, Germany) for 18S rDNA and Rex3 probes.
FISH experiments with the microsatellites d(CA)15 and d(GA)15 as probes were performed as described in Kubat et al. (2008), with slight modifications. These sequences were directly labeled with Cy3 at 5′ terminal during synthesis by Sigma (St. Louis, MO, USA). The chromosomes were counterstained with DAPI (1.2 μg/ml), mounted in antifade solution (Vector, Burlingame, CA, USA), and analyzed in an epifluorescence microscope Olympus BX50 (Olympus Corporation, Ishikawa, Japan).
Results
Karyotyping
The karyotype of the four populations displays the general features that distinguish Erythrinus from the other Erythrinidae fish, i.e., a great number of acrocentric chromosomes and few biarmed chromosomes. The samples from Barão de Melgaço (MT), Birigui (SP) and Resistência (AR), showed 2n = 54 chromosomes (6m + 2st + 46a), without differentiated sex chromosomes, which characterizes the karyomorph A (Fig. 2).
Karyotypes of males and females of karyomorphs A and C under different cytogenetic analyses. The karyotypes were probed with 5S rDNA, 18S rDNA and Rex3 retrotransposable element by FISH and counterstained with DAPI. The co-localization of the 5S and Rex3 sites are indicated by arrows in the metaphase plates. Note the significant spreading of the conjugated 5S rDNA/Rex3 sites in the karyomorph C. m metacentric chromosomes; sm submetacentric chromosomes; st subtelocentric chromosomes; a acrocentric chromosomes. Bar 5 μm
The sample from Manaus (AM) shows 2n = 52 chromosomes (6m + 2sm + 6st + 34a + X1X1X2X2) in the females and 2n = 51 chromosomes (6m + 2sm + 6st + 34a + X1X2Y) in the males This karyotype structure characterizes the karyomorph C of E. erythrinus. (Fig. 2). The X1 and X2 chromosomes were tentatively identified according to previous results for karyomorph D, which has the same X1X1X2X2/X1X2Y sex system.
Chromosomal mapping of 18S and 5S rDNAs and Rex3
The cytogenetic molecular markers displayed the same distribution pattern for all populations analyzed of karyomorph A. Thus, we are presenting only the results obtained from Barão de Melgaço population as representative of all other populations of this karyomorph. A similar distribution pattern for the 18S rDNA sites was found for karyomorphs A and C. Four acrocentric pairs bear 18S rDNA sites in the terminal region of the short or long arms, in addition to one chromosome pair with bitelomeric sites. In turn, the 5S rDNA shows a very distinct distribution (Fig. 2). Karyomorph A has only one acrocentric pair with 5S rDNA sites on the telomeric region of the short arms. This same chromosome pair also bears 18S rDNA sites on its long arms, thus evidencing an unusual syntenic condition concerning these two classes of ribosomal DNAs. This same synteny is shared by karyomorph C, although also found in an additional pair of chromosomes (Fig. 2). In addition, several acrocentric chromosomes show 5S DNA sites in this karyomorph, totalizing 14 pairs in the females and 12 pairs plus the X1, X2 and Y chromosomes in the males. In all chromosomes, the 5S DNA is localized in the telomeric region of the short arms, with exception of the Y chromosome where it is found on the centromeric region (Fig. 2). In both karyomorphs there is a clear co-localization of the transposable element Rex3 and the 5S rDNA sites, as demonstrated by the double-FISH mapping (Fig. 2).
Chromosomal mapping of the microsatellite repeats
The microsatellites (CA)15 and (GA)15 are abundantly distributed in all chromosomes of the karyomorph A. Conspicuous sites are found on the telomeric regions of the chromosomes, but clearly interstitial sites can also be observed (Fig. 3). A similar distribution pattern for these microsatellites is also found in karyomorph C, without significant differences between males and females. The metacentric Y presents conspicuous interstitial sites for both microsatellites, as well as other acrocentric chromosomes of the complement (Fig. 3).
Discussion
Chromosomal mapping of 18S and 5S rDNAs and Rex3
The in situ investigation of 5 classes of repetitive DNA sequences resulted in useful characteristics for comparative genomics at the chromosomal level, providing new insights into the karyoevolution of E. erythrinus. Although karyomorphs A and C share a similar 18S rDNA sites distribution, the last one shows a remarkably difference in the genomic constitution concerning the 5S rDNA and Rex3 sequences. Indeed, while karyomorph A presents only one acrocentric pair harboring 5S rDNA sites co-localized with the Rex3 retroelement, in the karyomorph C 27 chromosomes in the males and 28 chromosomes in the females were found to have such co-localized sites (Fig. 2). It is outstanding that this same scenario is found in the karyomorph D (Cioffi et al. 2010), although differing by the number of chromosomes harboring the 5S rDNA-Rex3 complex, i.e., 21/22 chromosomes in the karyomorph D and 27/28 chromosomes in the karyomorph C (Figs. 2, 4).
