Abstract
Ubiquitination is a eukaryotic post-translational protein modification, implicated in various cellular processes in higher plants, and E3 ligases play an essential role in this cascade. In this study, we identified an Skp1–Cullin–F-box (SCF) E3 ligase encoding gene, Arabidopsis phloem protein 2-B11 (AtPP2-B11; At1g80110), by searching drought-responsive element in the promoter regions of 1,488 putative E3s in the Arabidopsis genome. The expression of AtPP2-B11 in seedlings was remarkably induced with increased duration of drought treatment. Transgenic lines overexpressing AtPP2-B11 exhibited obvious hypersensitivity to drought stress during seed germination and in mature plants. Protein interaction assay with Arabidopsis Skp1 (ASK) proteins showed that AtPP2-B11 strongly interacted with ASK 7, ASK 18, and ASK 19, respectively, indicating that AtPP2-B11 may function as an F-box protein. An AtPP2-B11-interacting protein, AtLEA14, was identified by a yeast two-hybrid screening and bimolecular fluorescence complementation assay. Real-time quantitative RT-PCR and Western blot analysis showed that the transcript and protein levels of AtLEA14 decreased in 35S::AtPP2-B11 transgenic plants subjected to drought conditions. Transgenic Arabidopsis plants overexpressing AtPP2-B11 showed altered expression of abiotic stress response marker genes (COR15a, COR47, ERD10, KIN1, RAB18, and RD22) under both normal and drought conditions. Overall, these results suggested that AtPP2-B11, a SCF E3 ligase, played an important role in response to drought stress as a negative regulator in Arabidopsis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Water-deficit stress, one of the most adverse conditions for plants, can inflict fatal damage to plants and adversely affect crop productivity. Plants respond and adapt to water-deficit stress through physiological, biochemical processes, molecular, and cellular processes. A number of genes have been identified that respond to drought stress at the transcriptional level in plants. Their gene products are believed to play integral roles in protecting cells from water deficit (Shinozaki and Yamaguchi-Shinozaki 1996; Shinozaki et al. 2003; Thomashow 1999; Zhu 2002). Based on the functions of these drought-responsive genes, they have been classified into two major groups. One group encodes products that directly protect plant cells from stresses, such as antioxidants, chaperones, late embryogenesis abundant (LEA) proteins and enzymes for osmolyte biosynthesis (Goyal et al. 2005; Mittler 2002; Sun et al. 2002; Yoshiba et al. 1997). The gene products of the second group regulate gene expression and signal transduction during abiotic stress responses, including key metabolic enzymes, protein kinases, and transcription factors (Liu et al. 1998; Nakashima et al. 2009; Zhu 2002).
Genes that function in response to drought stress at the cellular level are regulated by different signaling pathways which have been classified into abscisic acid (ABA)-dependent and ABA-independent signaling pathways (Kang et al. 2011; Lee et al. 2009; Shinozaki and Yamaguchi-Shinozaki 2007; Shinozaki et al. 2003; Yamaguchi-Shinozaki and Shinozaki 2005; Zhang et al. 2004). Two major cis-acting elements, ABA-responsive element (ABRE) and drought-responsive element (DRE), have been previously demonstrated to function in ABA-dependent and ABA-independent gene expressions, respectively, in drought stress responses (Gomez-Porras et al. 2007; Shen and Ho 1995; Shinozaki et al. 2003; Yamaguchi-Shinozaki and Shinozaki 2005). In the ABA-dependent regulatory system, drought triggers the production of phytohormone ABA, which regulates stomata closure and induces the expression of many genes that may function in dehydration tolerance (Gomez-Porras et al. 2007; Hobo et al. 1999; Narusaka et al. 2003; Shen and Ho 1995; Shinozaki and Yamaguchi-Shinozaki 2007; Zhang et al. 2005). For the ABA-independent regulatory system, DRE is the key cis-element that regulates the expression of osmotic and cold stress response genes (Agarwal et al. 2007; Nakashima et al. 2009; Shinozaki and Yamaguchi-Shinozaki 2007). A 9-bp DRE motif, TACCGACAT, has first been identified in the promoter of the stress response gene RD29A in Arabidopsis (Yamaguchi-Shinozaki and Shinozaki 1994). Analysis of the DRE sequence in RD29A promoter reveals that a single copy of DRE is sufficient for the ABA-independent expression of RD29A, indicating that DRE functions in a different manner from ABRE and does not require other elements to facilitate stress-inducible gene expression. Previous studies have revealed that the DRE motif is also located in the promoter regions of marker genes such as KIN1, COR15a, and COR47 and involved in the stress response pathways (Lee et al. 2010; Yamaguchi-Shinozaki and Shinozaki 1994). In soybean, the DRE cis-element is enriched in the promoters of drought stress-responsive genes, suggesting the involvement of DRE in stress response to dehydration (Maruyama et al. 2012). Analysis of the promoters of F-box genes from maize has revealed that 13 ZmFBX genes contain the DRE motif in their promoters (Jia et al. 2013). The DRE core sequence CCGAC is also found in the promoter region of the cold-inducible gene BN115 in oilseed rape and crucially functions in stress response (Jiang et al. 1996). The wheat WCS120 gene, which contains a DRE motif in its promoter, encodes a protein believed to have important functions in plant acclimation to cold (Ouellet et al. 1998). The transcriptional regulator DRE-binding protein (DREB), harboring an ERF/AP2-type DNA-binding domain, specifically binds to the DRE cis-element to induce the expression of stress response genes (Liu et al. 1998; Nakashima et al. 2000; Sakuma et al. 2002). Several DREBs have been reported to be involved in the drought response pathway (Lee et al. 2010; Liu et al. 1998). DREB2A, a member of the DREB subfamily containing a single conserved DNA-binding domain, is a transcriptional activator that recognizes DRE in Arabidopsis (Liu et al. 1998). The DREB1A homologues DREB1B and DREB1C, which have also been isolated from Arabidopsis, play important roles in response to drought (Liu et al. 1998; Sakuma et al. 2002). Therefore, based on the abovementioned studies, it is critical to identify DRE-containing proteins that regulate drought response in Arabidopsis.
