Abstract
As a major component of the cell wall, lignin plays an important role in plant development and defense response to pathogens, but negatively impacts biomass processability for biofuels. Silencing the target lignin genes for greater biomass processability should not significantly affect plant development and biomass yield but also must not compromise disease resistance. Here, we report experiments to identify a set of lignin genes that may be silenced without compromising disease resistance. We profiled the expression of 32 lignin biosynthetic candidate genes by qRT-PCR in 17 wheat tissues collected at three developmental stages. Twenty-one genes were expressed at a much higher level in stems compared to sheaths and leaf blades. Expression of seven these genes significantly correlated with lignin content. The co-expression patterns indicated that these 21 genes are under strong developmental regulation and may play a role in lignin biosynthesis. Profiling gene expression of same tissues challenged by two fungal pathogens, Fusarium graminearum and Puccina triticina indicated that expression of 17 genes was induced by F. graminearum. Only PAL1, a non-developmental-regulated gene, was induced by P. triticina. Thus, lignin biosynthetic pathway overlaps defense response to F. graminearum. Based on these criteria, 17 genes, F5H1, F5H2, 4CL2, CCR2, COMT1, and COMT2 in particular that do not overlap with disease resistance pathway, may be the targets for downregulation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Lignin, a complex phenylpropanoid-derived polymer, is mainly present in the secondary thickened cell walls of plants and plays an important role in development, growth, and resistance to biotic and abiotic stress. However, when it comes to industrial uses of plant cell wall materials, lignin is negatively associated with biomass quality for forage, paper, and biofuels.
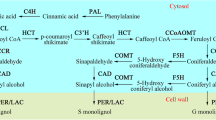
Based on the methoxylation level, three major monolignols (lignin precursors) have been recognized: non-methoxylated p-hydroxylphenyl (H), mono-methoxylated guaiacyl (G), and di-methoxylated syringyl (S) units. These monolignols are synthesized via phenylpropanoid pathway. The monolignol pathway has been well documented in the model plant Arabidopsis thaliana by induction and characterization of the knockout mutants (for review, see Humphreys and Chapple, 2002; Raes et al., 2003) and in dicot crops such as alfalfa, tobacco, and aspen by gene silencing (for review, see Li et al., 2008). Enzymes upstream of this pathway, including phenylalanine ammonia lyase (PAL), cinnamate 4-hydroxlase (C4H), and 4-coumarate CoA ligase (4CL), are also involved in the biosynthesis of other phenylpropanoids such as flavonoids, coumarins, and stilbenes. Most enzymes of this pathway exist as multiple forms of isoenzymes encoded by gene families with diversified members (Costa et al., 2003; Raes et al., 2003; Tuskan et al., 2006, Sibout et al., 2005) and metabolite fluxes among different branches are channeled (for review, see Winkel, 2004). In addition to a feed-back loop at the entry point of the general phenylpropanoid pathway (Blount et al., 2000), expression of many genes for monolignol synthesis is tightly and coordinately regulated at the transcriptional level (for review, see Rogers and Campbell, 2004). An important feature of this orchestral regulation is the presence of the AC elements (or H-boxes) that are rich in A and C in the promoter regions of lignin biosynthetic genes. These cis elements are thought to enhance expression of the genes in xylem and to prevent their expression in the adjacent phloem and cortical cells.
In Arabidopsis, one or more AC elements were found in the promoters of PAL, 4CL, C4H, p-coumaroyl shikimate/quinate 3′-hydroxylase (C3H), caffeoyl CoA O-methyltransferase (CCoAOMT), cinnamoyl CoA reductase (CCR), and cinnamyl alcohol dehydrogenase (CAD) (Mizutani et al., 1997; Raes et al., 2003). Upstream in the cascade of transcriptional factors, several secondary cell wall NAC domain proteins, including NST1, NST2, SND1, VNP6, and VNP7 play important roles in secondary cell wall thickening and lignification (for review, see Zhong and Ye, 2007). These secondary wall-associated NACs regulate secondary wall biosynthesis by targeting other transcription factors such as MYB46, MYB103, SND3, and KNT7 (Zhong et al., 2008). NST1 and NST2 are in turn regulated by MYB26 (Yang et al., 2007). All these studies indicate that lignin deposition and monolignol biosynthesis is tightly regulated during secondary wall development. This in turn suggests a possibility to identify lignin biosynthetic genes based on co-expression patterns of the candidate genes at different developmental stages and/or in tissues differing significantly in lignin content at the same developmental stage.
In contrast to the type-I cell walls of dicots and non-commelinoid monocots, the type-II cell walls of the commelinoid monocots including grasses have lower levels of pectin and higher content of phenylpropanoids (Carpita, 1996). The understanding of the monolignol pathway in grasses is still limited despite the long history of research. Early investigations of lignin in grasses dealt with the induction of the brown midrib (bm) mutants in maize (Jorgensen, 1931; Kuc and Nelson, 1964) and (bmr) sorghum (Porter et al., 1978) and gold hull (gh) and internode mutants in rice (Nagao and Takahashi, 1963; Iwata and Omura, 1971; 1977). Recently, molecular characterization of these mutants demonstrated that bm1 of maize (Halpin et al., 1998), gh2 of rice (Zhang et al., 2006) and bmr6 of sorghum (Sattler et al., 2009; Saballos et al., 2009) were caused by mutations in CAD. The bm3 of maize was caused by a mutation in caffeic acid/5-hydroxyferulic acid O-methyltransferase (COMT) gene (Vignols et al., 1995). The completion of the rice genome sequence and large collections of expressed sequence tags (ESTs) in other grasses have facilitated the documentation of the genes in the lignin biosynthetic pathway and their role in type-II cell wall development. The structural genes of the lignin biosynthetic pathway identified in Arabidopsis are also present in the genomes of rice (Yokoyama and Nishitani, 2004), maize (Guillaumie et al., 2007) and wheat (the present research).
