Abstract
Superoxide dismutases (SOD) play important roles in plant disease resistance. In this study, a manganese superoxide dismutase gene, designated RsrSOD, was isolated from oil radish (Raphanus sativus var. raphanistroides). RsrSOD was 696 bp in length predicted to encode 231 amino acids. The deduced amino acid sequence contained four putative Mn-binding sites. Under the control of the CaMV35S promoter, RsrSOD was introduced into broccoli (Brassica oleracea var. italica) via Agrobacterium tumefaciens-mediated transformation. Six transgenic lines were obtained out of 35 independent shoots. Both gene expression and enzyme activity of SOD increased significantly in transgenic lines when challenged with Hyaloperonospora parasitica. Three lines, L19, L23, and L25, exhibited the highest resistance against downy mildew with disease symptoms restricted completely. These highly resistant lines would serve as good broccoli breeding materials.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Broccoli (Brassica oleracea var. italica) is an important health-promoting vegetable crop and contains high concentrations of selenium, glucosinolates, flavonoids, hydroxycinnamic acids, carotenoids, tocopherols, ascorbic acid, and vitamin C (Kurilich et al. 1999; Vallejo et al. 2004; Rodriguez-Cantu et al. 2011). Broccoli is a rich source of inducers of enzymes that protect human against chemical carcinogens and has been regarded as a unique vegetable for mammalian heart protection through the redox cycling of the thioredoxin superfamily (Fahey et al. 1997; Mukherjee et al. 2008). Linhai is the main broccoli planting region in China with an average cultivated area of 6,500 hm2 and mean flower head yield of 1.3 × 108 kg per year. However, serious yield loss between 10% and 25% occurred in recent years due to downy mildew (Hyaloperonospora parasitica, formerly Peronospora parasitica) (Coelho and Monteiro 2003). Downy mildew infects cotyledons as well as mature leaves and inflorescences with irregular, yellow to orange necrotic lesions, resulting to quality decline and unmarketable.
Superoxide dismutases (SOD, EC 1.15.1.1) are enzymes that repair cells and reduce cellular damage caused by superoxide via catalyzing the dismutation of superoxide into oxygen and hydrogen peroxide (Dionisi et al. 1975). SODs localize in different cell compartments and play important roles in plant stress defense (Alscher et al. 2002). In higher plants, there are three major forms of SOD isozyme present in cells, grouped by the metal cofactor. CuZn-SOD is present in cytosol, chloroplasts, peroxisomes, and apoplast. Fe-SOD is found both in chloroplasts and peroxisomes, while manganese superoxide dismutase (Mn-SOD) localizes to the mitochondria and peroxisomes (Corpas et al. 2006). Physiological correlations between elevated SOD activity and disease resistance have been reported, suggesting that the upregulation of SOD levels may enhance plant disease resistance, and transgenic plants carrying SOD genes exhibiting increased tolerance to fungus were observed (Sahoo et al. 2007; Tertivanidis et al. 2004). Oil radish (Raphanus sativus var. raphanistroides) is a weed plant distributed mainly in the coastal areas of Japan, Korea, and China (Makino 1904). Oil radish is an excellent gene sources for important traits such as resistance to drought, low temperature, insects as well as plant diseases. In this paper, a Mn-SOD gene, designated RsrSOD, was isolated from oil radish and later introduced into broccoli. Overexpression of RsrSOD in broccoli significantly increased SOD activities and effectively enhanced downy mildew resistance.
Materials and Methods
Plant Materials
Oil radish seedlings were collected from Dachen Islands in Zhejiang Province and transplanted to a greenhouse. Tender leaves were used for DNA and RNA extraction. A broccoli inbred line, Bo113, was selected for genetic transformation.