Protein degradation is an important post-translational regulatory process that mediates cell responses to intracellular signals and changing environmental conditions. The ubiquitin/26S (Ub/26S) pathway is a common regulatory mechanism for protein degradation and is conserved in all eukaryotes. The Ub/26S pathway controls the protein quantity and stability by the covalent attachment of a 76-amino acid ubiquitin polypeptide to target proteins. Three enzymes that consecutively act are involved in the conjugation cascade, namely, ubiquitin-activating enzyme (E1), ubiquitin-conjugating enzyme (E2), and ubiquitin ligase (E3 ligase) (Glickman and Ciechanover 2002; Smalle and Vierstra 2004; Vierstra 2009). In Arabidopsis, more than 1,400 genes encode components of the Ub/26S pathway, and 90 % of these genes encode E3 ligases (Moon et al. 2004; Smalle and Vierstra 2004). Among the several classes of E3 proteins, the multiple subunits E3 Skp1–Cullin-F–box (SCF) complexes are essential to the post-translational regulation of many important factors involved in signal transduction (Craig and Tyers 1999; Deshaies 1999; Lechner et al. 2006). The SCF complex is composed of an Arabidopsis Skp1 (ASK) homolog, a Cullin homolog, an Rbx (RING box) homolog, and an F-box protein, in which the F-box protein is the most important and best understood subunit. The Arabidopsis genome encodes more than 700 putative F-box proteins (Gagne et al. 2002; Lechner et al. 2006). The F-box proteins contain at least one F-box motif, a protein structural motif consisting of approximately 50 conserved amino acids that mediates protein–protein interactions (Xiao and Jang 2000). F-box proteins bind to the ASK proteins through direct protein–protein interaction through the F-box domain. In addition to the F-box domain, the C-terminal domains of F-box proteins are specific to bind the substrates for degradation. Many F-box proteins reportedly have essential functions in plant developmental and physiological processes as post-translational regulators of many important factors involved in signal transduction (Baudry et al. 2011; Dharmasiri et al. 2005; Risseeuw et al. 2003; Sawa et al. 2007; Thines et al. 2007; Wang et al. 2009b; Zheng et al. 2011). The first F-box protein identified in plants is UFO which has an important function in floral meristem identity and floral organ development (Hepworth et al. 2006; Laufs et al. 2003; Levin and Meyerowitz 1995; Samach et al. 1999). AtTLP9 is an Arabidopsis TUBBY-like protein that interacts with ASK 1 and is likely to participate in the ABA signaling pathway (Lai et al. 2004). EID1-like protein 3 (EDL3) functions as a positive regulator in ABA-dependent signaling cascades that regulates germination induction, root growth, and flowering (Koops et al. 2011). DOR from Arabidpsis acts as a negative regulator of guard cell and inhibits the ABA-induced stomata closure under drought stress (Zhang et al. 2008). OsFbx352, a rice F-box gene, is involved in glucose-delayed seed germination (Song et al. 2012). In addition, many F-box proteins from Arabidopsis are known to be involved in plant hormone responses including TIR1, AFB 1-3, and COI1 (Gray et al. 2001; Sheard et al. 2010; Walsh et al. 2006). Several F-box proteins involved in drought response have already been identified, but the detailed relationship between F-box proteins and drought stress remains unclear (Earley et al. 2010; Koops et al. 2011; Lai et al. 2004; Yan et al. 2011; Zhang et al. 2008). The key function of DRE in response to drought has been identified. Thus, the mechanism underlying the ability of DRE-containing F-box proteins to regulate drought stress in Arabidopsis must be investigated.
In the present study, 23 DRE-containing putative E3s were identified in the Arabidopsis genome. Data showed that among the 23 E3 proteins, Arabidopsis phloem protein 2-B11 (AtPP2-B11) that contains an F-box domain at the N-terminal, was severely induced by drought stress. Moreover, transgenic lines overexpressing AtPP2-B11 exhibited obvious hypersensitivity to drought stress during seed germination and in mature plants. Interaction studies with ASKs indicated that AtPP2-B11 may function as a component of the SCF ubiquitin ligase complex. AtPP2-B11 was also found to interact with AtLEA14, suggesting a possible connection between AtPP2-B11 and drought response through an ABA-independent signaling pathway.
Materials and Methods
Plant Materials and Growth Conditions
Seeds of each genotype Arabidopsis thaliana from Columbia (Col-0) background were harvested at the same time from plants grown at the same condition. Seeds were surface sterilized by soaking in 70 % ethanol for 5 min and 2.6 % sodium hypochlorite for 10 min, and than washed five times with sterilized water. The seeds were plated on Murashige and Skoog (MS) medium. The plates were then kept in the dark at 4 °C for stratification for 3 days and transferred to a growth chamber with a light/dark cycle of 16-h light/8-h dark at 22 °C.
Analysis of Promoter Regions of Putative E3 Ligases in Arabidopsis
All the putative E3 genes in Arabidopsis genome were retrieved from PlantsUBQ database (http://plantsubq.genomics.purdue.edu/), including ten different types: 694 F-box genes, 470 RING finger genes, 220 PHD genes, 80 BTB genes, 60 U-BOX genes, 21 ASKs, 10 Cullins, 7 HECTs, 5 DDB genes, and 2 RBXs. In 1,569 putative genes, the E3s with the same IDs were conflated and the final database of putative E3 genes in Arabidopsis consisted of 1,488 candidate genes. A database of 1-kb upstream sequences of 1,488 E3s was retrieved from the TAIR (www.arabidopsis.org) genome annotation. The frequency matrix of 48 sequences containing DRE core motif (A/GCCGAC) was created as described in a previous study (Gomez-Porras et al. 2007). The promoter sequences of 1,488 putative E3s were screened for occurrences of the DRE using the frequency matrices of DRE motif; 155 candidate genes containing one or more DRE elements were obtained. Combining with the previously described DREB-targeting genes, 23 DRE-containing putative E3s were finally identified for further analysis.
Expression Pattern of 23 DRE-Containing E3s by Semiquantitative RT-PCR Analysis
Two-week-old plate-grown plants were harvested from MS agar medium, and then dehydrated on filter paper at room temperature and approximately 70 % humidity under dim light. Plants were subjected to drought stress for 0.5, 1, 2 h and were frozen in liquid nitrogen. Total RNA was extracted with Trizol reagent, and the RNA preparation was then treated with RNase-free DNase I. First-strand cDNA synthesis was performed using 2 μg of total RNA with oligo (dT) primer and M-MLV RT. Semiquantitative RT-PCR was performed as described previously (Zhang et al. 2007). The expression of EF1-α was used as an internal control. Gene-specific primers of 23 E3s were listed in Table S1. These experiments were independently replicated at least three times under identical conditions.
Protein Extraction and Fluorometric GUS Assay
Plant protein extraction and analysis for GUS activity were performed as previously describe (Jefferson et al. 1987). The protein concentration of the extract was measured with a nanodrop instrument, and fluorescence was determined with a Microplate Spectrofluorometer (Hitachi). For analysis of GUS activity in different samples, the data were obtained by subtracting the background 4-methyiumbelliferyl glucuronide of the transgenic plants. The average GUS activity was obtained from at least five independent transformants, and each assay was repeated three times.
Real-Time Quantitative RT-PCR Analysis
Total RNA was prepared as describe above. cDNA was synthesized using PrimeScript reverse transcriptase (RT) with oligo (dT) primer using the PrimeScript RT master mix kit (Takara). All samples were prepared to a final volume of 10 μL. A SYBR green real-time PCR master mix (Takara) and a Chromo 4 real-time PCR detector (Bio-Rad) were used. PCR amplification was performed with primers of specific genes shown in Table S1. Amplification of the GAPDH gene was used as an internal control, and three technical replicates were performed for each experiment.
Construction of 35S::AtPP2-B11 Transgenic Plants and Drought Phenotype Analysis
The full-length AtPP2-B11 cDNA was amplified by PCR using a EcoRI and BamHI primer set: 5′-GAATTCATGAATAATCTCCCAGAAGA-3′ and 5′-GGATCCTTAGGGCACTGGTCTAATCT-3′. The resulting PCR product was cloned into the EcoRI and BamHI sites of binary vector pBI121 under the control of a cauliflower mosaic virus 35S promoter and transformed into Arabidopsis using an Agrobacterium tumefaciens-mediated floral dip method. To obtain independent transgenic lines, the collected seeds were selected on MS plates containing 50 mg/L kanamycin. Homozygous T3 lines were obtained by further self-crossing and used for phenotypic analysis.
Seed germination assay was performed with 100 seeds and repeated three times. Seeds, 3 days after stratification, of wild-type and 35S::AtPP2-B11 lines were grown on MS medium supplemented with 2.5 % glycerin at 22 °C with a 16-h-light/8-h-dark cycle. Germination was defined as an obvious emergence of the radicle through the seed coat. Wild-type and 35S::AtPP2-B11 plants were grown for 4 weeks under normal growth conditions and then subjected to dehydration stress by ceasing irrigation for 12 days. Three days after re-watering, the surviving plants were counted.
