Abstract
Understanding the evolution of the maize B chromosome requires insight into the molecular organization of a large number of B clones, which can be potentially obtained by microdissection of the chromosome. Yet, the microdissection protocols currently available are ineffective for a large-scale isolation. In an attempt to improve its efficiency, a protocol was adopted to screen a microdissected B library with probes prepared from the degenerate oligonucleotide primed-PCR product of genomic DNA. This protocol resulted in 59 new B clones, most of which were highly repetitive sequences located in various B regions but mostly in the heterochromatic blocks of the long arm. They also appeared in A chromosomes. Twenty-four of these were retrotransposons, ten knob, 18 noncoding sequences, and seven unknown sequences. The implication of the new B sequences on the B evolution is discussed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The maize B chromosome is supernumerary as it carries none of the functional genes involved in plant development (Randolph 1941). Having a size larger than half of chromosome 10, the shortest of the maize complement, its existence in the maize genome remains intriguing. One approach to understanding its role is to study its molecular structure, and this requires analysis of many B sequences. However, this approach is hampered by the fact that B sequences cannot be cloned using conventional protocols due to the high homology between B and A chromosomes (Chilton and McCarthy 1973). Several studies have been attempted to overcome this hurdle (Alfenito and Birchler 1993; Stark et al. 1996; Peng et al. 2005). Due to inherent constraints, none of these approaches are capable of generating large numbers of B sequences required for studying problems associated with the evolution of the B chromosome. However, this can be potentially resolved by direct cloning of sequences from microdissected B fragments. This approach has been well established and successfully applied in isolation of B sequences from several plant species, including rye, Crepis capillaris, and Brachycome dichromosomatica (Sandery et al. 1991; Jamilena et al. 1995; Houben et al. 1996, 1997). The same technology has also been successfully applied to isolate 19 maize B sequences (Cheng and Lin 2003).
The overall goal of this study is to isolate and characterize a large number of maize B sequences using a modified protocol previously described by Cheng and Lin (2003). A total of 59 sequences distinct from those isolated previously by Cheng and Lin (2003) were obtained, and all were repetitive in nature. Most sequences (52 clones) were homologous to published sequences, and some (seven clones) had no similarity to any sequence in GenBank. These sequences were located not only in the B chromosome but also A chromosomes. The significance of the new sequences to the B evolution is discussed.
Materials and Methods
Plant Material
Two maize inbred lines (L289 and W22) were used in this study. Plants of the inbred line L289 carrying 2, 4, and 6 Bs (L289 + 2B, L289 + 4B, and L289 + 6B, respectively) were used for preparation of mitotic and meiotic spreads for fluorescence in situ hybridization (FISH) analysis. The inbred line W22 carrying 0, 4, 10, 14, and 16 Bs (W22 + 0B, W22 + 4B, W22 + 10B, W22 + 14B, and W22 + 16B, respectively) were utilized in Southern hybridization analysis.
Construction and Screening of λ Phage Library Carrying Microdissected B Fragments
A λ phage library was constructed from microdissected pachytene B fragments after being amplified twice by DOP-PCR as outlined by Cheng and Lin (2003). The first DOP-PCR products, provided by Cheng and Lin (2003), were used as template for the second DOP-PCR reaction. Sequentially, the final products were attached—by PCR amplification—at both ends to the degenerate oligonucleotide primer bearing the EcoRI cutting site (5′-ccggaattcccgactcgag-3′), cleaved by EcoRI, and cloned into lambda ZAPII/EcoRI vector following supplier’s instructions (Stratagene).
The library was screened with two different procedures to uncover phages carrying the B DNA. For the first procedure, aliquots of the library (~800 phages per 150 mm plate) were plated (first plating) and incubated until plaques were visible (1 mm). Then, the plaques were blotted onto nylon membranes, probed with the W22 + 16B DNA as outline by Cheng and Lin (2003), and washed first at 25°C with low stringency washing buffer (2× SSC, 0.1% SDS) twice for 10 min each and then by high stringency washing buffer (0.1× SSC, 0.1% SDS) for 1 h. All plaques, whether carrying B DNA or not, displayed similar signal intensity, independent of the washing condition. To identify the B-carrying phages, 64 plaques were randomly selected, transferred orderly with toothpicks onto a fresh bacterial lawn, incubated until the diameter of plaques reaching about 3 to 4 mm, blotted onto membranes, and hybridized with the labeled W22 + 16B DNA. Most plaques exhibited similar signal intensity, but six plaques with slightly more intense signal were then eluted and suspended in SM buffer. For preparation of probes for Southern analysis of genomic DNA, the phage insert was directly isolated from a PCR amplification of an aliquot of each phage suspension (2 μl), using M13 forward/reverse primers. The resulting products were used to probe the W22 + 10B DNA. Only a few B-carrying phages were isolated by this protocol (see “Results”). To enhance recovery of the B-carrying phages, the library was screened with an amplified genomic probe as described below.