DNA Isolation, Total RNA Extraction, and cDNA Synthesis
Genomic DNA was isolated using the CTAB method (Doyle and Doyle 1987). The total RNA of oil radish leaves was extracted by Trizol reagent (Gibco BRL, USA) and then treated with DNase (Promega, USA) to remove genomic DNA contamination. The first- and second-strand cDNA were synthesized according to the user manual of SMART™ PCR cDNA Synthesis Kit (Gibco BRL, USA).
RsrSOD Cloning
The complete coding sequence of RsrSOD was amplified using two specific primers (SODP1: 5′-TCTCCCGGGATGGCGATTC-3′; SODP2: 5′-TACGAGCTCATGACTTGCATTCC-3′) according to the SOD gene sequence of R. sativus published in NCBI database. PCR reactions were prepared in 20-μL volumes containing 30-ng template DNA, 25 pmol of each primer, 1.5 U of Taq DNA polymerase, 0.2 μM dNTPs, 50 mM Tris–HCl (pH 8.3), and 2.0 mM MgCl2. The PCR reaction was carried out as follows: 95°C for 5 min; 33 cycles of 95°C for 30 s, 58.2°C for 45 s and 72°C for 90 s; 72°C for 10 min. PCR products were electrophoresed in a 1% agarose gel. The band was excised and purified using QIAquick Gel Extraction Kit (QIAGEN, Germany). The purified PCR products then were ligated into pGEM-T EASY vector (Promega, UK) and sequenced on ABI Prism 3700 DNA Analyzer (PE Applied Biosystems, CA, USA).
Plant Expression Vector Construction
The PCR products were digested with both Sma I and Sac I, and then cloned into pBI121 vector digested by the same enzymes. The constructs were sequenced to verify the inserts at the cloning sites. The recombinant vector was then introduced into Agrobacterium tumefaciens strain LBA4404.
Broccoli Transformation
The pre- and co-culture mediums were Murashige and Skoog (MS) supplemented with 0.02 mg/L of naphthaleneacetic acid (NAA), 4.0 mg/L of 6-benzylaminopurine (6-BA), and 5.0 mg/L of AgNO3. The induction medium was MS plus 0.02 mg/L of NAA, 4.0 mg/L of 6-BA, 4.0 mg/L of AgNO3, and 50.0 mg/L of kanamycin (Km). Shoots were rooted in MS medium containing 0.2 mg/L of NAA and 50.0 mg/L of Km. The pH of the medium was adjusted to 5.8 with 1 N NaOH or HCl, and then autoclaved at 121°C for 20 min. Tender stems from 18-day-old seedlings were sterilized with 0.1% HgCl2 for 10 min. Stem segments of 1.0 cm in length were inoculated with an A. tumefaciens. The cultures were then incubated at 25 ± 2°C under 16-h photoperiod provided by white fluorescent light.
PCR Screening of Transgenic Plants
Total leaf RNA of positive plants was extracted with Trizol reagent, and cDNA was synthesized using SMART™ cDNA Synthesis Kit, following manufacturer’s instructions. Primers SODP3 (5′-CGATTCGTTCCGTAGCCA-3′) and SODP4 (5′-TCACTTGCATTCCTTCTCATAAAC-3′) were designed according to the RsrSOD sequence for both PCR screening of transformed plants and RT-PCR verification. Approximately 30-ng DNA and 45-ng cDNA were employed as PCR templates, respectively. The PCR conditions were as follows: initial denaturation at 95°C for 5 min; 33 cycles of 95°C for 30 s, 54.3°C for 30 s and 72°C for 45 s; extension at 72°C for 10 min.
Disease Assessment
Tissue culture was performed to propagate enough T0 plantlets using stems of six transgenic broccoli lines as explants, then acclimatized and transplanted in a greenhouse, and inoculated with H. parasitica isolate at the three-leaf stage. Disease assessment was done by using a six-point (0, 1, 3, 5, 7, and 9) scale that corresponds to the class of interaction phenotypes (Dickson and Petzoldt 1993) and disease indices were calculated based Williams’ formula (Williams 1985).