Overview of the evolutionary karyotypic pathways proposed for karyomorphs A–D of E. erythrinus on the basis of their karyotypic features and FISH mapping for karyomorphs A, C and D. The main chromosomal changes from the probable basal karyotype found in karyomorph A (2n = 54; 6m + 2st + 46a) are underlined (modified from Cioffi et al. 2010)
The heterochromatin is a complex composition of various types of repetitive sequences (Charlesworth et al. 1994; Kidwell 2002). Chromosomal repatterning may alter the number and position of these sequences, driving an intragenomic dynamism during the evolutionary process. Therefore, differentiation in the amount and distribution of the heterochromatic genomic fraction may usually occur, as in many fish species (Cioffi and Bertollo 2012). Under the selfish DNA hypothesis, inserted repetitive elements would accumulate in the heterochromatin because there are fewer genes in these regions (Horvath et al. 2001; Grewal and Jia 2007) and inserted elements are therefore less likely to be deleterious and more likely to be reproduced (Biémont and Vieira 2006). However, the dynamics of such accumulation, the specificities in targeting and location of different sequences, and the possible roles that repetitive elements might play within heterochromatin contravene this view (Dimitri and Junakovic 1999). Rather than representing the mere addition of ‘junk DNA’ to the genomic ‘ghost town’, the accumulation of repetitive DNAs in heterochromatin might reveal an important evolutionary interaction between these two ubiquitous and fluid components of the genome (Grewal and Jia 2007). Indeed, our findings regarding the mapping and distribution of the 5S and Rex3 sequences on the chromosomes of E. erythrinus show the dynamism of the rDNA clusters in this species, strongly supporting the view that these repetitive sequences are relevant genomic components mediating genetic differentiation during the evolutionary process.
Chromosomal mapping of the microsatellite repeats
Microsatellites are abundant repeated sequences present in all eukaryotic genomes studied thus far and are found either between the coding regions of structural genes or between other repetitive sequences (Tautz and Renz 1984). One of the most remarkable properties of these sequences is their ability to give rise to variants with a different number of repeats. In addition, microsatellite repeats may be organized in long stretches consisting of hundreds to several thousand tandem units, and associated with constitutive heterochromatin in many species (reviewed in Martins 2007). In the fish genomes, microsatellites are usually localized in the telomeres, centromeres, as well as in the sex chromosomes, where a significant fraction of repetitive DNA is also found (Cioffi and Bertollo 2012).
In E. erythrinus both microsatellites (CA)15 and (GA)15 probes generated evident signals on euchromatic and heterochromatic chromosomal regions, but with a preferential location in the heterochromatin (Fig. 3). Likewise, these microsatellite repeats were also found clustered in other fish species, as in Imparfinis schubarti, Steindachneridion scripta and Rineloricaria latirostris (Loricariidae), with a remarkable accumulation of the (GA)15 microsatellite in telomeric regions (Vanzela et al. 2002). Furthermore, in the zebrafish Danio rerio, and in the Characidae Triportheus auritus, both (CA)15 and (GC)15 repeats were preferentially located in centromeric and telomeric regions (Shimoda et al. 1999; Cioffi et al. 2012b). This preferential accumulation may indicate particular chromosomal regions where microsatellites are present as very large perfect or degenerate arrays.
In spite of the general similar distribution in both karyomorphs, (CA)15 and (GA)15, microsatellites showed conspicuous interstitial marks on the long arm of the Y chromosome in karyomorph C. However, these sites are also found in other acrocentric chromosomes of the chromosome set, being more frequent for microsatellite (GA)15 (Fig. 3). The comparative analysis between male and female karyotypes clearly indicated that the large metacentric Y chromosome, found in karyomorphs B-D, was originated by a centric fusion between two non homologous acrocentric chromosomes (Bertollo et al. 2004). Although we have no still specific markers to identify these acrocentric chromosomes, those ones carrying the interstitial microsatellite sites are good candidates. Indeed, there is a good correspondence between these sites and that one located on the Y chromosome (Fig. 3).