Grasses, including important forage and cereal crops predominate the earth and are the most abundant resource of potential feedstocks for conversion into biofuels. Lignin is a major recalcitrant factor for biomass processing by limiting the accessibility of hydrolytic enzymes to cellulose. The downregulation of lignin genes can significantly improve the biomass processability (Chen and Dixon, 2007). However, as demonstrated in Arabidopsis, most gene families are composed of multiple members with diverse functions. As a result, not all the members in a family are involved in lignin synthesis. At the same time, target genes for RNA silencing or mutagenesis must be carefully selected because some genes in lignin biosynthetic pathway also play an important role in defense response and their silencing must not compromise disease resistance. Similarly, silencing of the target genes should not significantly affect plant development and growth (biomass yield). As part of the effort to dissect the lignin biosynthetic pathway in grasses, we identified the candidate genes in wheat (Triticum aestivum L.) by database mining. Here, we report studies on expression profiling of candidate genes at different developmental stages and in different tissues at the same developmental stage in wheat using quantitative real-time polymerase chain reaction (qRT-PCR).
Expression patterns of the lignin biosynthetic pathway candidate genes were also investigated in tissues challenged by fungal pathogens Fusarium graminearum Schwabe (the wheat head blight pathogen) and Puccinia triticina Eriks (the wheat leaf rust pathogen) to evaluate the role of these genes in plant defense response. These results provide a base for further investigation on lignin biosynthesis and selection of targets for lignin modification in wheat and, possibly, other grasses as well.
Materials and Methods
Database Mining for Lignin Candidate Genes
Over one million wheat ESTs are deposited in NCBI (http://www.ncbi.nlm.nih.gov/dbEST/dbEST_summary.html) and assembled into 216,452 unigenes (91,464 tentative consensuses (TCs), 124,732 singleton ESTs, and 256 singleton mature transcripts) in the wheat gene index database TaGI (release 11.0, http://compbio.dfci.harvard.edu/tgi/cgi-bin/tgi/gimain.pl?gudb=wheat). TaGI releases 10.0 and 11.0 were searched by key words for the monolignol gene families except for those of C3H and hydroxycinnamoyl CoA:shikimate/quinate hydroxycinnamoyltransferase (HCT) that were searched by tBLAST using sequences of Arabidopsis protein CYP98A3 (Franke et al., 2002, GenPept accession: NP_850337) and tobacco HCT (Hoffmann et al., 2003, GenPept accession: CAD47830) as queries. The retrieved unigene sequences were grouped based on UniRef100 cluster (Suzek et al., 2007) accession numbers and representative TCs of major clusters with high coding region coverage (over 70% for most gene families) were used to design qRT-PCR primers.
Plant Material and Treatments
Wheat inbred line HR58, derived from a cross between wheat varieties Sumai3 and Stoa (Pumphrey et al., 2007), was used for this research. HR58 is a fast-growing genotype with a date-to-heading of 40 days. All the plants were grown in the greenhouse at 20-25°C and day-length period of minimum 16 h. Wheat plants at Feekes’ stages 2, 6, 10, and 11 were analyzed (Fig. 1; Large, 1954). Seven tissues were sampled at three developmental stages, i.e., whole shoot at Feekes’ stage 2 (2); leaf blade (6b) and leaf sheath (6sh) at Feekes’ stage 6; and flag leaf blade (10b), flag leaf sheath (10sh), inflorescence (10in), and stem (10st) at Feekes’ stage 10. At Feekes’ stage 10, the first (F-1st), second (F-2st), and third (F-3st) internode of the main stem under the flag leaf along with their leaf blades (F-1b, F-2b and F-3b) and leaf sheaths (F-1sh, F-2sh and F-3sh) were also collected to investigate tissue specificity of the expression profile of the lignin pathway genes (Fig. 2). At Feekes’ stage 10, the first internode (F-1st) of the main stem under the flag leaf was divided into lower half (F-1st-low) and upper half (F-1st-up) because the texture of the former was significantly tender as compared to the latter.
Developmental stages of wheat at which lignin biosynthetic expression was profiled and lignin content was measured. a Feekes’ stage 2.0 (tillering initiated); b Feekes’ stage 6.0 (the first node visible); c Feekes’ stage 10.0 (heads emerging through flag leaf sheaths) and D) Feekes’ stage 11.4 (matured). Figure according to Large (1954)
A wheat plant illustrating the tissues collected for lignin and gene expression studies. These include spike (SPK, rachis + glumes + awns), peduncle (PDC), flag leaf blade(F-b), flag leaf sheath (F-sh), blade (F-1b) and sheath (F-1sh) of the first leaf under flag, the first internode under flag (F-1st), blade (F-2b) and sheath (F-2sh) of the second leaf under flag, the second internode under flag (F-2st), blade (F-3b) and sheath (F-3sh) of the third leaf under flag, and the third internode under flag (F-3st)
Three independent biological replicates were performed for scab and leaf rust experiments. For scab infection, F. graminearum isolate GZ3639 was used. Inoculation of HR58 with isolate GZ3639 was performed according to Li et al. (2001). For each treatment, three spikes were inoculated. The inoculated spikes were wrapped with a moist plastic bag to maintain high humidity and placed in the greenhouse with temperature set at 22-26°C. The mock-inoculated spikes were used as controls. Spikelets of treated and control plants were collected 24 and 48 h post inoculation (HPI).