Determination of SOD Activity
T1 seeds of six transgenic lines, L08, L11, L19, L23, L25, and L33, were surface-sterilized with 0.1% HgCl2 for 15 min and germinated on MS medium containing 50 mg/L of kanamycin. Ten-day-old seedlings were transferred to soil in plastic pots with the same culture conditions as wild type (WT). Plants at three-leaf stage were challenged with H. parasitica isolate (approximately 1,000 spores/mL), and leaves were collected immediately (day 0) and 5 days after inoculated.
For assay of SOD activity, fresh leaf samples were ground in liquid nitrogen and homogenized in 1.5 mL of extraction buffer. The homogenate was filtered through four layers of cloth and centrifuged at 13,000 g for 20 min at 4°C for determination of SOD enzyme activities. SOD activity was determined as reported (Bayer and Fridovich 1987). Statistical analysis was performed with SPSS 11.5 software package. The data were evaluated by one-way analysis of variance.
Results
Cloning and Characteristics of RsrSOD
Gene-specific primers based on the sequence of R. sativus were designed and employed to amplify an oil radish SOD gene using leaf cDNA as well as DNA as template, respectively. The gene, designated RsrSOD, was isolated, cloned, and sequenced. Nucleotide sequence analysis revealed that the genomic DNA of RsrSOD was 1,302 bp in length with five introns. The RsrSOD gene shares 97% and 88% similarity with Mn-SOD sequences of R. sativus and Arabidopsis thaliana at exon level, respectively. The complete open reading frame is 696 bp in length encoding 231 amino acids. His55, His103, Asp192, and His196 were recognized as putative Mn-binding sites (Fig. 1).
Sequence Comparisons
To compare the predicted amino acid sequence of RsrSOD with those present in the GenBank, ten Mn-SOD sequences were downloaded from NCBI and aligned using ClusterX 1.81 (Fig. 2). The oil radish SOD protein was closely related to Mn-SOD sequences from various plant species. The highest identity was shared with the SOD sequence from R. sativus, and only one residue difference between them was observed with genetic distance of 0.0228 (data not shown). Identity to the remaining Mn-SOD so far studied ranged from 70% (Ginkgo biloba) to 95% (A. thaliana). The genetic distance between oil radish and G. biloba was 0.3151, revealing their distinct relationship.
Cluster analysis was performed by MEGA 3.1 based on the Poisson correction amino acid distance, the complete deletion model, and 1,000 bootstrap replications (Fig. 3). RsrSOD and four SODs from other Cruciferae plants, R. sativus, Brassica juncea, A. thaliana, and Thellungiella halophila, grouped together. Two Gramineae SODs from Oryza sativa and Saccharum officinarum joined together. However, G. biloba SOD was observed outside these groupings.
PCR Detection of Transgenic Plants
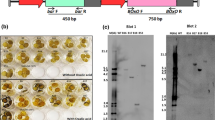
To analyze the potential function in downy mildew resistance, the complete coding region of RsrSOD was introduced into broccoli driven by the CaMV 35S promoter. Altogether, six independent transgenic lines, L08, L11, L19, L23, L25, and L33, were obtained out of 35 regenerated plants by PCR (data not shown). Semiquantitative RT-PCR was performed to detect the expression of RsrSOD in transgenic lines. RsrSOD was expressed in all six lines, and the lowest expression levels were observed in lines of 08 and 11 (Fig. 4).