Subcellular Localisation
The 35S::AtPP2-B11-GFP and 35S::GFP plasmids were constructed using pJIT163 transient expression vectors. The constructed fusion genes were transformed into wild-type Arabidopsis protoplasts by means of PEG treatment following Jen Sheen’s lab protocol (Yoo et al. 2007). The expressions of AtPP2-B11-GFP and GFP were monitored 12 h after transformation. Transformed protoplasts were placed on the slide glass and observed using a laser scanning confocal microscope.
Yeast Two-Hybrid Assay
Yeast two-hybrid assay was carried out using Matchmaker® Gold Yeast Two-Hybrid System (Clontech, Palo Alto, CA). The AtPP2-B11 and ASKs cDNAs were amplified and expressed as a BD fusion and AD fusion. Transformation of BD fusion and AD fusion into the Y2H Gold Yeast and Y187 cells, respectively, was carried out according to the manufacturer’s instruction (Clontech). Y2H Gold Yeast with BD fusion and Y187 with AD fusion were then mated. The mated culture was spread on double (DDO; SD/-Trp/-Leu) and quadruple dropout (QDO; SD/-Trp/-Leu/-His/-Ade) media and incubated at 30 °C for 5–14 days. Colonies on the QDO medium were considered as positive clones. To confirm the results, colonies obtained on QDO medium were streaked on quadruple dropout (QDO/X/A; SD/-Trp/-Leu/-His/-Ade supplemented with X-α-Gal and Aureobasidin A) medium to ensure the interplay of AtPP2-B11 and ASKs.
A drought-induced Arabidopsis cDNA library was prepared in the pGADT7 vector (MatchMaker® Gold Yeast Two-Hybrid System, Clontech) using mRNA isolated from 2-week-old seedlings under 2 h dehydration treatment. Yeast transformation was performed according to the supplier’s instructions (Clontech).
Bimolecular Fluorescence Complementation Assay
cDNA without a termination codon encoding AtPP2-B11 was cloned into pSPYCE-35S, and the cDNA encoding AtLEA14 was cloned into pSPYNE-35S. These vectors were introduced into A. tumefaciens strain GV3101. For infiltration of Nicotiana benthamiana, the P19 protein of tomato bushy stunt virus was used to suppress gene silencing (Voinnet et al. 2003). Co-infiltration of A. tumefaciens strains containing the bimolecular fluorescence complementation (BiFC) constructs and the P19 silencing plasmid was infiltrated into leaves of 4-week-old N. benthamiana plants as described previously (Walter et al. 2004). After 2 days, epidermal cell layers of tobacco leaves were assayed for fluorescence under a fluorescence microscope.
Immunoblot Assay
Total protein of wild-type and 35S::AtPP2-B11 plants under normal and drought stress conditions were extracted using Plant Protein Extraction Kit (CWBIO). Antibody specific for AtLEA14 (CWBIO) was used as a primary antibody. Thirty micrograms of total protein per lane was separated on 15 % SDS-PAGE. After electrophoresis, proteins were electrotransferred to PVDF membrane. After blocking for 3 h in Tris-buffered saline with Tween (TBST; 50 mM Tris-HCl, pH 7.5, 150 mM NaCl, and 0.05 % Tween 20) with 5 % nonfat dry milk at room temperature, membranes were incubated with antibody of AtLEA14 at 1:1,000 dilution for 2 h. Following three times washing with TBST, membranes were incubated with Anti-IgG from rabbit (Sigma-Aldrich) for 1 h. After three washings with TBST, the membranes were visualized using an enhanced Lumi-Light Western Blotting Substrate kit (Thermo Scientific) following the manufacturer’s instructions. Coomassie Brilliant Blue staining was used to show protein loading levels.
Results
Identification of the Drought-Induced F-box Protein AtPP2-B11 and Functions of the DRE Motif in AtPP2-B11 Induction Under Drought Stress
To investigate the functions of E3s in response to drought stress in Arabidopsis, all gene IDs of putative E3s were retrieved from the PlantsUBQ database (http://plantsubq.genomics.purdue.edu/). After conflating genes with the same IDs, 1,488 putative E3 genes were obtained from this database, and then we searched against the upstream promoter regions of 1,488 E3s for the DRE motif using a matrix-based in silico screening procedure (Gomez-Porras et al. 2007; Wang et al. 2009a) (Fig. 1a). Using this approach, 155 candidate E3s were identified. In a previous study, a genome-wide investigation of DREB-targeting genes was performed, and 474 candidate genes were identified in Arabidopsis (Wang et al. 2009a). By combining the two sets of data obtained using two different strategies, 23 DRE-containing putative E3s that potentially function in drought response were identified (Fig. 1b).
Identification of DRE-containing E3s responding to drought stress. a Sequence logos and matrices for DRE motif used in this study. Sequence logos (top) were created online using the Weblogo resource (http://weblogo.berkeley.edu/). The frequency matrix of DRE motif was created as described in a previous study (Gomez-Porras et al. 2007). b Putative DRE-containing E3s involved in drought stress identified through two different strategies. The graph was generated by Venny program (http://bioinfogp.cnb.csic.es/tools/venny/index.html). c Induction patterns of 23 DRE-containing E3s by drought stress. Total RNA was extracted from normal or drought-treated 2-week-old wild-type seedlings and semiquantitative RT-PCR was performed with three biological repeats. Elongation factor (EF1-α) transcript levels were used as loading controls. Plus sign indicated the degree of change observed in the semiquantitative RT-PCR analysis
To further elucidate the expression patterns of 23 DRE-containing putative E3s under drought stress, semiquantitative RT-PCR was used to determine the transcript levels of 23 DRE-containing putative E3s through a drought-treatment time course. As shown in Fig. 1c, 18 of 23 genes were detectable in semiquantitative RT-PCR analysis, and the expression levels of six putative E3 genes containing the DRE motif were found to be upregulated, indicating their potential involvement in drought response.
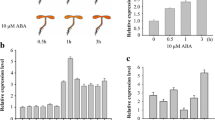
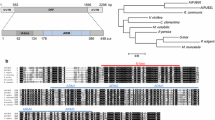
Among the six DRE-containing E3s, the expression of At1g80110 was significantly induced by drought stress (Fig. 1c). At1g80110 encodes AtPP2-B11, a member of the phloem protein 2 family proteins (Dinant et al. 2003) (Fig. 2a). Multiple sequence alignment of AtPP2-B11 with proteins such as Arabidopsis SON, UFO, yeast (Saccharomyces cerevisiae) CDC4, rice DRF, and human Skp2, which have been previously identified as F-box proteins, was performed. Results showed that some residues within the F-box domain of AtPP2-B11 were highly conserved revealing that the AtPP2-B11 protein contained a putative F-box domain (Fig. S1). Real-time quantitative RT-PCR revealed that AtPP2-B11 exhibited apparent drought stress response, in agreement with semiquantitative RT-PCR analysis (Fig. 2c). The expression of AtPP2-B11 in all tissues of Arabidopsis was detected, including roots, stems, rosette leaves, stem leaves, flowers, and siliques. The expression levels of AtPP2-B11 in flowers and rosette leaves were higher than those in other organs (Fig. 2d). To examine the function of DRE motif in AtPP2-B11 promoter, AtPP2-B11 native or DRE-deleted promoter fused with the β-glucuronidase (GUS) coding sequence was constructed (Fig. 2b). GUS activities of the two kinds of transgenic seedlings under normal condition had no significant differences (Fig. 2e). However, under drought stress conditions, GUS activities of seedlings carrying the AtPP2-B11 native promoter increased by almost 70 %, whereas only a 10 % increase in GUS activities was observed in seedlings when the 9-bp DRE motif was deleted (Fig. 2f). These results indicated that the DRE motif in the promoter region of AtPP2-B11 may have an important function in inducing AtPP2-B11 expression under drought stress and that our screening strategy based on promoter cis-element was efficient in identifying genes involved in drought stress.