For leaf rust infection, P. triticina isolate PRTUS6 was used. For each treatment, HR58 wheat plants grown in 4-in pots filled with potting soil (16 plants per pot) at the Feekes’ stage 2 were inoculated with urediospore suspension in light mineral oil. The control plants were sprayed with mineral oil only. The inoculated plants were incubated overnight in a mist chamber (100% relative humidity) and then transferred to the greenhouse. Leaf tissues were sampled at 24 and 48 HPI.
cDNA Synthesis and qRT-PCR
Gene-specific primers with Tm ranging from 50°C to 60°C and amplifying PCR products ranging from 75 to 200 bp in length were designed using Beacon Designer software (PREMIER Biosoft International, Palo Alto, CA, USA). All the designed primers were tested for full efficiency before use for expression profiling of target genes.
For all tissues tested, at least two biological replicates were pooled for RNA isolation. Total RNA was isolated using Trizol™ Reagent (Invitrogen, Carlsbad, CA, USA) and purified using RNeasy Mini Kit (Qiagen, Valencia, CA, USA). The concentration and quality of isolated RNA was determined by NanoDrop 1000 Spectrophotometer (Thermo Scientific, Wilmington, DE, USA) and 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). The first strand cDNA was synthesized using the Superscript® III Reverse Transcriptase (Invitrogen) following the manufacturer’s instructions and used as a template for qRT-PCR. The qRT-PCR assays were performed on the iCycler iQ Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA) by using the SyberGreen MasterMix (Applied Biosystems, Foster City, CA, USA). The wheat housekeeping gene Actin (GenBank accession CV778724) was used as a reference to normalize the cDNA concentration among all samples and to compare data from different PCR runs. The constitutive expression of reference gene (Actin) in different wheat tissues was validated as described in Czechowski et al. (2005). A pBluescript plasmid LjNEST3f1 containing the Lotus japonicus LjLb2 full-length cDNA sequence (GenBank accession: BI416412) was obtained from Dr. Michael Udvardi (The Samuel R. Noble Foundation, Ardmore, OK, USA) and in vitro transcribed using the T7 MegaScript Kit (Ambion, Austin, TX, USA). Five microgram of total RNA was mixed with 30 pg of LjLb2 mRNA before use in a reverse transcription experiment. The resultant cDNA was subjected to qRT-PCR analysis of Actin against LjLb2.
Acetyl Bromide Soluble Lignin Measurements
Acetyl bromide soluble lignin (ABSL) was determined using the method of Fukushima and Hatfield (2001) except 30 mg of cell wall material was used. An extinction coefficient of 17.2 was used for calculating ABSL. For the first experiment, three replicate samples were analyzed for each tissue; for the second experiment, more than three biological replicates were pooled for analysis.
Data Normalization and Statistical Analyses
All the genes were analyzed by qRT-PCR in triplicate and the mean values were used in subsequent calculations. Relative transcription levels among the tissues were determined by 2−−ΔΔCt method as described by Livak and Schmittgen (2001). Correlation coefficients (r) between the lignin content and expression levels and their significance probabilities were estimated using a web-based test service (http://www.stattucino.com/calculate/correlations.html). For significance test, t value was calculated for each r according to the formula t = r/sqrt[(1 − r 2)/(N − 2)], where N is sample size. For cluster analysis, dendrogram was created by DendroUPGMA (http://www.tinet.org/∼debb/UPGMA) that uses Pearson coefficients and the UPGMA algorithm and displayed by TreeViewX (Page, 1996; http://darwin.zoology.gla.ac.uk/∼rpage/treeviewx/).
Results
Lignin Content among Tissues at Different Developmental and Growth Stages
The wheat tissues collected for lignin content and gene expression studies are illustrated in Fig. 2. ABSL lignin content (Table 1) was measured for the tissues collected at Feekes’ stages 2, 6, 10.0, and 11.4 (Fig. 1). Because the tissues collected at different developmental stages were exposed to different environments, comparison should be made between different tissues at the same developmental stage rather than between the same tissue types at different developmental stages. Among the different tissues at the same development stage, lignin content was highest in stem, next highest in the sheath and lowest in leaf blade (Table 1, Fig. 3). This pattern is consistent in Feekes’ stage 6.0, 10.0, and 11.4. At Feekes’ stage 11.4, wheat plant is fully mature and has golden hue following senescence (Fig. 1). At this stage, lignin content was the highest in stem at 24.7%, followed by inflorescence (peduncle + rachis) at 22.8%, sheath at 16.8%, and leaf blade at 14.9% (Table 1).
Charts showing lignin content (%) of same tissues of different age at Feekes’ stage 10.0. Top lignin content of flag leaf blade (F-b), and blade of the first (F-1b), second (F-2b) and third (F-3b) leaf under flag; middle: sheath of flag leaf (F-sh), and sheath of the first (F-1sh), second (F-2sh) and third (F-3sh) leaf under flag; bottom: lower half (F-1st-low) and upper half (F-1st-up) of the first internode under the flag, and the second (F-2st) and third (F-3st) internode under the flag
Among the stem tissues of different age at the same growth stage, lignin content variation was also observed. At Feekes stage 10, stem internode 3 represents the oldest tissue and has lignin content of 20.7% as compared to 16% in the tender F-1st-low (Fig. 3 bottom). However, unlike stem internodes, lignin content in leaf and sheath was the highest in younger tissues at the top than the older tissues at the basal position (Fig. 3 top and middle). Lignin content increased by one third in the flag leaf (topmost and also the youngest in age) as compared to the oldest leaf number 3 at Feekes’ stage 10 (Fig. 3 top).
