Abstract
Genetic diversity of 56 radish accessions, representing nearly all the typical types and origins of cultivated radish germplasms conserved in the National Mid-term Genebank for Vegetables of China, was assessed with amplified fragment length polymorphism (AFLP) and random amplified polymorphic DNA (RAPD) markers. A total of 72 and 128 polymorphic bands were generated by the 12 selected RAPD primers and eight AFLP primer combinations respectively. A moderate correlation with the value of r = 0.66 was observed between AFLP and RAPD markers. The total 200 polymorphic bands were integrated to assess the genetic diversity of 56 radish accessions. The Jaccard similarity coefficients between the accessions varied from 0.30 to 0.83 with the mean of 0.54. Cluster analysis classified the germplasms into three groups of var. hortensis Becker, var. sativus, and var. niger Kerner. The three-dimensions scatter plot of principle coordinate analysis (PCA) further divided var. hortensis Becker germplasms into two separate groups. The results indicated that the genetic diversity harbored among var. hortensis Becker germplasms was very abundant, which could be further exploited for radish genetic improvement.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Radish, Raphanus sativus L., 2n = 2× = 18, an edible root vegetable of the Brassicaceae family, is one of the staple vegetable crops in Asia, especially in China, Japan, and Korea. Cultivated radish has been classified into many varieties according to the morphology of its edible root and different usages, such as R. sativus var. sativus (European small radish), var. hortensis Becker (East Asian big long radish), var. niger Kerner (black radish), var. chinensis Gallizioli (Chinese oil radish), and var. caudatus Hooler and Anderson (tail-podded radish) (Lu et al. 2008). The open pollination habit helped the species to accumulate abundant variations. It was reported that even the flora morphology exhibited great variations among the radish accessions (Kobayashi et al. 2006). Those variations offered abundant genetic resources for radish genetic enhancement. As a result, appraisal on the genetic diversity of radish will be greatly conducive to the utilization and improvement of radish germplasm.
Many molecular markers, such as random amplified polymorphic DNA (RAPD) (Yamagishi et al. 1998; Matveeva et al. 2002; Huh and Ohnishi 2003; Madhou et al. 2005) and amplified fragment length polymorphism (AFLP) (Huh and Huh 2001; Huh and Ohnishi 2002), have been applied respectively to estimate the genetic diversity of radish. Multiple types of makers were also employed simultaneously to examine genetic diversity of radish germplasm. Morphological traits and RAPD markers were utilized to survey the genetic variations of radish (Rabbani et al. 1998; Pradhan et al. 2004). RAPD, inter simple sequence repeat (ISSR) and sequence-related amplified polymorphism (SRAP) makers were employed to investigate the genetic diversity of late-bolting radish accessions (Liu et al. 2008). Chloroplast and mitochondrial DNA sequence polymorphisms in combination of polymerase chain reaction–restriction fragment length polymorphism (PCR-RFLP) technique were used to elucidate the diversity and evolution of wild and cultivated radish (Yamagishi 2004; Yamane et al. 2005; Lu et al. 2008). The combination of different markers provided more comprehensive information for genetic diversity evaluation.
The genetic resource of radish is very abundant in China. Up to now, more than 2,100 radish accessions have been conserved in the National Mid-term Genebank for Vegetables located in the Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences. Evaluation and exploitation of the diversity of those germplasms are crucial for radish genetic resources management and breeding programs.
In this study, RAPD and AFLP markers were employed to estimate the genetic diversity of radish germplasms conserved in National Mid-term Genebank for Vegetables of China, with the aims of offering useful information for efficient utilization and conservation of radish genetic resources.
Materials and Methods
Plant Materials
To make the tested radish samples can represent the genetic resources conserved in National Mid-term Genebank for Vegetables at large, the sampling strategy was adopted that only the germplasms with typical or representative phenotypes from each province in China and other countries were sampled. However, the germplasms of var. chinensis and var. caudatus were not conserved in the Mid-term Genebank. Consequently, a total of 56 radish accessions, representing the types of R. sativus var. sativus L., var. hortensis Becker, and var. niger Kerner, were selected in the study. In which, “Fekete” was the only germplasm belonging to var. niger that conserved in the Mid-term Genebank. Most of those radish germplasms were landraces collected in 1980s. The names, accession numbers (ID), and origins of the tested materials are listed in Table 1.
Molecular Marker Analysis
Young leaves from 10 individuals of each accession were randomly collected and mixed for genomic DNA isolation. The genomic DNA was isolated with the method of 2% CTAB as described by Tang et al (2007).