Schematic structure, expression pattern, and DRE deletion analysis of AtPP2-B11. a Schematic representation of AtPP2-B11 protein. c Induction investigation of AtPP2-B11 by dehydration stress. Total RNA from 2-week-old wild-type plants were obtained and analyzed by real-time quantitative RT-PCR. d Expression patterns of AtPP2-B11 in different organs. Total RNA from root, stem, rosette leaf, stem leaf, flower, and silique were extracted and analyzed by real-time quantitative RT-PCR. b, e Activity analysis of AtPP2-B11 full-length promoter and DRE-deleted promoter by GUS assay. Two-week-old seedlings were dehydrated for 2 h, and GUS activities of different transgenic lines were measured using a fluorimetric assay with 4-methylumbelliferyl β-d-glucuronide as substrate
Overexpression of AtPP2-B11 Attenuated Drought Tolerance in Arabidopsis
To test the biological function of AtPP2-B11 in response to drought stress in plants, transgenic Arabidopsis constitutively expressing AtPP2-B11 cDNA fused with 35S promoter (35S::AtPP2-B11) were generated. As shown in Fig. 3a, AtPP2-B11 was highly expressed in the transgenic lines OE10 and OE11 under normal growth conditions, and these two independent transgenic lines were selected for further analysis. Figure 3b shows that during seed germination, no obvious difference of germination percentage was observed between wild-type plants (100 %) and the transgenic plants OE10 (99 %) and OE11 (99 %) under normal growth conditions. However, under drought stress conditions, the wild-type seeds germinated earlier than the transgenic seeds. After 2 days of drought stress, 77 % of wild-type seeds germinated, whereas only 41 and 23 % of the OE10 and OE11 seeds germinated, respectively (Fig. 3b, c). This finding suggested that transgenic lines overexpressing AtPP2-B11 were more sensitive to drought stress than the wild- type plants. Considering that seed germination in the OE11 transgenic line was lower than the OE10 line under drought conditions and that the AtPP2-B11 expression level in the OE11 line was higher than expression levels in the OE10 line, we suggested that drought hypersensitivity in these AtPP2-B11 overexpressing lines was correlated with AtPP2-B11 expression levels (Fig. 3a).
Phenotypic and drought stress tolerance studies of wild-type and AtPP2-B11 overexpressing transgenic plants. a Expression of AtPP2-B11 in wild-type and transgenic plants. Two-week-old plants were investigated by real-time quantitative RT-PCR analysis. b Drought response of wild-type and 35S::AtPP2-B11 transgenic lines in germination stage. c Seed germination of wild-type and transgenic plants grown on MS medium containing 2.5 % glycerin which imitated drought stress. Germination rates in terms of an obvious emergence of the radicle through the seed coat were determined after the end of stratification. d Drought tolerance of wild-type, 35S::AtPP2-B11 transgenic lines OE10 and OE11. Four-week-old plants were grown for 12 days without watering. Survival rates were determined after 5 days of re-watering. Fifty plants were tested in each experiment
To investigate whether AtPP2-B11 affected the tolerance to drought stress in mature plants, 4-week-old transgenic and wild-type plants were used to test their response to water deficit. Under normal growth conditions, the transgenic lines and wild-type plants showed no significant morphological or developmental abnormalities. However, when subjected to drought stress by ceasing irrigation for 12 days, both AtPP2-B11 overexpressing lines exhibited obvious leaf rolling and wilting phenotype, whereas the wild-type plants did not exhibit significant wilting (Fig. 3d). After re-watering for 3 days, 84 % (42 of 50) of the wild-type plants were able to survive and resume growth. However, the survival rates of the AtPP2-B11 overexpressing lines were only 48 (24 of 50, OE10) and 26 % (13 of 50, OE11), respectively. These results indicated that AtPP2-B11 may have an important function in plant response to drought stress.
AtPP2-B11 Functioned as an F-box Protein by Interacting with ASK Proteins
F-box proteins are known to interact with the ASK subunit of SCF complexes (Risseeuw et al. 2003). To determine whether AtPP2-B11 actually functioned as an F-box protein, the interactions between AtPP2-B11 and ASK proteins were examined by yeast two-hybrid analysis. In this assay, AtPP2-B11 cDNA was fused with the DNA-binding domain of GAL4 in the pGBKT7 vector as bait. The coding sequences of eight ASK proteins (ASK 4, ASK 5, ASK 7, ASK 13, ASK 16, ASK 17, ASK 18, and ASK 19) in Arabidopsis were fused with the DNA activating domain of GAL4 in the pGADT7 vector as prey. The BD and AD fusion vectors were then transformed into Gold Yeast and Y187, respectively. Results showed that AtPP2-B11 selectively interacted with ASK 7, ASK 18, and ASK 19 proteins, respectively. Therefore, AtPP2-B11 functioned as an F-box protein by interacting with specific ASK proteins (Fig. 4).
Interaction analysis between AtPP2-B11 and ASKs by yeast two-hybrid assay. BD pGBKT7 vector, AD pGADT7 vector. BD-AtPP2-B11 as bait was transformed into Gold Yeast strain, and different ASKs fused with AD as prey were transformed into Y187 strains. Y2H Gold Yeast with BD fusion and Y187 with AD fusion were then mated, and mated yeast cells were then spread on the selective medium: DDO, QDO, and QDO/X/A. BD-53 BD fused with murine p53, BD-LAM BD fused with lamin, AD-T7 AD fused with SV40 large T-antigen. Mating of BD-53 and AD-T7 were severed as positive control and mating of BD-53 and AD-T7 as negative control
Subcellular Localisation of AtPP2-B11
To investigate the cellular localisation of AtPP2-B11, an in vivo localisation experiment was conducted. The AtPP2-B11 coding region was fused with the GFP gene driven by the CaMV 35S promoter and transiently transformed into Arabidopsis leaf protoplasts using a polyethylene glycol-mediated method (Yoo et al. 2007). Confocal microscopy revealed a strong fluorescence signal in the cytoplasm attributed to the AtPP2-B11-GFP fusion protein compared with the localisation pattern of the control (35S::GFP), indicating that AtPP2-B11 mainly functioned in the cytoplasm (Fig. 5).
Subcellular localization of AtPP2-B11 protein. AtPP2-B11 cDNA was fused to green fluorescent protein (GFP) and the 35S::GFP and 35S::AtPP2-B11-GFP constructs were transformed into Arabidopsis leaf protoplasts by a PEG-mediated method. Localization of fusion proteins was visualized by fluorescence microscopy
Identification of AtLEA14 as an AtPP2-B11 Interacting Protein
To identify the proteins that interact with AtPP2-B11 and are potentially involved in the drought response signal transduction pathway, yeast two-hybrid screening of drought-induced cDNA library from Arabidopsis was carried out using AtPP2-B11 as bait. After screening on high-stringency QDO/X/A (SD/-Trp/-Leu/-His/-Leu supplemented with X-α-Gal and Aureobasidin A) medium and sequencing of isolated clones, a cDNA clone corresponding to the Arabidopsis At1g01470 gene encoding AtLEA14 protein was identified as a strong AtPP2-B11 interactor. The full-length AtLEA14 was further fused with the pGADT7 vector to confirm its interaction with the full-length AtPP2-B11. As shown in Fig. 6a, in the high-stringency medium, the protein–protein interactions of full-length AtPP2-B11 and AtLEA14 appeared very strong, similar to the RecT-53 positive control. This finding indicated physical interaction between AtPP2-B11 and AtLEA14 (Fig. 6a).