Selection of Candidate Genes by Mining Wheat EST Database
Searches of TaGI retrieved 651 unigene sequences for the ten lignin gene families, which were organized into 116 UniRef100 clusters. Except for ferulate 5-hydroxylase (F5H), 71 TCs of 31 UniRef100 clusters had over 70% coverage of the coding regions. After merging the highly homologous unigenes with over 96% of identity, the unique sequences were used for designing qRT-PCR primers. Primers were also designed for two unigenes for F5H with low coverage of the coding regions. A total of 82 pairs of qRT-PCR primers were designed and 32 pairs showed high PCR efficiency. The primer sequences, corresponding TCs/ESTs and Tm are listed in Table 2.
Expression Profile at the Different Developmental Stages and in Different Tissues
Because lignin deposition is largely developmentally regulated, it is expected that the genes involved in lignin biosynthesis will also be developmentally regulated and show the same or similar expression pattern. To investigate the developmental regulation patterns of genes, 17 samples collected at three Feekes’ stages (see Materials and Methods) were investigated by qRT-PCR. As the first step of qRT-PCR analysis, we conducted reference validation using LjLb2 as internal control. In the first experiment, the Actin expression level was similar in samples 2, 6b, 6sh, 10in, and 10st, but approximately 2.5-fold lower in 10b and 10sh. In the second experiment at Feekes’ stage 10, Actin transcription level was very similar in most tissues except for F-1st-low (the highest) and F-3st (the lowest). Actin expression level in F-1st-low was 2.5-fold higher as compared to F-3st. The Actin expression level was adjusted accordingly to make it virtually constitutive. The adjusted Actin expression was used to normalize lignin gene expression. For Feekes’ stage 10.0, although we sampled the tissues in different ways from two different batches of plants in experiments 1 and 2, high correlation was observed for gene expression in the stem (r = 0.80199 to 0.93078, P < 1.234e-07) and leaf samples (r = 0.73478 to 0.89097, P< 3.001e-06) between these experiments, indicating a high reproducibility for these tissues and effectiveness of experimental error control.
Transcription profiling by qRT-PCR revealed over twofold variation for many of the 32 genes among the different tissue samples (Tables 3 and 4). Based on expression patterns, these 32 genes can be divided into three groups as illustrated in Fig. 4. The first group consists of 9 genes, 4CL1, C4H, C3H1, CAD4, CCR2, F5H1, F5H2, PAL6, and PAL8. The expression pattern of the group 1 genes parallels the variation in the lignin content. Their transcription levels increase in the same tissue type as developmental stages proceed (Table 3). At the same developmental stage (Feekes’ stage 10.0), they are expressed at much higher levels in stems compared to the inflorescence, sheath, and leaf blade (Table 3). The inflorescence and sheath rank number two and three in expression levels and leaf blades have the lowest expression. This pattern of expression modulates lignin content variation. At Feekes’ stage 10.0, the expression of the nine genes within this group is highly correlated among all members (r > 0.9101, P < 0.000128, Supplementary Tables s1). Statistically significant correlation was observed between lignin content and expression levels for seven of the nine genes (r > 0.5652, P < 0.0443105). The remaining two, CAD4 and PAL8, showed positive correlation with lignin content that was not statistically significant (r > 0.5285, P < 0.058134).
The second group contains 12 genes as shown in Tables 3 and 4 and Fig. 4. Their expression was also highly correlated among their members (r > 0.8512, P < 0.0008925, Supplementary Tables s1). Similar to group 1 genes, group 2 genes are expressed at higher levels in stem as compared to leaf blade and leaf sheath. Unlike the group 1 genes, the group 2 genes are expressed highest in F-1st-low (younger stem tissue half) and also significantly higher than in F-1st-up (older stem half). This specific pattern was also observed in four genes (CAD1, PAL3, PAL4, and PAL5) of group 3 as shown in Fig. 4, where they are close to the group 2. However, these four genes were placed in group 3 because they are expressed at lower levels in other parts of the stem as compared to sheath and leaf blade. Within the group 1, only CAD4 and PAL8 were expressed approximately twofold higher in F-1st-low than in F-1st-up. Compared to F-1st-up, F-1st-low is soft and elongating at Feekes’ stage 10.0. This may indicate that the group 2 genes play an important role in the early stages of the secondary wall development, and the group 1 genes function throughout the whole process of secondary wall thickening.
The third group includes the remaining 11 genes (Tables 3 and 4). They either did not show any specific pattern related to development and lignin content (CAD1, CAD2, CAD3, CCR1, CCR4, PAL2, PAL3, PAL4, and PAL5) or were expressed at much higher levels in leaf and sheath as compared to those in stem (CCoAOMT2 and PAL1).
In hexaploid wheat, each enzyme in the monolignol pathway is encoded by three or more members but only one or two genes always predominate in a specific tissue or a developmental stage. Generally, the expression levels of group 1 genes were higher than group 2, and group 2 genes were higher than that of the group 3 genes (Supplementary Table 2s). One exception to this is CAD2, which is a group 2 gene but showed highest expression level in all the tissues as compared to other CAD members. The variation in expression levels was also observed among members within the same group. For example, in the PAL family in the stem, the abundance of PAL8 transcript was about threefold to sevenfold higher than PAL6 across the tissues (Supplementary Table 2s). In the CCoAOMT family, all tissues at Feekes’ stage 10 showed the same order of relative expression, i.e., CCoAOMT1> CCoAOMT4 > CCoAOMT5 > CCoAOMT3. Similarly, 4CL1, F5H2, and HCT1 were always expressed more than 4CL2, F5H1, and HCT2 (Supplementary Table 2s). Most probably, 4CL1, CCoAOMT1 and CCoAOMT4, F5H2, HCT1, and PAL8 play a greater role in lignin biosynthesis than their other homologues.