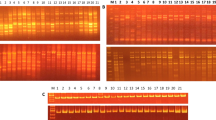
RAPD reactions were performed in 10-μl volumes containing 30 ng genomic DNA, 1.5 mM MgCl2, 125 μM each of dNTPs, 1.2 μM random primer, and 0.65 U Taq polymerase. Amplification was carried out with the program of one cycle of 94°C for 2 min, followed by 40 cycles of 94°C for 40 s, 36°C for 90 s, 72°C for 90 s, at last, 5 min extension at 72°C. PCR products were resolved by electrophoresis on 1.5% agarose gel, stained with ethidium bromide and photographed under UV light.
AFLP analyses were conducted according to Vos et al. (1995) with minor modifications. One hundred nanograms of DNA was digested with 3 U MseI and 1.5 U EcoRI restriction enzyme and ligated with adaptors by 1.5 U T4 ligase under the reaction condition of 37°C for 4 h, followed by 22°C for 4 h and then 65°C for 10 min, at last, stored at 4°C. Preamplification was carried out with the primer combination of M02 and E00. Selective amplifications were performed in 20 μl volume containing 1.25 mM MgCl2, 2 mM each of dNTP, 1.2 μM MseI and EcoRI primer pair, 0.65 U Taq polymerase, and 2 μl preamplification product. Amplification products were resolved on 6% denaturing PAGE gel and visualized by silver staining as described in the Gene Print® STR Systems (Promega).
Data Analysis
Polymorphic markers were manually scored as binary data with presence as “1” and absence as “0.” Only clear and unambiguous bands were included in the analysis. Data analyses were conducted using the NTSYS-pc2.10e software according to its manual (Rohlf 2000). Dendrograms were constructed with the method of unweighted pair group method with arithmetic mean (UPGMA) based on the similarity matrices calculated with Jaccard coefficient. Correlation between the AFLP and RAPD similarity matrices was assessed by the Mantel test, which assumes that the two matrices were obtained independently. Principal coordinate analysis (PCA) was performed to further elucidate the relationship among the tested radish accessions.
Results
RAPD and AFLP Polymorphisms
In the RAPD analysis, 79 random primers were initially screened and 12 RAPD primers yielding sharp, polymorphic, and reproducible bands patterns were selected to evaluate radish genetic diversity. The number of bands and the degree of polymorphism revealed by each primer are given in Table 2. A total of 109 distinct bands were generated in the RAPD analysis, with 72 bands being polymorphic. The polymorphic bands for each primer varied from two to 13, with an average of 6.0 polymorphic bands. The proportions of polymorphic bands produced by the selected primers ranged from 40.0% to 88.9%, with the mean polymorphic proportion of 66.1%.
Among the 30 AFLP primer combinations tested on radish accessions, distinct and polymorphic products were obtained from eight primer combinations that produced 327 DNA fragments, 128 of which were polymorphic (Table 2). The total number of DNA bands per primer combination ranged from 32 to 51, while the number of polymorphic bands per primer combination varied between 10 and 32. On average, 16.0 polymorphic bands were detected for each AFLP primer combination. The polymorphic rates produced by the selected primer combinations varied from 27.0% to 62.7% with the mean polymorphism of 39.1%.
Genetic Relationships among Radish Germplasms
The binary data matrices yielded by RAPD and AFLP were used to estimate genetic similarity of the tested germplasms, respectively. The pairwise similarity coefficient detected by RAPD markers varied from 0.32 to 0.90 with the mean of 0.61, while the pairwise similarity coefficient detected by AFLP markers ranged from 0.23 to 0.85, with a mean of 0.50. The matrices of similarity coefficients calculated by AFLP and RAPD analyses were compared using regression analysis performed by Mantel test. A moderate correlation with the value of r = 0.66** was observed between AFLP and RAPD markers.
The total 200 polymorphic bands produced by RAPD and AFLP were integrated into one matrix to assess the genetic diversity of 56 radish accessions. The pairwise Jaccard coefficients varied from 0.30 to 0.83, with a mean of 0.54. The similarity coefficient matrices obtained by the integration of AFLP and RAPD, AFLP, and RAPD were compared using the Mantel test. A high correlation coefficient was observed between the integrated data with AFLP (r = 0.96**) and RAPD (r = 0.85**), respectively.
Dendrogram constructed by UPGMA separated the 56 radish accessions into three groups (Fig. 1). Group I was primarily comprised of 45 East Asian big long radish accessions mainly from China, Japan, and Korea, except for “Kerik” and “Onisaier” from Russia. Group II consisted of 10 European small radish accessions from Europe, with the exception of “Dangdishui” collected from Xinjiang, China. The only black radish germplasm of “Fekete” collected from Hungary was assigned to group III.