Assessment of Downy Mildew Resistance
Tissue culture was performed to propagate enough T0 plantlets using stems of six transgenic broccoli lines as explants, then acclimatized and transplanted in a greenhouse, and inoculated with H. parasitica isolate at the three-leaf stage. With the aid of a stereo microscope, flecks and sporulation were checked at magnifications up to × 65 and assigned to reaction phenotypes, and disease indices were calculated (Table 1). All the six lines demonstrated higher resistance against downy mildew than the control plants. The control broccoli exhibited a susceptible reaction, while L19, L23, L25, L11, L33 as well as L08 had the resistance ratings in very resistant (VR), moderately resistant (MR), and low resistance (LR) classes, respectively. Flecks and sporulation were observed on leaves of L08, L11 as well as L33. However, there were some differences between L08, and L11 and L33. The lesions of L11 as well as L33 were smaller and less abundant than L08 (Fig. 5). Semiquantitative RT-PCR was performed to evaluate RsrSOD expression, and higher levels of RsrSOD gene in leaves of broccoli plants challenged with H. parasitica were observed (Fig. 6).
SOD Activity Assay
Challenging with H. parasitica caused increases of SOD activity in transgenic plants as in WT, and the values were higher as compared to control plants after 5 days of inoculation (Fig. 7). The lowest activity of this enzyme was observed in WT, and the highest was found in L25. The mean activity of WT plants was 4.56 U/g FW, while in lines L19, L23, and L25, the values were 15.74, 13.88, and 14.64 U/g FW, respectively. The SOD levels of L19, L23, and L25 were nearly two to threefolds higher than those in control leaves. However, the enzyme activities of L08, L11, and L33 lines increased slightly; their activities were 5.98, 7.30, and 7.69 U/g FW, respectively.
Discussion
Plants are continuously challenged by pathogens, and disease resistance is associated with the activation of a wide range of defense genes that serve to prevent pathogen infection (Feng et al. 2009). Introducing disease-resistant-related genes will provide an alternative approach to improve plant tolerance to disease and minimize yield loss. In recent years, a number of disease-related genes, including antioxidative enzyme, transcriptional factors, and secondary metabolites, have been identified from various plant species such as Musa spp., Zingiber zerumbet, Helianthus annuus, Gossypium barbadense, and Triticum aestivum (Sahoo et al. 2007; Huang et al. 2010; Aswati Nair et al. 2010; Carmen et al. 2011; Meng et al. 2010; Bi et al. 2011; Zhang et al. 2011). Among them, the antioxidative system operates with a sequential and simultaneous action of many enzymes such as SOD, peroxidase, and catalase to overcome excessive production of reactive oxygen species while challenged by pathogens (Mittler 2002; Chen et al. 2011).
SODs are members of a ubiquitous family of metalloenzymes that function to catalyze the dismutation of superoxide anions into hydrogen peroxide and dioxygen (Vajragupta et al. 2003). Mn-SOD exists as a tetramer and is initially synthesized containing a leader peptide, which targets this manganese-containing enzyme exclusively to the mitochondrial spaces (Zelko et al. 2002). Mn-SOD genes were isolated from various plants, including Thallassiosira weissflogii (Ken et al. 2005), Hevea brasiliensis (Miao and Gaynor 1993), A. thaliana (Kliebenstein et al. 1998), H. annuus (Fernández-Ocaña et al. 2011), Nelumbo nucifera (Dong et al. 2009), Prunus persica (Bagnoli et al. 2002), R. sativus (Kwon and An 2003), and Capsicum annuum (Lee et al. 2002).
In this paper, we isolated an Mn-SOD gene from oil raddish which was 1,302 bp in length with five introns. The length of complete coding sequence was the same as RsMnSOD from R. sativus (Kwon and An 2003). Four putative Mn-binding sites and the regions adjacent to them were well conserved (Parker and Blake 1988). RsrSOD was grouped with Cruciferae species from R. sativus, B. juncea, A. thaliana, and T. halophila, while O. sativa and S. officinarum comprised another group, confirming the agreement between traditional taxonomy and molecular evolution (Kliebenstein et al. 1998).