Interactions and differentiation of the karyomorphs
The main steps demonstrating the differentiation and interrelationships among karyomorphs A–D of E. erythrinus is summarized in the Fig. 4. It was previously considered that the chromosomal characteristics found in karyomorph A (2n = 54 chromosomes in males and females and absence of heteromorphic sex chromosomes) represent the probable basal karyotype from which the other karyomorphs were differentiated. Thus, some chromosomal rearrangements, such as centric fusions and pericentric inversions, gave rise to the X1X2Y sex chromosome system that occurs in karyomorphs B-D, as well as their different karyotypic formulas (Bertollo et al. 2004).
Although we have no further information regarding the karyomorph B, the occurrence of chromosomal markers shared by karyomorphs A, C and D allowing infer about the ancestry of these characteristics within the group. This is the case of the 18S rDNA bi-telomeric sites (present in both telomeric regions of a same chromosome), the conjugated sites of 5S rDNA + Rex3 elements, as well as the syntenic condition of the 18S + 5S/Rex3 sites. In turn, the spreading of the 5S/Rex3 sites through the genome is found in karyomorphs C and D, thus representing a synapomorphic condition for these karyomorphs. However, the karyomorph C has also unique characteristics, such as the additional spreading of 5S/Rex3 sites in three chromosome pairs and the additional synteny 18S + 5S/Rex3 sites in one chromosome pair.
It is clear that the spreading of associated transposable elements and the 5S ribosomal DNA in the genome is strongly associated with the evolution of both karyomorphs C and D. In both karyomorphs all 5S rDNA sites are co-located with the Rex3 elements, showing a surprising increase of these associated sites in contrast with karyomorph A. Considering that karyomorphs C and D represent derivative forms with respect to karyomorph A, the results clearly demonstrate a large dispersion of these elements in the centromeric region of the acrocentric chromosomes. Thus, it is likely that retroelement Rex3 sequences have been inserted in the 5S rRNA and that the complex rDNA 5S/Rex3 has been dispersed in karyotype (Cioffi et al. 2010). In addition, the centromeric 5S/Rex3 site in the large metacentric Y chromosome reinforces its origin by centric fusion between two acrocentric chromosomes.
Conclusions
Besides classical chromosomal rearrangements, repetitive DNAs played an important role in the differentiation of E. erythrinus karyomorphs, as evidenced by the presence and distribution of marker sequences on the chromosomes. The scenario found in E. erythrinus enhances the usefulness of the repetitive sequences in revealing the large amount of diversity found among fishes, improving the comprehension of their genome structure and evolution. In fact, the mapping of these sequences highlights unique or shared features among karyomorphs, indicating evolutionary pathways during their differentiation. The similar chromosomal features shared by populations of karyomorph A reinforce the view that they constitute an evolutionary unit, despite their isolation in different river basins. In addition, although karyomorphs C and D also share several features, exclusive chromosomal markers show the derivative evolutionary pathway between them, as well as from karyomorph A. Thus, although there are no conspicuous morphological differences easily detectable between the distinct populations, the set of chromosomal characteristics strongly corroborate that E. erythrinus corresponds to a species complex instead of a single biological entity.
References
Bertollo LAC (2007) Chromosome evolution in the Neotropical Erythrinidae fish family: an overview. In: Pisano E, Ozouf-Costaz C, Foresti F, Kapoor BG (eds) Fish cytogenetics. Science Publishers, Enfield, NH, pp 195–211
Bertollo LAC, Takahashi CS, Moreira-Filho O (1978) Cytotaxonomic considerations on Hoplias lacerdae (Pisces, Erythrinidae). Brazil J Genet 1:103–120
Bertollo LAC, Oliveira C, Molina WF, Margarido VP, Fontes MS, Pastori MS, Falcão JN, Fenocchio AS (2004) Chromosome evolution in the erythrinid fish, Erythrinus erythrinus (Teleostei: Characiformes). Heredity 93:228–233
Biémont C, Vieira C (2006) Junk DNA as an evolutionary force. Nature 443:521–524