The second procedure is as follows: the phage plating and blotting were as described above. The blots were then hybridized with the DOP-PCR products amplified from the W22 + 16B DNA (300 ng). After hybridization, the blots were washed as described above, except that the high stringency washing (0.1× SSC, 0.1% SDS) was conducted at 43°C. Phages of positive plaques were eluted, suspended, and amplified as described above. The phage suspensions containing multiple fragments in their amplification products were discarded.
Sequence Analysis
Sequence analyses were performed using RepeatMasker (http://repeatmasker.org) and Blastn software (http://www.ncbi.nlm.nih.gov). Sixty-two sequences have been deposited in GenBank (accession nos. EI394604 to EI394665).
Southern Analysis
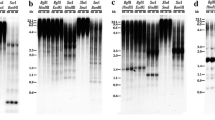
The procedures of Southern analyses were those outlined by Lin et al. (1997) with modifications. Exposure time in Fig. 1 was variable: 2 days for pBK31 and pBK111 but 2 weeks for the remaining clones.
Fluorescence In Situ Hybridization of Pachytene B Chromosome
The procedure of FISH analyses followed Cheng and Lin (2003). The hybridization signal was detected with either anti-digoxigenin-rhodamine (Roche) or Streptavidin-FITC (Invitrogen), and chromosomes were counterstained with 4′,6-diamidine-2′-phenylindole dihydrochloride (Roche). The B-specific ZmBs fragment (Alfenito and Birchler 1993) was cloned from PCR products of + B genomic DNA using two primers (5'-tttccctaactcaaaaacctaagcc-3' and 5'-ctggttagggcttgttatcttaaag-3') which were kindly provided by Dr. Ya-ming Cheng.
Results
Isolation of the Clones Derived from Microdissected B Fragments
In an attempt to further understand the molecular structure of the maize B chromosome, a phage library carrying the B DNA was constructed to isolate a large number of B sequences. The inserts of the library were the amplified products of DOP-PCR of pachytene B fragments that had been microdissected by Cheng and Lin (2003).
The library was first screened with the genomic probe (the first procedure). The screening resulted in two B-carrying clones (pBN10 and pBN15), an efficiency of 3.1% (2/64; for details, see “Materials and Methods”). To achieve 100 B-carrying phages, for example, this screening strategy would require to transfer individually 3,225 phages (100/0.031) onto new bacterial lawns and to perform 302 (3,225 × 6/64) Southern analyses, an impossible task for a laboratory with limited resources. These handicaps forced us to develop an alternative screening strategy which enabled us to identify positive plaques at the first plating.
The alternative screening involved a procedure identical to the first one with the exception of the probe, which was prepared from the labeled DOP-PCR product of + B genomic DNA (the second procedure). Using this procedure, a total of 45,000 phages were screened, 266 of which displayed clear signals, 3–5 positive plaques per plate. Of these, confluent positive plaques were discarded, and the remaining 153 clones were used as probes to hybridize the W22 + 10B blot. After eliminating those phages showing slightly smeared signals (26 phages) and those requiring further purification (62 phages), 65 were proven to carry the B DNA. Five phages were subsequently found to be duplicates. In other words, 60 new clones were obtained.
Analysis of the Fragments Derived from Microdissection
The 60 new clones had an average of 390 (116–1,406) bp in length. Fifty-two of these showed homology to the maize genome: 24 to retrotransposons including Ty3/gypsy-type retrotransposon, Grande, Xilon-1, Tekay, CRM, PREM-2, Reiver and Opie-2; ten to maize 180-bp knob repeat; 18 to those deposited in GenBank but with unknown nature. Seven clones had no similarity to any published sequence. One clone (pBK13) was homologous to the 23S ribosomal RNA genes of Herbaspirillum putei, exhibiting no FISH signal on either the A nor B genome and no dosage response in Southern hybridization with 4B and 14B DNA (data not shown). Its property is similar to those contaminated sequences (either chloroplast or exotic sequences) reported previously by Cheng and Lin (2003). Thus, a total of 59 B clones were identified in this paper.