Plant MnSODs were found to be highly inducible by various environmental stresses, including ethylene, salicylic acid, pathogenic infection, sugar, osmotic stress, drought, phytohormones, and water stress (Bowler et al. 1989; Wu et al. 1999; Kwon and An 2003; Brou et al. 2007). Overexpression of MnSOD genes were reported to obtain significant improvements of oxidative stress tolerance in Nicotiana tabacum, Medicago sativa, Zea mays, and O. sativa (Samis et al. 2002; Kingston-Smith and Foyer 2000; Wang et al. 2005). In our study, transgenic broccoli harboring RsrSOD gene exhibited enhanced resistance to downy mildew, and SOD enzyme activity increased when inoculated with H. parasitica. SOD activities in WT, L08, L11, and L33 increased insignificantly or slightly, indicating their weak resistance against downy mildew (Babitha et al. 2002). The control broccoli exhibited a susceptible reaction to downy mildew, while the six transgenic lines had the resistance ratings in VR, MR, and LR classes, respectively. Three transgenic lines, L19, L23, and L25, exhibited the highest resistance against downy mildew, and no visible lesion was observed on leaves. These transgenic broccoli lines would serve as good breeding material for downy mildew resistance.
Abbreviations
- CaMV35S:
-
Cauliflower mosaic virus 35S promoter
- 6-BA:
-
6-Benzylaminopurine
- NAA:
-
1-Naphthaleneacetic acid
- Km:
-
Kanamycin
- RT-PCR:
-
Reverse transcription PCR
- SOD:
-
Superoxide dismutase
- WT:
-
Wild type
Reference
Alscher RG, Erturk N, Heath LS (2002) Role of superoxide dismutases (SODs) in controlling oxidative stress in plants. J Exp Bot 53(372):1331–1341
Aswati Nair R, Kiran AG, Sivakumar KC, Thomas G (2010) Molecular characterization of an oomycete-responsive PR-5 protein gene from Zingiber zerumbet. Plant Mol Biol Rep 28(1):128–135
Babitha MP, Bhat SG, Prakash HS, Shetty HS (2002) Differential induction of superoxide dismutase in downy mildew-resistant and susceptible genotypes of pearl millet. Plant Pathol 51:480–486
Bagnoli F, Giannino D, Caparrini S, Camussi A, Mariotti D, Racchi ML (2002) Molecular cloning, characterisation and expression of a manganese superoxide dismutase gene from peach (Prunus persica [L.] Batsch). Mol Genet Genomics 267(3):321–328
Bayer WF, Fridovich I (1987) Assaying for superoxide dismutase activity: some large consequences of minor changes in condition. Annals Biochem 161:559–566
Bi C, Chen F, Jackson L, Gill BS, Li WL (2011) Expression of lignin biosynthetic genes in wheat during development and upon infection by fungal pathogens. Plant Mol Biol Rep 29(1):149–161
Bowler C, Alliotte T, De Loose M, Van Montagu M, Inze D (1989) The induction of manganese superoxide dismutase in response to stress in Nicotiana plumbaginifolia. EMBO J 8:31–38
Brou YC, Zézé A, Diouf O, Eyletters M (2007) Water stress induces overexpression of superoxide dismutases that contribute to the protection of cowpea plants against oxidative stress. Afr J Biotechnol 6(17):1982–1986
Carmen M, Jens HK, Claudio C (2011) Analysis of differential transcript expression reveals time-dependent leaf responses to Sclerotinia sclerotiorum in wild and cultivated sunflower. Plant Mol Biol Rep 29(3):597–608
Chen XP, Wang ML, Holbrook C, Culbreath A, Liang XQ, Brenneman T, Guo BZ (2011) Identification and characterization of a multigene family encoding germin-like proteins in cultivated peanut (Arachis hypogaea L.). Plant Mol Biol Rep 29(2):389–403