Identification of interaction between AtPP2-B11 and AtLEA14 via yeast two-hybrid and BiFC assay. a AtPP2-B11 interacted with AtLEA14 in yeast two-hybrid assay. AtPP2-B11 was fused with BD vector and AtLEA14 was fused with AD vector. Mated yeast cells were grown on the highest selective medium QDO/X/A. Mating of BD-53 and AD-T7 were indicated as positive control and mating of BD-LAM and AD-T7, BD-AtPP2-B11, and AD as negative control. b BiFC interaction assay using N. benthamiana leaves. Epidermal cell layers of tobacco leaves were assayed for fluorescence at 2 days after transfection. The YFP fluorescence of the positive control vectors, SPYCE-bZIP63 and SPYNE-bZIP63, was detected only in the nucleus. Bar = 10 μm
A BiFC assay was performed to further confirm the interaction between AtPP2-B11 and AtLEA14 in plants. The SPYCE::AtPP2-B11 and SPYNE::AtLEA14 pair was co-transformed into N. benthamiana leaves, with SPYCE::AtPP2-B11 and SPYNE pairs serving as negative controls, and SPYCE::bZIP63 and SPYNE::bZIP63 as positive controls. The strong YFP fluorescence signals were visible when AtPP2-B11 and AtLEA14 were co-expressed, similar to the positive control. By contrast, the negative controls SPYCE::AtPP2-B11 and SPYNE pairs exhibited no YFP signal (Fig. 6b). The above results indicated that AtPP2-B11 can interact with AtLEA14 in plant cells.
Overexpression of AtPP2-B11 Reduced AtLEA14 Expression at Both Transcript and Protein Levels
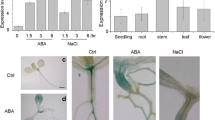
Considering the potential function of AtPP2-B11 as an F-box protein and its possible interaction with AtLEA14, we presumed that transgenic lines overexpressing AtPP2-B11 may exhibit reduced tolerance to drought stress by decreasing the abundance of AtLEA14, which might contribute to cellular dehydration tolerance. Therefore, we tested whether AtPP2-B11 affected AtLEA14 expression under drought condition. Real-time quantitative RT-PCR was firstly performed to detect the transcript levels of AtLEA14 using 2-week-old seedlings of wild-type and transgenic lines OE10 and OE11. As shown in Fig. 7a, the expression levels of AtLEA14 were lower in the OE10 and OE11 lines than that in the wild type under normal growth conditions. After drought treatment, the transgenic lines especially the OE11 line exhibited significantly reduced expression of LEA14 transcripts compared with the wild type, indicating that AtPP2-B11 overexpression markedly decreased the transcript levels of AtLEA14 under drought stress conditions.
AtLEA14 was downregulated in transgenic plants by drought stress at both transcript and protein levels. a AtLEA14 was downregulated in transgenic plants by drought stress at transcript level. Total RNA was prepared from 2-week-old 35S::AtPP2-B11 and wild-type seedlings. Induction patterns of AtLEA14 under normal or drought stress condition were investigated by real-time quantitative RT-PCR. b AtLEA14 was downregulated in transgenic plants by drought stress at protein level. Levels of AtLEA14 in 2-week-old 35S::AtPP2-B11 and wild-type seedlings were determined in the total protein extracts by immunoblot analysis using anti-AtLEA14 antibody (top). Coomassie blue staining was used to confirm equal loading (bottom)
To further determine whether AtPP2-B11 regulated AtLEA14 stability, the protein levels of AtLEA14 under normal and drought stress conditions were analyzed. As shown in Fig. 7b, the AtLEA14 protein levels in transgenic lines were similar to those in the wild type under normal growth conditions. Interestingly, after drought treatment, the AtLEA14 protein levels in the OE11 transgenic line significantly decreased compared with the wild type. These results suggested that AtPP2-B11 might regulate AtLEA14 stability by modulating AtLEA14 abundance at both transcript and protein levels.
Overexpression of AtPP2-B11 Altered Transcript Accumulation of Drought Stress-Induced Genes
Considering that 35S::AtPP2-B11 overexpressing lines altered plant tolerance to drought, the transcription levels of several drought relative marker genes were further investigated, including COR15a, COR47, ERD10, KIN1, RAB18, and RD22. As shown in Fig. 8, under drought stress, the transcript levels of these marker genes were considerably lower in transgenic lines, especially in the OE11 line, than the wild type. These results suggested that AtPP2-B11 may play a key role in drought-response pathway by directly decreasing the AtLEA14 levels and simultaneously affecting the transcriptions of drought-inducible marker genes.
Real-time quantitative RT-PCR analysis of transcript profiles of drought inducible genes. Two-week-old wild-type and 35S::AtPP2-B11 plants were dehydrated for 2 h. Total RNA was obtained from treated plants and analyzed by real-time quantitative RT-PCR using the gene-specific primers listed in Table S1. The graphs indicated the induction fold of the COR15a, COR47, ERD10, KIN1, RAB18, and RD22 in response to drought stress as compared with the control. Mean values from three independent technical replicates were normalized to the levels of an internal control, GAPDH. Error bar indicates SD (n = 3)
Discussion
DRE Motif in the Promoter Region of AtPP2-B11
Cis-acting regulatory elements are important molecular switches involved in the transcriptional regulation of a dynamic network of gene activities that control various biological processes. DRE is a major cis-acting regulatory element in ABA-independent gene expression under abiotic stress conditions (Yamaguchi-Shinozaki and Shinozaki 1994, 2005). Considering the crucial role of DRE in gene stress response, it is urgent to know, in Arabidopsis, weather the DRE motifs in E3 ligase promoters also perform the equivalent function under drought stress. In other words, future studies must focus on determining whether some E3 ligase genes involved in stress response are regulated by the DRE cis-element. In the present study, 23 DRE-containing putative E3 ligases from 1,488 putative E3 encoding genes were identified in the Arabidopsis genome. Semiquantitative RT-PCR analysis revealed that 6 out of the 23 DRE-containing E3s exhibited altered expression levels under drought stress condition. These results raise the possibility of the crucial function of DRE motif in E3 promoter activity, thereby providing new insight into the DRE sequence function under drought stress.
E3 ubiquitin ligases, which are known to have multiple cellular functions, usually function as regulators at the post-translation level through the Ub/26S pathway (Moon et al. 2004). Many isoforms of E3 ligases are involved not only in normal growth and development processes as well as in signaling pathways response to abiotic environmental stresses (Dreher and Callis 2007; Lee and Kim 2011; Smalle and Vierstra 2004; Vierstra 2009). However, the transcriptional regulation of E3 ligase itself remains largely uncharacterised. In the present study, an F-box-containing gene AtPP2-B11 which harbored a DRE motif in the promoter region was identified and the regulation of its expression was examined. Semiquantitative RT-PCR and real-time quantitative RT-PCR analysis suggested that AtPP2-B11 was strongly induced by drought stress (Figs. 1c and 2c). The DRE motif was demonstrated to be critical to the drought-inducible expression of AtPP2-B11 by promoter-GUS analysis (Fig. 2b, e). Therefore, we suggested that a novel ABA-independent pathway may exit via regulating the expression of an F-box gene through DRE motif in its promoter region under drought stress conditions (Fig. 9).