The molecular nature of these clones was studied first by Southern hybridization with genomic DNA (Fig. 1). Since the knob sequence had been previously reported, the ten knob-carrying clones were excluded from the current analysis. The remaining 49 clones, when used as probes, presented either multiple discrete or various degrees of smeared signals, indicative of multiple copies, and can be roughly classified into two general types. Type I, including 15 clones (pBK2, pBK5, pBK12, pBK79, pBK108, pBK118, pBK129, pBK131, pBK155, pBK199, pBK202, pBK206, pBK215, pBK222, and pBK248), had discrete signals, some of which were clearly proportional to the B number. Five (pBK5, pBK108, pBK118, pBK131, and pBK206) of this group manifested faint signals in 0B DNA but intense signals in + B DNA, indicating extensive amplification in the B chromosome. Type II includes the remaining 34 clones, with similar signal pattern and intensity in 0B and + B DNA (pBK31 and pBK111, Fig. 1).
The location of these clones in pachytene and somatic chromosomes was investigated with FISH analysis (Fig. 2a,b). With exception of three clones (pBK5, pBK129, pBK248) which exhibited no FISH, all Type I clones had signals of variable intensity in diverse B regions, and their signal on the A chromosomes was likewise variable. For example, pBK118, a clone with high degree of B specificity, had a distinct signal in the heterochromatic H3 and a less distinctive one in H4 region of the long arm of pachytene B chromosome. It also had barely visible signal on 18 root-tip A chromosomes as would be expected from its Southern picture (Fig. 1). Another clone, pBK108, displayed comparable signal pattern, with FISH on PE (proximal euchromatic region), H1 to H3, and 14 A chromosomes, although its A signals were much weaker than pBK118. Signals of pBK222 concentrated in H1 and H3 with some very weak signals in PE, H2, and H4 on the B long arm and on all A chromosomes. Its A signals were slightly more intense than that of pBK118. The next clone, pBK79, had a distinctive signal pattern, covering the entire distal heterochromatic regions and some in the proximal euchromatic region close to the centromeric knob on the B long arm and on every A chromosome. The remaining Type I clones were, by and large, in parallel with pBK79. The FISH pattern of Type II clones, pBK31 as an example, were more uniformed; heavily spreading on the B chromosome, euchromatic and heterochromatic region alike, and all A chromosomes.
FISH analysis of new B sequences. a Pachytene B chromosomes were hybridized with rhodamine-labeled B clones. Each B clone had its distinct FISH pattern in the B chromosome. b Metaphase spreads of root-tip cells were probed by rhodamine-labeled ZmBs and FITC-labeled B clones. The B centromere displayed prominent ZmBs signal in red in each cells, but B and A chromosomes manifested variable green signals of new B clones, each with its distinctive pattern. Scale bar 10 μm
Discussion
The most effective way for attending a large scale of B cloning is microdissection. Cheng and Lin (2003) successfully adopted this strategy to obtain sequences from pachytene B chromosome. Using plasmid as vector, they constructed a microdissected library, screened it with a genomic probe, and observed no signal on the blots (personal communication). They finally achieved signals by a cumbersome procedure (Cheng and Lin 2003), including growing each colony in liquid medium overnight before DNA extraction and hybridization with the probe. It is conceivable that their protocol is not suitable for a large-scale isolation of B sequences in a laboratory with a limited resource. In this paper, this problem was resolved by an improvement on the probe preparation; that is, using products from amplification of genomic DNA by DOP-RCR.
To hybridize the B-carrying library, the DOP-PCR generated probe was more efficient than the genomic probe. It was due to the fact that the probe prepared from DOP-PCR products carried more B sequences than the genomic probe. Data generated from maize B chromosome cloning of our laboratory—and some from other laboratory—indicated that most, if not all, B sequences are highly repetitive and, therefore, are expected to be amplified predominately by DOP-PCR. When used as probe to screen a library (microdissected B DNA) blot, such probe would be expected to result in more positive plaques than the genomic probe.