The germplasm in group I could be further divided into six subgroups. Subgroup I consisted of 13 accessions collected from China, with the exception of “Kerike” from Russia. Subgroup II included 22 accessions from China with the exception of “Huaye” from Korea and “Onishaier” from Russia. Subgroup III contained three accessions, with the distinguishable trait of heat tolerance. Subgroup IV comprised four newly introduced germplasms (“Chunbai 2,” “YR baichun,” “R1010,” and “Baiguang”) with the known agronomic trait of late-bolting in spring. “Guoguang” and “Yizhehengding” were assigned into subgroup V, indicating the underlying distinct genetic variations harbored by them. Subgroup VI only included the accession of “Xinlimei” with the distinct trait of pink flesh.
PCA was employed to further elucidate the genetic relationships among the tested germplasms. The first, second, and third PCA accounted for 15.9%, 8.0%, and 5.3% of the total variations, respectively. The three-dimensions scatter plot divided the 56 radish accessions into four groups (Fig. 2), which was similar with the results of UPGMA cluster analysis. All the accessions of East Asian big long radish, collected from Japan, Korea, Russia, and China, were classified into group I-A and group I-B, which spread along the third PCA, showing remarkable genetic diversity harbored among them. Group II comprised 10 accessions, which were all European small radish. The other three accessions of European small radish (“Zhuajiaza,” “Yanghua,” and “Shui”) originated in China were classified into East Asian big long radish but not into European small radish, which was probably due to the frequent gene flow with big long varieties by natural or artificial hybridizations. “Fekete” from Hungry with black skin of root was also distinguished from the other varieties and formed group III.
Discussions
In this study, 66.1% bands generated by RAPD assay were polymorphic, which was lower than the polymorphic proportion of 78.2% detected by RAPD among Pakistan radish germplasms (Rabbani et al. 1998), 88.5% among Australian radish cultivars (Pradhan et al. 2004), 85.4% among late-bolting radish cultivars (Liu et al. 2008), and 82% in wild radish population (Raphanus raphanistrum L.) (Madhou et al. 2005). As for AFLP marker, the polymorphic rates of 58.4% and 76.5% were detected among wild radishes (Huh and Huh 2001) and cultivated radish varieties (Muminovic et al. 2005), respectively. However, in this study, the average polymorphic proportion yielded by AFLP was only 39.1%, which was significantly lower than that of the previous researches. The relatively low polymorphism acquired in this study probably resulted from the different primers selected and sampling strategy. The qualities of amplification products were given more attention than the polymorphism during the process of primer screening. Meanwhile, the genetic variations extensively existed among the individuals of the accession for conserved germplasms. The randomly mixed sampling strategy could represent the different individual genotypes in the accession, however, at the cost of decreasing polymorphism among the accessions. Consequently, the relatively low polymorphism detected by molecular markers did not totally mean the low degree of genetic diversity harbored in the tested radish germplasms.
Furthermore, it was reported that the average genetic similarities of 0.70 and 0.78 were detected by AFLP in 68 cultivated radish varieties (Muminovic et al. 2005) and by RAPD in 35 late-bolting radish cultivars (Liu et al. 2008), respectively. Those data were higher than that acquired by AFLP (0.50) and RAPD (0.61), respectively, in this study, indicating more abundant genetic diversity harbored in the tested radish germplasms.
The combination of polymorphic information derived from different marker systems was expected to decrease the effect of their independent inaccuracies. In this study, the RAPD and AFLP data were integrated to elucidate the genetic relationships among the radish germplasms.
Dendrogram constructed by the integration of RAPD and AFLP data clearly classified the radish germplasm into groups of var. hortensis Becker, var. sativus, and var. niger Kerner. Moreover, the var. hortensis accessions could be further divided into six subgroups. In the subgroups, the accessions with the known trait of heat tolerance and late-bolting in spring were distinguished from the other East Asian big long radish gemplasms and formed separate subgroups, respectively. Xinlimei, a well-known landrace collected from Beijing, China, with the unique trait of pink flesh, formed separate subgroup, indicating it has different genetic background in comparison with the other East Asian big long radish accessions. In addition, “Guoguang” and “Yizhehengding,” with unavailable data of their genetic backgrounds up to now, exhibited distant relationships with the other East Asian big long radish accessions. As a result, more attentions should be paid on the two germplasms for traits identification and valuable gene mining.
PCA exhibited similar genetic relationships among the germplasms as UPGMA cluster analysis. Furthermore, PCA distinctly classified the accessions of var. hortensis Becker into two groups, exhibiting abundant diversity harbored among the accessions of var. hortensis Becker.