Coelho PS, Monteiro AA (2003) Inheritance of downy mildew resistance in mature broccoli plants. Euphytica 131(1):65–69
Corpas FJ, Fernández-Ocaña A, Carreras A, Valderrama R, Luque F, Esteban FJ, Rodríguez-Serrano M, Chaki M, Pedrajas JR, Sandalio LM, del Río LA, Barroso JB (2006) The expression of different superoxide dismutase forms is cell-type dependent in olive (Olea europaea L.) leaves. Plant Cell Physiol 47(7):984–994
Dickson MH, Petzoldt R (1993) Plant age and isolate source affect expression of downy mildew resistance in broccoli. HortSci 28(7):730–731
Dionisi O, Galeotti T, Terranova T, Azzi A (1975) Superoxide radicals and hydrogen peroxide formation in mitochondria from normal and neoplastic tissues. Biochim Biophys Acta 403(2):292–300
Dong C, Li G, Li Z, Zhu H, Zhou M, Hu Z (2009) Molecular cloning and expression analysis of an Mn-SOD gene from Nelumbo nucifera. Appl Biochem Biotechnol 58(3):605–614
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19:11–15
Fahey JW, Zhang Y, Talalay P (1997) Broccoli sprouts: an exceptionally rich source of inducers of enzymes that protect against chemical carcinogens. Proc Natl Acad Sci U S A 94(19):10367–10372
Feng DS, Li Y, Wang HG, Li XF, Gao JR (2009) Isolation and evolution mode analysis of NBS-LRR resistance gene analogs from hexaploid wheat. Plant Mol Biol Rep 27(3):266–274
Fernández-Ocaña A, Chaki M, Luque F, Gómez-Rodríguez MV, Carreras A, Valderrama R, Begara-Morales JC, Hernández LE, Corpas FJ, Barroso JB (2011) Functional analysis of superoxide dismutases (SODs) in sunflower under biotic and abiotic stress conditions. Identification of two new genes of mitochondrial Mn-SOD. J Plant Physiol 168(11):1303–1308
Huang X, Lu XY, Zhao JT, Chen JK, Dai XM, Xiao W, Chen YP, Chen YF, Huang XL (2010) MaSERK1 gene expression associated with somatic embryogenic competence and disease resistance response in banana (Musa spp.). Plant Mol Biol Rep 28(2):309–316
Ken CF, Hsiung TM, Huang ZX, Juang RH, Lin CT (2005) Characterization of Fe/Mn-superoxide dismutase from diatom Thallassiosira weissflogii: cloning, expression, and property. J Agric Food Chem 53(5):1470–1474
Kingston-Smith AH, Foyer CH (2000) Overexpression of Mn-superoxide dismutase in maize leaves leads to increased monodehydroascorbate reductase, dehydroascorbate reductase and glutathione reductase activities. J Exp Bot 51(352):1867–1877
Kliebenstein DJ, Monde RA, Last RL (1998) Superoxide dismutase in Arabidopsis: an eclectic enzyme family with disparate regulation and protein localization. Plant Physiol 118:637–650
Kurilich AC, Tsau GJ, Brown A, Howard L, Klein BP, Jeffery EH, Kushad M, Wallig MA, Juvik JA (1999) Carotene, tocopherol, and ascorbate contents in subspecies of Brassica oleracea. J Agric Food Chem 47(4):1576–1581
Kwon SI, An CS (2003) Cloning and expression of mitochondrial MnSOD from the small radish (Raphanus sativus L.). Mol Cells 16(2):194–200
Lee JH, Yoon YH, Kim HY, Shin DH, Kim DU, Lee IJ, Kim KU (2002) Cloning and expression of squalene synthase cDNA from hot pepper (Capsicum annuum L.). Mol Cells 13(3):436–443
Makino T (1904) Observations on the flora of Japan. Bot Mag Tokyo 18:31–35
Meng XP, Li FG, Liu CL, Zhang CJ, Wu ZX, Chen YJ (2010) Isolation and characterization of an ERF transcription factor gene from cotton (Gossypium barbadense L.). Plant Mol Biol Rep 28:176–183