Induced Expression of Lignin Biosynthetic Genes by Fungal Pathogens
Reinforcement of the cell wall by lignin formation is an important component of the plant defense response to pathogen invasion. Many of the lignin biosynthetic genes are induced by fungal pathogens. We comparatively investigated the expression of 32 candidate genes in wheat leaves infected by P. triticina and spikelets infected by F. graminearum and mock-inoculated controls. Of the 32 genes studied, only PAL1 expression was induced by more than threefold by P. triticina infection 24 HPI and 3.8-fold 48 HPI; no significant change was observed for the remaining 31 genes (Table 5). Because PAL1 is a group 3 gene and may not be involved lignin biosynthesis, therefore, lignin deposition may not be required for defense against P. triticina.
Unlike the response to P. triticina, the expression of 17 genes (4CL1, C3H1, C4H1, CAD1, CAD4, CCoAOMT1, CCoAOMT3, CCoAOMT5,CCR3, HCT1, HCT2, PAL1, PAL2, PAL3, PAL4, PAL5, and PAL7) was induced by threefold or more by F. graminearum infection (Table 5). Different members within a gene family responded differentially. In the PAL family, all the members except PAL6 and PAL8 were induced more than tenfold by F. graminearum infection. In the CCR family, only CCR3 was induced. Of the two 4CLs, 4CL1 was induced by F. graminearum infection, but 4CL2 was not. Of the five CCoAOMT genes, four were induced by F. graminearum infection, whereas CCoAOMT2 was expressed constitutively. Upon F. graminearum infection, CAD1 and CAD4 were induced, whereas no significant change was observed in CAD2 and CAD3 transcript levels. Of the induced genes, C4H, C3H1, CCoAOMT3, CCoAOMT5, PAL1, PAL2, PAL3, and PAL5 reached their peak at 24 HPI but the induced expression of others had yet to peak at 48 HPI. No significant induction was observed in the S-lignin pathway-specific genes COMTs and F5Hs, indicating that the S-lignin pathway is not mobilized for the defense-response to this specific pathogen.
Discussion
Because lignification mainly occurs in the secondary walls of the xylem, lignin biosynthetic genes are abundant in the publicly available genomics databases. The lignin biosynthetic enzymes are encoded by multiple gene families but not every member of a gene family is involved in or contributes equally to lignin biosynthesis as seen in Arabidopsis (Raes et al., 2003). Therefore, identification of the most appropriate targets for further characterization is the first step towards the dissection of the lignin biosynthetic pathway. Lignin deposition is largely developmentally regulated and varies at different developmental stages and in different tissues. Therefore, investigating expression patterns of the candidates in various tissues and at different developmental stages has been an efficient approach to identify the bona fide lignin biosynthetic genes (Raes et al., 2003; Guillaumie et al., 2007; Koutaniemi et al., 2007).
In the present research, we profiled 32 candidate genes in 17 samples collected from three Feekes’ stages and identified 21 genes under strong developmental regulation. These genes were expressed at much higher levels in the stems as compared to the leaf sheaths and leaf blades. This suggests that these genes may play a role in lignin biosynthesis in wheat. These 21 genes were located in two groups, i.e., the group 1 and group 2. Expression of the genes within each group is highly correlated. Group 1 genes are expressed at higher levels compared to their group 2 homologues. Their expression levels correlated with lignin content. The group 2 genes are expressed at the highest level in younger stem tissues than older tissues (F-1st-low vs. F-1st-up). This suggests that they may play an important role in the early development of secondary cell walls. The co-expression patterns in the two groups strongly suggested that they are under coordinated regulation by different transcription factors due to different cis elements in their promoters. In dicots, promoters of most lignin biosynthetic genes harbor one or more AC elements. In pine (Patzlaff et al., 2003) and Eucalyptus (Goicoechea et al., 2005), xylem-associated MYB transcription factors can bind to the AC elements and activate the transcription of the lignin biosynthetic genes. The search of the annotated rice genome sequences (http://rice.plantbiology.msu.edu) revealed ACII motif in the promoters of PAL, 4CL, C4H, C3H, CCoAOMT, CCR, and CAD and an ACIII motif in the promoter of an HCT gene. These rice genes showed the highest homology to wheat genes PAL6, PAL8, 4CL1, C4H, HCT1, C3H1, CCoAOMT1, CCoAOMT3, CCR2, and CAD4. Currently, no one has characterized the coordinated regulation network in grasses, including AC elements of lignin biosynthetic genes. However, several lignin biosynthetic genes have been functionally characterized in grasses, i.e., bm1 of maize (Halpin et al., 1998), gh2 of rice (Zhang et al., 2006), bmr6 of sorghum (Sattler et al., 2009; Saballos et al., 2009) which encode CAD and showed high homology to wheat CAD4. The maize bm3 codes for COMT (Vignols et al., 1995) and is homologous to wheat COMT1 and COMT2. These data support our findings that expression profiling of organs differing in lignin content is an efficient approach to identify candidate genes for lignin pathway.
Metabolite flux continuously passes the pathway from upstream to downstream. If regulation at the protein level is not involved, the transcription level of the upstream genes should be higher than those downstream. In our research, expression levels of PALs and 4CLs were the highest, and CAD4 was the lowest among the group 1 genes. However, the expression level of CCoAMOT1 was higher than those of its upstream genes C4H and C3Hs. The F5H1 and F5H2, S-lignin pathway-specific genes, showed higher expression than upstream genes CCoAOMTs and CCR2 (Supplementary Table 2s). The regulation at the protein level may be required for catalytic activity of CCoAOMT and F5H. Alternately, more candidates for C4H, C3H, CCoAOMT, and CCR remain to be identified in wheat.