A model of AtPP2-B11 involving in drought stress response by regulating AtLEA14. Within the known ABA-independent pathway, plant tissues respond to drought stress by inducing DREBs and subsequently upregulating downstream drought response genes. In this study, a novel probable drought response pathway was identified. It was possible that DRE motif in the promoter region of AtPP2-B11 contributed to the severe induction under drought stress. AtPP2-B11, a putative F-box protein, may play a negative role in drought tolerance by interacting with AtLEA14 directly and downregulating the AtLEA14 at both transcript and protein levels
AtPP2-B11 Functions as an F-box Protein
Nearly 700 F-box proteins are predicted to be encoded in the Arabidopsis genome (Gagne et al. 2002; Kuroda et al. 2002). This large family reflects the diverse biological functions of F-box proteins, indicating that protein degradation is a prevalent developmental control mechanism in plants. A number of F-box proteins have been shown to be involved in plant development, such as floral development, shoot branching, leaf senescence, root branching, pollen self-incompatibility, as well as signaling transduction pathway of light, circadian clock, and plant hormones (Lechner et al. 2006; McClellan and Chang 2008; Mockaitis and Estelle 2008; Schwechheimer and Willige 2009; Zhang et al. 2008). Functional F-box proteins are usually proven by the interaction assay with ASK genes. Using the yeast two-hybrid system, many uncharacterized F-box proteins are found to interact with ASK homologues, thereby providing evidence that they are components of SCF E3 complexes (Gagne et al. 2002; Risseeuw et al. 2003). In this study, AtPP2-B11 was demonstrated to specifically interact with ASK 7, ASK 18, or ASK 19 proteins in yeast. The interaction of AtPP2-B11 with ASK proteins strongly indicated that AtPP2-B11 functioned as a component of an SCF complex. A conical F-box protein always contained a protein–protein interaction domain besides the F-box domain, such as WD40, LRR, and Kelch-like repeat (Gagne et al. 2002; Jain et al. 2007; Kuroda et al. 2002). A bioinformatics analysis of AtPP2-B11 revealed the presence of a PP2 domain in the C-terminal, a conserved domain in the phloem protein 2 family (Dinant et al. 2003), which can function as a protein–protein interaction domain. Therefore, we concluded that AtPP2-B11 may function as an F-box protein by cooperation of F-box domain and PP2 domain.
AtPP2-B11 Plays a Key Role as a Negative Regulator in the Drought Response Signaling Pathway
Several isoforms of E3 ligases have been characterized and found to be involved in drought response pathways. For example, a U-box-type E3 ubiquitin ligase AtPUB19 negatively regulates ABA and drought responses in Arabidopsis by enhancing ABA-induced stomata closing (Liu et al. 2011). The RING finger E3 ligase RHA2b additively acts with RHA2a in regulating ABA signaling and drought response (Li et al. 2011). The DWD protein DWA3 negatively regulates ABA responses and is involved in protein degradation mediated by CRL4 (Lee et al. 2011). Overexpression of an E3 ubiquitin ligase AtSAP5 containing A20 and AN1 zinc finger motifs leads to increased tolerance to water deficit stress, indicating that AtSAP5 has an important function as a positive regulator of stress responses in Arabidopsis (Kang et al. 2011). Several functional F-box proteins are also reportedly involved in the drought response pathways, including TLP9, DOR, and EDL3 from Arabidopsis (Koops et al. 2011; Lai et al. 2004; Zhang et al. 2008). Although some F-box proteins have been implicated in water-deficit stress response, more F-box genes involved in drought stress needs to be identified. In the present study, we reported the novel F-box gene AtPP2-B11 that has important functions as a negative regulator in drought signaling in Arabidopsis. First, AtPP2-B11 was strongly and rapidly induced by drought stress (Figs. 1c and 2c). Second, overexpressing AtPP2-B11 led to considerable hypersensitivity to drought stress during seed germination and in mature plants (Fig. 3). Third, yeast two-hybrid assay demonstrated that AtPP2-B11 was a component of SCF complex by interacting with ASK 7, ASK 18, and ASK 19, respectively (Fig. 4). Finally, the AtPP2-B11-interacting protein AtLEA14, a downstream regulator involved in drought signaling pathway, was identified (Fig. 6). The transcript and protein levels of AtLEA14 were significantly decreased in the transgenic line OE11 under drought stress conditions (Fig. 7). Some factors could explain the reason that downregulation of AtLEA14 at transcriptional level was much more prominent than that at the protein level. Two hours of drought stress treatment could be enough to exchange the transcriptional level of AtLEA14. However, the accumulation of AtLEA14 in protein level was later than the transcription of AtLEA14, and the degradation rate of mRNA was higher than that of proteins. Moreover, the fact that downregulation of AtLEA14 at the transcriptional level was much more prominent than that at the protein level reflected the diverse of post-transcriptional regulation in plant.
Moreover, to further understand the cellular functions and regulatory mechanism of F-box proteins, it is very important to identify its substrates. To date, the direct targets of F-box proteins involved in drought stress response, such as TLP9, DOR, and EDL3, are unidentified. Compared with these three F-box proteins, AtPP2-B11 played a definite role in drought response and the AtPP2-B11-interacting protein AtLEA14 was identified through a yeast two-hybrid screening. It is the first time to identify a LEA protein interacting with an F-box protein, as LEA proteins are well characterized and have always been related to desiccation tolerance (Hundertmark and Hincha 2008). The detailed relationship between AtPP2-B11 and LEA14 in the regulation of plant response to drought stress needs to be investigated in the future.
Taken together, a model was generated according to our results (Fig. 9). AtPP2-B11 contained a DRE motif in the promoter region and was strongly induced by drought stress. AtPP2-B11 could form an SCF E3 ligase complex by interacting with ASK 7, ASK 18, or ASK 19. Elevated expression of AtPP2-B11 resulted in a decreased in the transcriptional and protein levels of the stress response gene AtLEA14. In addition, AtPP2-B11 overexpression altered the expression of stress-responsive genes, which led to increased sensitivity to drought stress in seedlings and water deficit in mature plants. Overall, the present study indicated that AtPP2-B11, which interacts with LEA14, may play an important role as a negative regulator in the drought response signaling pathway.