Generally, the molecular markers of RAPD and AFLP classified the radish accessions into varieties of var. hortensis Becker, var. sativus, and var. niger Kerner, which were in agreement with the relationships determined by AFLP and ISSR analyses (Muminovic et al. 2005) and chloroplast DNA sequence variations of trnK/matK (Lu et al. 2008). Moreover, the results showed diverse genetic differences among these germplasms, which could be exploited for radish genetic improvement. The study offered primary information for core collection and utilization of radish germplasms conserved in National Mid-term Genebank for Vegetables of China.
References
Huh MK, Huh HW (2001) Genetic diversity of Raphanus sativus var. hortensis f. raphanistroides in Korea using AFLP markers. Kor J Genet 23:45–53
Huh MK, Ohnishi O (2002) Genetic diversity and genetic relationships of East Asian natural populations of wild radish revealed by AFLP. Breeding Sci 52:79–88
Huh MK, Ohnishi O (2003) Genetic diversity and relationships among natural and cultivated populations of radish in Korea revealed by RAPD. Kor J Genet 25:119–125
Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences (1992) The catalog of vegetable germplasm resources in China. (Series one). Wanguo Academic Press, Beijing
Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences (1998) The catalog of vegetable germplasm resources in China. (Series two). Meteorology Press, Beijing
Kobayashi K, Horisaki A, Niikura S, Ohsawa R (2006) Inter-accession variation in floral morphology in radish (Raphanus sativus L.). Euphytica 152:87–97
Liu LW, Zhao LP, Gong YQ, Wang MX, Chen LM, Yang JL, Wang Y, Yu FM, Wang LZ (2008) DNA fingerprinting and genetic diversity analysis of late-bolting radish cultivars with RAPD, ISSR and SRAP markers. Sci Hortic 116:240–247
Lu N, Yamane K, Ohnishi O (2008) Genetic diversity of cultivated and wild radish and phylogenetic relationships among Raphanus and Brassica species revealed by the analysis of trnK/matK sequence. Breeding Sci 58:15–22
Madhou P, Wells A, Pang ECK, Stevenson TW (2005) Genetic variation in populations of Western Australian wild radish. Aust J Agr Res 56:1079–1087
Matveeva TV, Simonova AV, Lutova LA (2002) Molecular markers of inbred radish (Raphanus sativus var. radicola Pers.) lines. Cell Mol Biol Lett 7:845–848
Muminovic J, Merz A, Melchinger AE, Lubberstedt T (2005) Genetic structure and diversity among radish varieties as inferred from AFLP and ISSR analyses. J Am Soc Hortic Sci 130:79–87
Pradhan A, Yan G, Plummer JA (2004) Development of DNA fingerprinting keys for the identification of radish cultivars. Aus J Exp Agr 44:95–102
Rabbani MA, Murakami Y, Kuginuki Y, Takayanagi K (1998) Genetic variation in radish (Raphanus sativus L.) germplasm from Pakistan using morphological traits and RAPDs. Genet Resour Crop Evol 45:307–316
Rohlf FJ (2000) NTSYS-pc: numerical taxonomy and multivariate analysis system, version2.1. Exeter Software, Setauket, New York
Tang SQ, Bin XY, Peng YT, Zhou JY, Wang L, Zhong Y (2007) Assessment of genetic diversity in cultivars and wild accessions of Luohanguo (Siraitia grosvenorii [Swingle] A. M. Lu et Z. Y. Zhang), a species with edible and medicinal sweet fruits endemic to southern China, using RAPD and AFLP markers. Genet Resour Crop Evol 54:1053–1061
Vos P, Hogers R, Bleeker M, Reijans M, Lee T, Hornes M, Frijters A, Pot J, Peleman J, Kuiper M, Zabeau M (1995) AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res 23:4407–4414
Yamagishi H (2004) Assessment of cytoplasmic polymorphisms by PCR-RFLP of the mitochondrial orfB region in wild and cultivated radishes (Raphanus). Plant Breed 123:141–144
Yamagishi H, Tateishi M, Terachi T, Murayama S (1998) Genetic relationships among Japanese wild radishes (Raphanus sativus f. raphanistroides Makino), cultivated radishes and R-raphanistrum revealed by RAPD analysis. J Jpn Soc Hortic Sci 67:526–531
Yamane K, Lu N, Ohnishi O (2005) Chloroplast DNA variations of cultivated radish and its wild relatives. Plant Sci 168:627–634
Acknowledgements
This work was funded by National Key Technology R&D Program of China (Grant No. 2006BAD13B06) and by Key Laboratory of Horticultural Crops Genetic Improvement, Ministry of Agriculture.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kong, Q., Li, X., Xiang, C. et al. Genetic Diversity of Radish (Raphanus sativus L.) Germplasm Resources Revealed by AFLP and RAPD Markers. Plant Mol Biol Rep 29, 217–223 (2011). https://doi.org/10.1007/s11105-010-0228-7
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11105-010-0228-7