The phenylpropanoid pathway has long been recognized to be involved in biotic and abiotic stress response (for review, see Dixon et al., 2002). A more recent study in Arabidopsis showed that accumulation of intermediate metabolite due to silencing of COMT affected its interaction with an array of pathogens (Quentin et al., 2009). Our previous research found that several genes of this pathway were induced by F. graminearum (Hill-Ambroz et al., 2006). In this study, 17 genes, four group 1, seven group 2, and six group 3 genes were induced by F. graminearum. In contrast to the expression pattern induced by F. graminearum, only the PAL1 gene was significantly induced by P. triticina. P. triticina is an obligate fungus that requires living plant cells to survive and the young leaves were inoculated at Feekes’ stage 2. F. graminearum is a facultative fungus that can survive on dead plant tissue and was inoculated to the florets at Feekes’ stage 10.0. These distinctive induction patterns suggest that infection of different types of fungal pathogens trigger different sets of plant genes or/and plants mobilize different set genes for defense at different developmental stages and in different tissues. PAL1 was induced by both pathogens and most probably is a part of general defense response of wheat plant to pathogen challenge.
The remaining five genes in group 3, CAD2, CAD3, CCoAOMT2, CCR1, and CCR4 did not show developmental-regulated expression pattern and were not induced by pathogen infections either. Of these five genes, CAD2 was nearly constitutively expressed in all the samples but was at the highest level among the CAD members. CAD2 is homologous to a CAD-like protein of Arabidopsis (UniRef100_Q8L9G4 Cluster). Most probably, CAD2 does not belong to CAD family. In addition, CCR1 and CCR4 were selected from TaGI release 10.0 and showed weak homology to the annotated rice CCR genes.
Of the 21 group 1 and group 2 genes, PAL7 was induced by more than tenfold and 4CL1, C4H and CCoAMOT3 were induced approximately by fivefold by F. graminearum. On the contrary, CCR2, F5H1, and F5H2 of the first group, and 4CL2, COMT1, and COMT2 of the second group were not induced. The remaining 11 gene were induced by more than threefold. Based solely on the pathogen-induced expression, 4CL1, C4H, CCoAOMT3, and PAL7 are not recommended as targets for gene silencing. The remaining 17 genes, especially F5H1 and F5H2 of group 1 and 4CL2, CCR2, COMT1, and COMT2 of group 2 may be the targets of choice for gene silencing. Because the damaging effect of down regulating the lignin pathway genes also modulated by genetic background, the lignin-reducing genes (transgenes for RNA silencing or point mutations) should be incorporated in multiple genetic backgrounds to minimize the risk as suggested by Pedersen et al. (2005).
In summary, we profiled the transcription of 32 putative lignin candidate genes by qRT-PCR in 17 samples collected from tissues at different developmental stages and found 21 genes under strong developmental regulation. These genes are expressed at much higher level in stem compared to in the leaf sheath and blades and are most probably involved in lignin biosynthesis. They are potential targets for downregulating lignin in wheat, and possibly in other grasses, for biofuel production. Profiling the pathogen-challenged tissues shed light on the role of the monolignol pathway in the defense response and provided an additional dimension for the target selection.
Abbreviations
- 4CL:
-
4-coumarate CoA ligase
- ABSL:
-
Acetyl bromide soluble lignin
- C3H:
-
p-coumaroyl shikimate/quinate 3′-hydroxylase
- C4H:
-
cinnamate 4-hydroxlase
- CCoAOMT:
-
caffeoyl CoA O-methyltransferase
- CCR:
-
cinnamoyl CoA reductase
- CAD:
-
cinnamyl alcohol dehydrogenase
- COMT:
-
caffeic acid/5-hydroxyferulic acid O-methyltransferase
- EST:
-
expressed sequence tag
- F5H:
-
ferulate 5-hydroxylase
- HCT:
-
hydroxycinnamoyl CoA:shikimate/quinate hydroxycinnamoyltransferase
- PAL:
-
phenylalanine ammonia lyase
- qRT-PCR:
-
quantitative real-time PCR
- TaGI:
-
Triticum aestivum gene index database
- TC:
-
tentative consensus
References
Blount JW, Korth KL, Masoud SA, Rasmussen S, Lamb C, Dixon RA (2000) Altering expression of cinnamic acid 4-hydroxylase in transgenic plants provides evidence for a feedback loop at the entry point into the phenylpropanoid pathway. Plant Physiol 122:107–116
Carpita NC (1996) Structure and biogenesis of the cell wall of grasses. Annu Rev Plant Physiol Plant Mol Biol 47:445–476
Chen F, Dixon RA (2007) Lignin modification improves fermentable sugar yields for biofuel production. Nat Biotechnol 25:759–761
Costa MA, Collins RE, Anterola AM, Cochrane FC, Davin LB, Lewis NG (2003) An in silico assessment of gene function and organization of the phenylpropanoid pathway metabolic networks in Arabidopsis thaliana and limitations thereof. Phytochemistry 64:1097–1112
Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139:5–17
Dixon RA, Achnine L, Kota P, Liu CJ, Reddy MSS, Wang L (2002) The phenylpropanoid pathway and plant defence—a genomics perspective. Mol Plant Pathol 3:371–390
Franke R, Humphreys JM, Hemm MR, Denault JW, Ruegger MO, Cusumano JC, Chapple C (2002) The Arabidopsis REF8 gene encodes the 3-hydroxylase of phenylpropanoid metabolism. Plant J 30:33–45
Fukushima RS, Hatfield RD (2001) Extraction and isolation of lignin for utilization as a standard to determine lignin concentration using the acetyl bromide spectrophotometric method. J Agric Food Chem 49:3133–3139
Goicoechea M, Lacombe E, Legay S, Mihaljevic S, Rech P, Jauneau A, Lapierre C, Pollet B, Verhaegen D, Chaubet-Gigot N, Grima-Pettenati J (2005) EgMYB2, a new transcriptional activator from Eucalyptus xylem, regulates secondary cell wall formation and lignin biosynthesis. Plant J 43:553–567
Guillaumie S, San-Clemente H, Deswarte C, Martinez Y, Lapierre C, Murigneux A, Barrière Y, Pichon M, Goffner D (2007) MAIZEWALL. Database and developmental gene expression profiling of cell wall biosynthesis and assembly in maize. Plant Physiol 143:339–363
Halpin C, Holt K, Chojecki J, Oliver D, Chabbert B, Monties B, Edwards K, Barakate A, Foxon GA (1998) Brown-midrib maize (bm1)—a mutation affecting the cinnamyl alcohol dehydrogenase gene. Plant J 14:545–553
Hill-Ambroz K, Webb CA, Matthews AR, Li W, Gill BS, Fellers JP (2006) Expression analysis and physical mapping of a cDNA library of Fusarium head blight infected wheat spikes. Plant Genome, a supplement to Crop Sci 46:S14–S26
Hoffmann L, Maury S, Martz F, Geoffroy P, Legrand M (2003) Purification, cloning, and properties of an acyltransferase controlling shikimate and quinate ester intermediates in phenylpropanoid metabolism. J Biol Chem 278:95–103
Humphreys JM, Chapple C (2002) Rewriting the lignin roadmap. Curr Opin Plant Biol 5:224–229
Iwata N, Omura T (1971) Linkage analysis by reciprocal translocation method in rice plants (Oryza sativa L.). II. Linkage groups corresponding to the chromosomes 5, 6, 8, 9, 10 and 11. Sci Bull Fac Agric Kyushu Univ 25:137–153
Iwata N, Omura T (1977) Linkage studies in rice (Oryza sativa L.) on some mutants derived from chronic gamma irradiation. J Fac Agric Kyushu Univ 21:117–127
Jorgensen LR (1931) Brown midrib in maize and its linkage relations. J Am Soc Agron 23:549–557
Koutaniemi S, Warinowski T, Kärkönen A, Alatalo E, Fossdal CG, Saranpää P, Laakso T, Fagerstedt KV, Simola LK, Paulin L, Rudd S, Teeri TH (2007) Expression profiling of the lignin biosynthetic pathway in Norway spruce using EST sequencing and real-time RT-PCR. Plant Mol Biol 65:311–328
Kuc J, Nelson OE (1964) The abnormal lignins produced by the brown-midrib mutants of maize. I. brown midrib mutant. Arch Biochem Biophys 105:103–113
Large EG (1954) Growth stages in cereals: illustration of the Feeke’s scale. Plant Pathol 3:128–129
Li W, Faris JD, Muthukrishnan S, Liu DJ, Chen PD, Gill BS (2001) Isolation and characterization of novel cDNA clones encoding chitinases and β-1, 3-glucanases from wheat spikelets infected by Fusarium graminearum. Theor Appl Genet 102:353–362
Li X, Weng JK, Chapple C (2008) Improvement of biomass through lignin modification. Plant J 54:569–581
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−∆∆Ct method. Methods 25:402–408
Mizutani M, Ohta D, Sato R (1997) Isolation of a cDNA and a genomic clone encoding cinnamate 4-hydroxylase from Arabidopsis and its expression manner in planta. Plant Physiol 113:755–763
Nagao S, Takahashi M (1963) Trial construction of twelve linkage groups in Japanese rice: genetical studies on rice plant, XXVII. J Fac Agric Hokkaido Univ 53:72–130
Page RD (1996) TREEVIEW: an application to display phylogenetic trees on personal computers. Comput Applic Biosci 12:357–358
Patzlaff A, McInnis S, Courtenay A, Surman C, Newman LJ, Smith C, Bevan MW, Mansfield S, Whetten RW, Sederoff RR, Campbell MM (2003) Characterisation of a pine MYB that regulates lignification. Plant J 36:743–754
Pedersen JF, Vogel KP, Funnell DL (2005) Impact of reduced lignin on plant fitness. Crop Sci 45:812–819
Porter KS, Axtell JD, Lechtenberg VL, Colenbrander VF (1978) Phenotype, fiber composition, and in vitro dry matter disappearance of chemically induced brown midrib (bmr) mutants of sorghum. Crop Sci 18:205–208
Pumphrey MO, Bernardo R, Anderson JA (2007) Validating the Fhb1 QTL for Fusarium head blight resistance in near-isogenic wheat lines developed from breeding populations. Crop Sci 47:200–206
Quentin M, Allasia V, Pegard A, Allais F, Ducrot PH, Favery B, Levis C, Martinet S, Masur C, Ponchet M, Roby D, Schlaich NL, Jouanin L, Keller H (2009) Imbalanced lignin biosynthesis promotes the sexual reproduction of homothallic oomycete pathogens. PLoS Pathog 5:e1000264