Charlesworth B, Snlegowski P, Stephan W (1994) The evolutionary dynamics of repetitive DNA in eukaryotes. Nature 371:215–220
Cioffi MB, Bertollo LAC (2012) Chromosomal distribution and evolution of repetitive DNAs in fish. In: Garrido-Ramos MA (ed) Repetitive DNA Genome Dynamics v 7. Karger, Basel, pp 197–221
Cioffi MB, Martins C, Centofante L, Jacobina U, Bertollo LAC (2009) Chromosomal variability among allopatric populations of Erythrinidae fish Hoplias malabaricus: mapping of three classes of repetitive DNAs. Cytogenet Genome Res 125:132–141
Cioffi MB, Martins C, Bertollo LAC (2010) Chromosomal spreading of associated transposable elements and ribosomal DNA in the fish Erythrinus erythrinus. Implications for genome change and karyoevolution in fish. BMC Evol Biol 10:217
Cioffi MB, Sánchez A, Marchal JA, Kosyakova N, Liehr T, Trifonov V, Bertollo LAC (2011) Cross-species chromosome painting tracks the independent origin of multiple sex chromosomes in two cofamiliar Erythrinidae fishes. BMC Evol Biol 11:186
Cioffi MB, Molina WF, Artoni RF, Bertollo LAC (2012a) Chromosomes as tools for discovering biodiversity. The case of Erythrinidae fish family. In: Padma Tirunilai (ed) Recent trends in cytogenetic studies−Methodologies and applications, 1st edn, InTech, pp. 125–146
Cioffi MB, Kejnovsky E, Marquioni V, Poltronieri J, Molina WF, Diniz D, Bertollo LAC (2012b) The key role of repeated DNAs in sex chromosome evolution in two fish species with ZW sex chromosome system. BMC Mol Cytogenet 5:28
Dimitri P, Junakovic N (1999) Revising the selfish DNA hypothesis: new evidence on accumulation of transposable elements in heterochromatin. Trends Genet 15:123–124
Grewal SIS, Jia S (2007) Complexities of heterochromatin in fungi, ciliates, plants and mammals. Nat Rev Genet 8:35–46
Horvath JE, Bailey JA, Locke DP, Eichler EE (2001) Lessons from the human genome: transitions between euchromatin and heterochromatin. Hum Mol Genet 10:2215–2223
Jurka J, Kapitonov VV, Pavlicek A, Klonowski P, Kohany O, Walichiewicz J (2005) Repbase update, a database of eukaryotic repetitive elements. Cytogenet Genome Res 110:462–467
Kidwell MG (2002) Transposable elements and the evolution of genome size in eukaryotes. Genetica 115:49–63
Kubat Z, Hobza R, Vyskot B, Kejnovsky E (2008) Microsatellite accumulation in the Y chromosome of Silene latifolia. Genome 51:350–356
Levan A, Fredga K, Sandberg AA (1964) Nomenclature for centromeric position on chromosomes. Hereditas 52:201–220
López-Flores I, Garrido-Ramos MA (2012) The repetitive DNA content in eukaryotic genomes. In: Garrido-Ramos MA (ed) Repetitive DNA Genome Dynamics v 7. Karger, Basel, pp 1–28
Martins C (2007) Chromosomes and repetitive DNAs: a contribution to the knowledge of fish genome. In: Pisano E, Ozouf-Costaz C, Foresti F, Kapoor BG (eds) Fish cytogenetics. Science Publishers, Enfield, NH, pp 421–453
Martins C, Ferreira IA, Oliveira C, Foresti F, Galetti PM Jr (2006) A tandemly repetitive centromeric DNA sequence of the fish Hoplias malabaricus (Characiformes: Erythrinidae) is derived from 5S rDNA. Genetica 127:133–141
Pinkel D, Straume T, Gray J (1986) Cytogenetic analysis using quantitative, high sensitivity, fluorescence hybridization. Proc Natl Acad Sci USA 83:2934–2938
Shimoda N, Knapik EW, Ziniti J, Sim C, Yamada E et al (1999) Zebrafish genetic map with 200 microsatellite markers. Genomics 58:219–232
Tautz D, Renz M (1984) Simple sequences are ubiquitous repetitive components of eukaryotic genomes. Nucleic Acids Res 12:4127–4138
Vanzela ALL, Swarça AC, Dias AL, Stolf R, Ruas PM et al (2002) Differential distribution of (GA)9 + C microsatellite on chromosomes of some animal and plant species. Cytologia 67:9–13
Acknowledgments
This work was supported by the Brazilian agencies FAPESP (Fundação de Amparo à Pesquisa do Estado de São Paulo), CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico), CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior), FAPEMAT (Fundação de Amparo à Pesquisa do Estado de Mato Grosso).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Martins, N.F., Bertollo, L.A.C., Troy, W.P. et al. Differentiation and evolutionary relationships in Erythrinus erythrinus (Characiformes, Erythrinidae): comparative chromosome mapping of repetitive sequences. Rev Fish Biol Fisheries 23, 261–269 (2013). https://doi.org/10.1007/s11160-012-9292-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11160-012-9292-4