Miao Z, Gaynor JJ (1993) Molecular cloning, characterization and expression of Mn-superoxide dismutase from the rubber tree (Hevea brasiliensis). Plant Mol Biol 23(2):267–277
Mittler R (2002) Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci 7(9):405–410
Mukherjee S, Gangopadhyay H, Das DK (2008) Broccoli: a unique vegetable that protects mammalian hearts through the redox cycling of the thioredoxin superfamily. J Agr Food Chem 56(2):609–617
Parker MW, Blake CCF (1988) Crystal structure of manganese superoxide dismutase from Bacillus stearothermophilus at 2.4 Å resolution. J Mol Biol 199:649–661
Rodriguez-Cantu LN, Gutierrez-Uribe JA, Arriola-Vucovich J, Diaz-De La Garza RI, Fahey JW, Serna-Saldivar SO (2011) Broccoli (Brassica oleracea var. italica) sprouts and extracts rich in glucosinolates and isothiocyanates affect cholesterol metabolism and genes involved in lipid homeostasis in hamsters. J Agric Food Chem 59(4):1095–1103
Sahoo MR, DasGupta M, Kole PC, Bhat JS, Mukherjee A (2007) Antioxidative enzymes and isozymes analysis of taro genotypes and their implications in Phytophthora blight disease resistance. Mycopathologia 163(4):241–248
Samis K, Bowley S, McKersie B (2002) Pyramiding Mn-superoxide dismutase transgenes to improve persistence and biomass production in alfalfa. J Exp Bot 53(372):1343–1350
Tertivanidis K, Goudoula C, Vasilikiotis C, Hassiotou E, Perl-Treves R, Tsaftaris A (2004) Superoxide dismutase transgenes in sugarbeets confer resistance to oxidative agents and the fungus C. beticola. Transgenic Res 13(3):225–233
Vajragupta O, Boonchoong P, Sumanont Y, Watanabe H, Wongkrajang Y, Kammasud N (2003) Manganese-based complexes of radical scavengers as neuroprotective agents. Bioorg Med Chem 11(10):2329–2337
Vallejo F, Tomas-Barberan FA, Ferreres F (2004) Characterisation of flavonols in broccoli (Brassica oleracea L. var. italica) by liquid chromatography-UV diode-array detection-electrospray ionisation mass spectrometry. J Chromatogr A 1054(1–2):181–193
Wang FZ, Wang QB, Kwon SY, Kwak SS, Su WA (2005) Enhanced drought tolerance of transgenic rice plants expressing a pea manganese superoxide dismutase. J Plant Physiol 162(4):465–472
Williams PH (1985) Crucifer genetics cooperative resource book. University of Wisconsin, Madison, pp 5–7
Wu G, Wilen RW, Robertson AJ, Gusta LV (1999) Isolation, chromosomal localization, and differential expression of mitochondrial manganese superoxide dismutase and chloroplastic copper/zinc superoxide dismutase genes in wheat. Plant Physiol 120(2):513–520
Zelko IN, Mariani TJ, Folz RJ (2002) Superoxide dismutase multigene family: a comparison of the CuZn-SOD (SOD1), Mn-SOD (SOD2), and EC-SOD (SOD3) gene structures, evolution, and expression. Free Radic Biol Med 33(3):337–349
Zhang H, Hu YG, Wang CY, Ji WQ (2011) Gene expression in wheat induced by inoculation with Puccinia striiformis West. Plant Mol Biol Rep 29(2):458–465
Acknowledgments
This study was supported by Zhejiang Provincial Natural Science Foundation of China (no. Y3080081) and Taizhou Science and Technology Project (no. 08XH02).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Jiang, M., Miao, Lx. & He, C. Overexpression of an Oil Radish Superoxide Dismutase Gene in Broccoli Confers Resistance to Downy Mildew. Plant Mol Biol Rep 30, 966–972 (2012). https://doi.org/10.1007/s11105-011-0407-1
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11105-011-0407-1