Raes J, Rohde A, Christensen JH, Van de Peer Y, Boerjan W (2003) Genome-wide characterization of the lignification toolbox in Arabidopsis. Plant Physiol 133:1051–1071
Rogers L, Campbell MM (2004) The genetic control of lignin deposition during plant growth and development. New Phytol 164:17–30
Saballos A, Ejeta G, Sanchez E, Kang C, Vermerris W (2009) A genomewide analysis of the cinnamyl alcohol dehydrogenase family in sorghum [Sorghum bicolor (L.) Moench] identifies SbCAD2 as the brown midrib6 gene. Genetics 181:783–795
Sattler SE, Saathoff AJ, Haas EJ, Palmer NA, Funnell-Harris DL, Sarath G, Pedersen JF (2009) A nonsense mutation in a cinnamyl alcohol dehydrogenase gene is responsible for the Sorghum brown midrib6 phenotype. Plant Physiol 150:584–595
Sibout R, Eudes A, Mouille G, Pollet B, Lapierre C, Jouanin L, Séguin A (2005) CINNAMYL ALCOHOL DEHYDROGENASE-C and -D are the primary genes involved in lignin biosynthesis in the floral stem of Arabidopsis. Plant Cell 17:2059–2076
Suzek BE, Huang H, McGarvey P, Mazumder R, Wu CH (2007) UniRef: comprehensive and non-redundant UniProt reference clusters. Bioinformatics 23:1282–1288
Tuskan GA, Difazio S, Jansson S, Bohlmann J, Grigoriev I, Hellsten U, Putnam N, Ralph S, Rombauts S, Salamov A, Schein J, Sterck L, Aerts A, Bhalerao RR, Bhalerao RP, Blaudez D, Boerjan W, Brun A, Brunner A, Busov V, Campbell M, Carlson J, Chalot M, Chapman J, Chen GL, Cooper D, Coutinho PM, Couturier J, Covert S, Cronk Q, Cunningham R, Davis J, Degroeve S, Déjardin A, Depamphilis C, Detter J, Dirks B, Dubchak I, Duplessis S, Ehlting J, Ellis B, Gendler K, Goodstein D, Gribskov M, Grimwood J, Groover A, Gunter L, Hamberger B, Heinze B, Helariutta Y, Henrissat B, Holligan D, Holt R, Huang W, Islam-Faridi N, Jones S, Jones-Rhoades M, Jorgensen R, Joshi C, Kangasjärvi J, Karlsson J, Kelleher C, Kirkpatrick R, Kirst M, Kohler A, Kalluri U, Larimer F, Leebens-Mack J, Leplé JC, Locascio P, Lou Y, Lucas S, Martin F, Montanini B, Napoli C, Nelson DR, Nelson C, Nieminen K, Nilsson O, Pereda V, Peter G, Philippe R, Pilate G, Poliakov A, Razumovskaya J, Richardson P, Rinaldi C, Ritland K, Rouzé P, Ryaboy D, Schmutz J, Schrader J, Segerman B, Shin H, Siddiqui A, Sterky F, Terry A, Tsai CJ, Uberbacher E, Unneberg P, Vahala J, Wall K, Wessler S, Yang G, Yin T, Douglas C, Marra M, Sandberg G, Van de Peer Y, Rokhsar D (2006) The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313:1596–1604
Vignols F, Rigau J, Torres MA, Capeliades M, Puigdomenech P (1995) The brown midrib3 (bm3) mutation in maize occurs in the gene encoding caffeic acid O-methyltransferase. Plant Cell 7(407):416
Winkel BS (2004) Metabolic channeling in plants. Annu Rev Plant Biol 55:85–107
Yang C, Xu Z, Song J, Conner K, Barrena GV, Wilson ZA (2007) Arabidopsis MYB26/MALE STERILE35 regulates secondary thickening in the endothecium and is essential for anther dehiscence. Plant Cell 19:534–548
Yokoyama R, Nishitani K (2004) Genomic basis for cell-wall diversity in plants. A comparative approach to gene families in rice and Arabidopsis. Plant Cell Physiol 45:1111–1121
Zhang K, Qian Q, Huang Z, Wang Y, Li M, Hong L, Zeng D, Gu M, Chu C, Cheng Z (2006) GOLD HULL AND INTERNODE2 encodes a primarily multifunctional cinnamyl-alcohol dehydrogenase in rice. Plant Physiol 140:972–983
Zhong R, Ye ZH (2007) Regulation of cell wall biosynthesis. Curr Opin Plant Biol 10:564–572
Zhong R, Lee C, Zhou J, McCarthy RL, Ye ZH (2008) A battery of transcription factors involved in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell 20:2763–2782
Acknowledgments
We thank Dr. Robert L. Bowden for inoculating P. triticina, Dr. Michael O. Pumphrey for supplying seeds of wheat line HR58 and conidia spores of F. graminearum and Dr Michael Udvardi for providing the LjLb2 cDNA clone. This project was funded by the USDA/DOE Feedstock Genomics Program. This paper is contribution No. 10-341-J from the Kansas Agricultural Experiment Station (Manhattan, KS, USA).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Table S1
Correlation coefficients between the expression levels of monolignol biosynthetic candidate genes in the internodes, sheath and leaf blades at Feeks’ Stage 10.0 (XLS 37 kb)
Table S2
XLS 34 kb
Table S3
XLS 34 kb
Table S4
XLS 33 kb
Table S5
XLS 33 kb
Table S6
XLS 32 kb
Table S7
Expression levels of monoligol biosynthetic candidate genes at Feeks’ stage 2, 6, and 10.0 (XLS 30 kb)
Table S8
Expression levels of monolignol biosynthetic candidate genes in internodes, sheaths and leaves at Feeks’ stage 10.0 (XLS 32 kb)
Table S9
XLS 32 kb
Rights and permissions
About this article
Cite this article
Bi, C., Chen, F., Jackson, L. et al. Expression of Lignin Biosynthetic Genes in Wheat during Development and upon Infection by Fungal Pathogens. Plant Mol Biol Rep 29, 149–161 (2011). https://doi.org/10.1007/s11105-010-0219-8
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11105-010-0219-8