In several aspects, the new sequences uncovered from the current procedure are comparable to those isolated by Cheng and Lin (2003): no coding sequence is evident; most clones are highly homologous—in terms of Southern signals—to the A genome; their presence in various regions of the distal heterochromatic region on the B long arm; and they are either mobile elements or unknown sequences. Yet, the new sequences possessed several distinctive features: first, they carried retroelements Opie, Reiver, and CRM, but none in Cheng and Lin’s clones. Second, they had less clones responsive to the B dosage than Cheng and Lin’s clones (25.4% vs. 57.9%) and those of Peng et al. (2005; 25.4% vs. 76.9%). The basis for this dissimilarity is not clear.
Study of a large number of the B sequences enhances our understanding on the origin of the B chromosome. Cheng and Lin (2003) isolated 19 B sequences, and ten of these hybridized to the large genomic clones located individually in chromosomes 1, 4, 7, and 9. Peng et al. (2005) obtained 13 sequences from ten different B regions and six of these displayed sequence similarity to chromosomes 3, 4, 7, 9, and 10. In this paper, 20 of the 59 new B sequences showed homology to chromosomes 1, 3, 4, 7, 9, and 10. All together, of the 91 B sequences isolated up to this day, 36 have homology to six different A chromosomes. Yet, these should not be taken to mean that they are fully consistent with Page et al. (2001)’s hypotheses—the B chromosome might be a “glomerate” heterochromatin from several chromosomes, for the map position of at least two of these genomic clones situated in 7S and 9S, which carried a short region homologous to a B sequence in addition to 19-kD zein and bz1, respectively, are not located in the heterochromatic region detectable by microscopic observation. Furthermore, some B sequences matched to more than two locations in a single chromosome. Four genomic clones, each of which carried a region with similarity to the B sequence in addition to the following well-known genes: 22-kD zein and tga1 in 4S, 19-kD zein locus in 7S, and Locus9009 in 7L, respectively. In short, the maize B chromosome is composed of many A sequences located in various regions of the maize genome, independent of chromatin structure.
References
Alfenito MR, Birchler JA (1993) Molecular characterization of a maize B chromosome centric sequence. Genetics 135:589–97
Cheng YM, Lin BY (2003) Cloning and characterization of maize B chromosome sequences derived from microdissection. Genetics 164:299–310
Chilton MD, McCarthy BJ (1973) DNA from maize with and without B chromosomes: a comparative study. Genetics 74:605–14
Houben A, Kynast RG, Heim U et al (1996) Molecular cytogenetic characterisation of the terminal heterochromatic segment of the B-chromosome of rye (Secale cereale). Chromosoma 105:97–103. doi:10.1007/BF02509519
Houben A, Leach CR, Verlin D et al (1997) A repetitive DNA sequence common to the different B chromosomes of the genus Brachycome. Chromosoma 106:513–9
Jamilena M, Garrido-Ramos M, Ruiz Rejon M et al (1995) Characterisation of repeated sequences from microdissected B chromosomes of Crepis capillaris. Chromosoma 104:113–20. doi:10.1007/BF00347693
Lin BY, Peng SF, Chen YJ et al (1997) Physical mapping of RFLP markers on four chromosome arms in maize using terminal deficiencies. Mol Gen Genet 256:509–16. doi:10.1007/s004380050595
Page BT, Wanous MK, Birchler JA (2001) Characterization of a maize chromosome 4 centromeric sequence: evidence for an evolutionary relationship with the B chromosome centromere. Genetics 159:291–302
Peng SF, Lin YP, Lin BY (2005) Characterization of AFLP sequences from regions of maize B chromosome defined by 12 B-10 L translocations. Genetics 169:375–88. doi:10.1534/genetics.104.032417
Randolph LF (1941) Genetic characteristics of the B chromosomes in maize. Genetics 26:608–31
Sandery MJ, Forster JW, Macadam SR et al (1991) Isolation of a sequence common to A- and B-chromosomes of rye (Secale cereale) by microcloning. Plant Mol Biol Rep 9:21–30. doi:10.1007/BF02669286
Stark EA, Connerton I, Bennett ST et al (1996) Molecular analysis of the structure of the maize B-chromosome. Chromosome Res 4:15–23. doi:10.1007/BF02254939
Acknowledgments
The authors would like to thank Dr. W.-H. Hsu and the Ministry of Education, Taiwan, R. O. C. under the ATU plan for financial support on part of this study and H.-H. Yang for technical assistance.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Lo, Kl., Lin, Yp., Chen, Lj. et al. Isolation and Characterization of New Maize B Sequences from a Microdissected Library. Plant Mol Biol Rep 27, 350–354 (2009). https://doi.org/10.1007/s11105-009-0092-5
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11105-009-0092-5