References
Agarwal P, Agarwal PK, Nair S, Sopory SK, Reddy MK (2007) Stress-inducible DREB2A transcription factor from Pennisetum glaucum is a phosphoprotein and its phosphorylation negatively regulates its DNA-binding activity. Mol Genet Genomics 277:189–198
Baudry A, Ito S, Song YH, Strait AA, Kiba T, Lu S, Henriques R, Pruneda-Paz JL, Chua NH, Tobin EM, Kay SA, Imaizumi T (2011) F-box proteins FKF1 and LKP2 act in concert with ZEITLUPE to control Arabidopsis clock progression. Plant Cell 22:606–622
Craig KL, Tyers M (1999) The F-box: a new motif for ubiquitin dependent proteolysis in cell cycle regulation and signal transduction. Prog Biophys Mol Biol 72:299–328
Deshaies RJ (1999) SCF and Cullin/Ring H2-based ubiquitin ligases. Annu Rev Cell Dev Biol 15:435–467
Dharmasiri N, Dharmasiri S, Estelle M (2005) The F-box protein TIR1 is an auxin receptor. Nature 435:441–445
Dinant S, Clark AM, Zhu Y, Vilaine F, Palauqui JC, Kusiak C, Thompson GA (2003) Diversity of the superfamily of phloem lectins (phloem protein 2) in angiosperms. Plant Physiol 131:114–128
Dreher K, Callis J (2007) Ubiquitin, hormones and biotic stress in plants. Ann Bot 99:787–822
Earley K, Smith M, Weber R, Gregory B, Poethig R (2010) An endogenous F-box protein regulates ARGONAUTE1 in Arabidopsis thaliana. Silence 1:15
Gagne JM, Downes BP, Shiu SH, Durski AM, Vierstra RD (2002) The F-box subunit of the SCF E3 complex is encoded by a diverse superfamily of genes in Arabidopsis. Proc Natl Acad Sci U S A 99:11519–11524
Glickman MH, Ciechanover A (2002) The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev 82:373–428
Gomez-Porras JL, Riano-Pachon DM, Dreyer I, Mayer JE, Mueller-Roeber B (2007) Genome-wide analysis of ABA-responsive elements ABRE and CE3 reveals divergent patterns in Arabidopsis and rice. BMC Genomics 8:260
Goyal K, Walton LJ, Tunnacliffe A (2005) LEA proteins prevent protein aggregation due to water stress. Biochemical Journal 388:151–157
Gray WM, Kepinski S, Rouse D, Leyser O, Estelle M (2001) Auxin regulates SCF(TIR1)-dependent degradation of AUX/IAA proteins. Nature 414:271–276
Hepworth SR, Klenz JE, Haughn GW (2006) UFO in the Arabidopsis inflorescence apex is required for floral-meristem identity and bract suppression. Planta 223:769–778
Hobo T, Asada M, Kowyama Y, Hattori T (1999) ACGT-containing abscisic acid response element (ABRE) and coupling element 3 (CE3) are functionally equivalent. Plant J 19:679–689
Hundertmark M, Hincha DK (2008) LEA (late embryogenesis abundant) proteins and their encoding genes in Arabidopsis thaliana. BMC Genomics 9:118
Jain M, Nijhawan A, Arora R, Agarwal P, Ray S, Sharma P, Kapoor S, Tyagi AK, Khurana JP (2007) F-box proteins in rice. Genome-wide analysis, classification, temporal and spatial gene expression during panicle and seed development, and regulation by light and abiotic stress. Plant Physiol 143:1467–1483
Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6:3901–3907
Jia F, Wu B, Li H, Huang J, Zheng C (2013) Genome-wide identification and characterisation of F-box family in maize. Mol Genet Genomics 288(11):559–577. doi:10.1007/s00438-013-0769-1
Jiang C, Iu B, Singh J (1996) Requirement of a CCGAC cis-acting element for cold induction of the BN115 gene from winter Brassica napus. Plant Mol Biol 30:679–684
Kang M, Fokar M, Abdelmageed H, Allen RD (2011) Arabidopsis SAP5 functions as a positive regulator of stress responses and exhibits E3 ubiquitin ligase activity. Plant Mol Biol 75:451–466
Koops P, Pelser S, Ignatz M, Klose C, Marrocco-Selden K, Kretsch T (2011) EDL3 is an F-box protein involved in the regulation of abscisic acid signalling in Arabidopsis thaliana. J Exp Bot 62:5547–5560
Kuroda H, Takahashi N, Shimada H, Seki M, Shinozaki K, Matsui M (2002) Classification and expression analysis of Arabidopsis F-box-containing protein genes. Plant Cell Physiol 43:1073–1085
Lai CP, Lee CL, Chen PH, Wu SH, Yang CC, Shaw JF (2004) Molecular analyses of the Arabidopsis TUBBY-like protein gene family. Plant Physiol 134:1586–1597
Laufs P, Coen E, Kronenberger J, Traas J, Doonan J (2003) Separable roles of UFO during floral development revealed by conditional restoration of gene function. Development (Cambridge, England) 130:785–796
Lechner E, Achard P, Vansiri A, Potuschak T, Genschik P (2006) F-box proteins everywhere. Curr Opin Plant Biol 9:631–638
Lee JH, Kim WT (2011) Regulation of abiotic stress signal transduction by E3 ubiquitin ligases in Arabidopsis. Mol Cells 31:201–208
Lee HK, Cho SK, Son O, Xu Z, Hwang I, Kim WT (2009) Drought stress-induced Rma1H1, a RING membrane-anchor E3 ubiquitin ligase homolog, regulates aquaporin levels via ubiquitination in transgenic Arabidopsis plants. Plant Cell 21:622–641
Lee SJ, Kang JY, Park HJ, Kim MD, Bae MS, Choi HI, Kim SY (2010) DREB2C interacts with ABF2, a bZIP protein regulating abscisic acid-responsive gene expression, and its overexpression affects abscisic acid sensitivity. Plant Physiol 153:716–727
Lee JH, Terzaghi W, Deng XW (2011) DWA3, an Arabidopsis DWD protein, acts as a negative regulator in ABA signal transduction. Plant Sci 180:352–357
Levin JZ, Meyerowitz EM (1995) UFO: an Arabidopsis gene involved in both floral meristem and floral organ development. Plant Cell 7:529–548
Li H, Jiang H, Bu Q, Zhao Q, Sun J, Xie Q, Li C (2011) The Arabidopsis RING finger E3 ligase RHA2b acts additively with RHA2a in regulating abscisic acid signaling and drought response. Plant Physiol 156:550–563
Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaguchi-Shinozaki K, Shinozaki K (1998) Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 10:1391–1406
Liu YC, Wu YR, Huang XH, Sun J, Xie Q (2011) AtPUB19, a U-box E3 ubiquitin ligase, negatively regulates abscisic acid and drought responses in Arabidopsis thaliana. Mol Plant 4:938–946
Maruyama K, Todaka D, Mizoi J, Yoshida T, Kidokoro S, Matsukura S, Takasaki H, Sakurai T, Yamamoto YY, Yoshiwara K, Kojima M, Sakakibara H, Shinozaki K, Yamaguchi-Shinozaki K (2012) Identification of cis-acting promoter elements in cold- and dehydration-induced transcriptional pathways in Arabidopsis, rice, and soybean. DNA Res 19:37–49
McClellan CA, Chang C (2008) The role of protein turnover in ethylene biosynthesis and response. Plant Sci 175:24–31
Mittler R (2002) Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci 7:405–410
Mockaitis K, Estelle M (2008) Auxin receptors and plant development: a new signaling paradigm. Annu Rev Cell Dev Biol 24:55–80
Moon J, Parry G, Estelle M (2004) The ubiquitin-proteasome pathway and plant development. Plant Cell 16:3181–3195
Nakashima K, Shinwari ZK, Sakuma Y, Seki M, Miura S, Shinozaki K, Yamaguchi-Shinozaki K (2000) Organization and expression of two Arabidopsis DREB2 genes encoding DRE-binding proteins involved in dehydration- and high-salinity-responsive gene expression. Plant Mol Biol 42:657–665
Nakashima K, Fujita Y, Kanamori N, Katagiri T, Umezawa T, Kidokoro S, Maruyama K, Yoshida T, Ishiyama K, Kobayashi M, Shinozaki K, Yamaguchi-Shinozaki K (2009) Three Arabidopsis SnRK2 protein kinases, SRK2D/SnRK2.2, SRK2E/SnRK2.6/OST1 and SRK2I/SnRK2.3, involved in ABA signaling are essential for the control of seed development and dormancy. Plant Cell Physiol 50:1345–1363
Narusaka Y, Nakashima K, Shinwari ZK, Sakuma Y, Furihata T, Abe H, Narusaka M, Shinozaki K, Yamaguchi-Shinozaki K (2003) Interaction between two cis-acting elements, ABRE and DRE, in ABA-dependent expression of Arabidopsis rd29A gene in response to dehydration and high-salinity stresses. Plant J 34:137–148
Ouellet F, Vazquez-Tello A, Sarhan F (1998) The wheat wcs120 promoter is cold-inducible in both monocotyledonous and dicotyledonous species. FEBS Lett 423:324–328
Risseeuw EP, Daskalchuk TE, Banks TW, Liu E, Cotelesage J, Hellmann H, Estelle M, Somers DE, Crosby WL (2003) Protein interaction analysis of SCF ubiquitin E3 ligase subunits from Arabidopsis. Plant J 34:753–767
Sakuma Y, Liu Q, Dubouzet JG, Abe H, Shinozaki K, Yamaguchi-Shinozaki K (2002) DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem Biophys Res Commun 290:998–1009
Samach A, Klenz JE, Kohalmi SE, Risseeuw E, Haughn GW, Crosby WL (1999) The UNUSUAL FLORAL ORGANS gene of Arabidopsis thaliana is an F-box protein required for normal patterning and growth in the floral meristem. Plant J 20:433–445
Sawa M, Nusinow DA, Kay SA, Imaizumi T (2007) FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science (New York, N.Y 318: 261–265
Schwechheimer C, Willige BC (2009) Shedding light on gibberellic acid signalling. Curr Opin Plant Biol 12:57–62
Sheard LB, Tan X, Mao H, Withers J, Ben-Nissan G, Hinds TR, Kobayashi Y, Hsu FF, Sharon M, Browse J, He SY, Rizo J, Howe GA, Zheng N (2010) Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature 468:400–405
Shen Q, Ho TH (1995) Functional dissection of an abscisic acid (ABA)-inducible gene reveals two independent ABA-responsive complexes each containing a G-box and a novel cis-acting element. Plant Cell 7:295–307
Shinozaki K, Yamaguchi-Shinozaki K (1996) Molecular responses to drought and cold stress. Curr Opin Biotechnol 7:161–167
Shinozaki K, Yamaguchi-Shinozaki K (2007) Gene networks involved in drought stress response and tolerance. J Exp Bot 58:221–227
Shinozaki K, Yamaguchi-Shinozaki K, Seki M (2003) Regulatory network of gene expression in the drought and cold stress responses. Curr Opin Plant Biol 6:410–417
Smalle J, Vierstra RD (2004) The ubiquitin 26S proteasome proteolytic pathway. Annu Rev Plant Biol 55:555–590
Song S, Dai X, Zhang WH (2012) A rice F-box gene, OsFbx352, is involved in glucose-delayed seed germination in rice. J Exp Bot 63:5559–5568
Sun W, Van Montagu M, Verbruggen N (2002) Small heat shock proteins and stress tolerance in plants. Biochim Biophys Acta 1577:1–9
Thines B, Katsir L, Melotto M, Niu Y, Mandaokar A, Liu G, Nomura K, He SY, Howe GA, Browse J (2007) JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 448:661–665
Thomashow MF (1999) PLANT COLD ACCLIMATION: freezing tolerance genes and regulatory mechanisms. Annu Rev Plant Physiol Plant Mol Biol 50:571–599
Vierstra RD (2009) The ubiquitin-26S proteasome system at the nexus of plant biology. Nat Rev Mol Cell Biol 10:385–397
Voinnet O, Rivas S, Mestre P, Baulcombe D (2003) An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J 33:949–956
Walsh TA, Neal R, Merlo AO, Honma M, Hicks GR, Wolff K, Matsumura W, Davies JP (2006) Mutations in an auxin receptor homolog AFB5 and in SGT1b confer resistance to synthetic picolinate auxins and not to 2,4-dichlorophenoxyacetic acid or indole-3-acetic acid in Arabidopsis. Plant Physiol 142:542–552
Walter M, Chaban C, Schutze K, Batistic O, Weckermann K, Nake C, Blazevic D, Grefen C, Schumacher K, Oecking C, Harter K, Kudla J (2004) Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J 40:428–438
Wang S, Yang S, Yin Y, Guo X, Wang S, Hao D (2009a) An in silico strategy identified the target gene candidates regulated by dehydration responsive element binding proteins (DREBs) in Arabidopsis genome. Plant Mol Biol 69:167–178
Wang X, Kong H, Ma H (2009b) F-box proteins regulate ethylene signaling and more. Genes Dev 23:391–396
Xiao W, Jang J (2000) F-box proteins in Arabidopsis. Trends Plant Sci 5:454–457
Yamaguchi-Shinozaki K, Shinozaki K (1994) A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell 6:251–264
Yamaguchi-Shinozaki K, Shinozaki K (2005) Organization of cis-acting regulatory elements in osmotic- and cold-stress-responsive promoters. Trends Plant Sci 10:88–94
Yan YS, Chen XY, Yang K, Sun ZX, Fu YP, Zhang YM, Fang RX (2011) Overexpression of an F-box protein gene reduces abiotic stress tolerance and promotes root growth in rice. Mol Plant 4:190–197
Yoo SD, Cho YH, Sheen J (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2:1565–1572
Yoshiba Y, Kiyosue T, Nakashima K, Yamaguchi-Shinozaki K, Shinozaki K (1997) Regulation of levels of proline as an osmolyte in plants under water stress. Plant Cell Physiol 38:1095–1102
Zhang JZ, Creelman RA, Zhu JK (2004) From laboratory to field. Using information from Arabidopsis to engineer salt, cold, and drought tolerance in crops. Plant Physiol 135:615–621
Zhang W, Ruan J, Ho TH, You Y, Yu T, Quatrano RS (2005) Cis-regulatory element based targeted gene finding: genome-wide identification of abscisic acid- and abiotic stress-responsive genes in Arabidopsis thaliana. Bioinformatics (Oxford, England) 21:3074–3081
Zhang Y, Yang C, Li Y, Zheng N, Chen H, Zhao Q, Gao T, Guo H, Xie Q (2007) SDIR1 is a RING finger E3 ligase that positively regulates stress-responsive abscisic acid signaling in Arabidopsis. Plant Cell 19:1912–1929
Zhang Y, Xu W, Li Z, Deng XW, Wu W, Xue Y (2008) F-box protein DOR functions as a novel inhibitory factor for abscisic acid-induced stomatal closure under drought stress in Arabidopsis. Plant Physiol 148:2121–2133
Zheng X, Miller ND, Lewis DR, Christians MJ, Lee KH, Muday GK, Spalding EP, Vierstra RD (2011) AUXIN UP-REGULATED F-BOX PROTEIN1 regulates the cross talk between auxin transport and cytokinin signaling during plant root growth. Plant Physiol 156:1878–1893
Zhu JK (2002) Salt and drought stress signal transduction in plants. Annu Rev Plant Biol 53:247–273
Acknowledgments
This work was supported by the National Basic Research Program (grant no. 2012CB114200), the National Natural Science Foundation (grant no. 31000121 and 30970225) in China.
Author information
Authors and Affiliations
Corresponding author
Additional information
Yanze Li and Fengjuan Jia contributed equally to this study.
Rights and permissions
About this article
Cite this article
Li, Y., Jia, F., Yu, Y. et al. The SCF E3 Ligase AtPP2-B11 Plays a Negative Role in Response to Drought Stress in Arabidopsis . Plant Mol Biol Rep 32, 943–956 (2014). https://doi.org/10.1007/s11105-014-0705-5
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11105-014-0705-5