Abstract
Aims
To identify the rhizobia nodulating Astragalus sinicus in the acid soils at Xinyang, China and to estimate the effects of fertilization strategies on the rhizobial community.
Methods
Soil samples were collected from treatments submitted to different fertilization strategies (chemical fertilizers, A. sinicus green manure, various input levels). Nodules obtained from the A. sinicus trapping plants grown in the soil samples were used for rhizobial isolation. The isolates were grouped into PCR-RFLP genotypes based on analyses of 16S rRNA gene and IGS, and representatives for each genotype were further subjected to phylogenetic analyses of housekeeping genes (16S rRNA gene, atpD, glnII, and rpoB), as well as symbiotic genes (nodC and nifH). Soil physicochemical characteristics were also estimated and redundancy analysis was performed to examine the relations between soil features and distribution of rhizobial genotypes across different fertilization treatments.
Results
A total of 257 rhizobial isolates were obtained and discriminated into 12 IGS types in a single 16S rRNA type by RFLP analyses. Further 16S rRNA gene sequence-based phylogeny and multilocus sequence analysis (MLSA) distinguished a minor group (10 strains, 4 IGS types) affiliated to Mesorhizobium huakuii and a main group (247 strains, 8 IGS types) to Mesorhizobium jarvisii, for which a novel symbiovar, sv. astragali, was described. The long-term input of A. sinicus green manure increased rhizobial diversity and modified rhizobial community structures, via the increase of organic matter and decrease of soil pH, available phosphate, available potassium, and total salt.
Conclusions
M. jarvisii sv. nov. astragali was the predominant microsymbiont of A. sinicus in the tested acidic soils. The long-term input of green manure modified the soil features, enhancing the genetic diversity of rhizobia associated with A. sinicus and alternating the rhizobial community structure.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Appropriate fertilizer application is important to improve the soil fertility (Fang et al. 2015). In the past decades, due to the rapid population increase and continuous decline of cultivated land area in China, the dose of fertilizer supply raised to increase the crop yields and production (Fang et al. 2015). However, the long-term inappropriate fertilization caused serious environmental challenges like acidification and low nutrient availability in soil (Chen et al. 2009). For reducing the negative effects of chemical fertilizer application, organic manures might be an alternative approach for maintaining both the high production and soil fertility.
As a herbaceous legume, Chinese milk vetch (Astragalus sinicus Linn.) is a traditional and the most popular green manure in wintry fallow paddy fields in China, Japan, and Korea (Chen et al. 2017; Gao et al. 2017; Zhang et al. 2000; Zheng et al. 2013; Murooka et al. 1993) due to its abundant biomass, adaptation to wet paddy soils, as well as its high N-fixing potential (Fang et al. 2015). It is also used to fertilize/restore soils (Chen et al. 2017; Lin and Gu 1998) and to reduce the ferric ion reduction in the red paddy soils in Southern China (Gao et al. 2017). Naturally, this plant only grew in South areas of the Huaihe River (latitude 31°–36° N) in China.
Astragalus sinicus forms nitrogen-fixing root nodules with specific symbiotic rhizobia in the species Mesorhizobium huakuii and Mesorhizobium qingshengii (Chen et al. 1991; Murooka et al. 1993; Nuswantara et al. 1999; Zheng et al. 2013), both originating from acidic soils in South (Hunan Province) and Southeast (Fujian, Jiangxi and Anhui Provinces) China. Nodulation (nod) genes are specific and conserved among the A. sinicus-nodulating rhizobia (Guo et al. 1999; Murooka et al. 1993; Zhang et al. 2000).
Xinyang City of Henan Province is the north border of A. sinicus cultivation, which covers an area of 189,000 km2 lying on 114°01′–114°06′ E and 31°46′–31°52′ N. Although A. sinicus has long been cultivated in Xinyang, its symbiotic rhizobia have never been examined in this area so far. In addition, the A. sinicus cultivated soils have been subjected to different long-term fertilization treatments, and thus constitute ideal samples for investigating the effects of fertilization strategy on rhizobial populations. This study was therefore conducted to evaluate the rhizobial diversity associated with A. sinicus in soils subjected to different fertilization strategies in paddy fields at Xinyang, China.
Materials and methods
Soil sampling, test, and statistical analysis and rhizobial trapping
The soils were sampled from a paddy field (32°7′ N, 114°05′ E) at an experimental area of Henan Academy of Agricultural Sciences. In this area, eight fertilizing treatments in triplicate were applied for the last 9 years, including CK (without chemical or green fertilizer), CF (only with chemical fertilizers of 225 kg CO(NH2)2, 35 kg P2O5, 135 kg K2O per hectare per year, which are the standard doses applied by the local farmers), and six treatments with A. sinicus as green manure (F1–F6 with 22.5 × 103 to 60 × 103 kg/ha/year with an increment of 7.5 × 103 kg at each level) (Suppl. Table S1). The fertilizers were supplied each year, after the rice harvest (in September or October). The size of experiment field for this study was 35 m × 15 m. Three replicates, each with the size of 3 m × 3 m, were employed for each treatment. In addition, the experimental plots were arranged by a complete random design. The neighbor treatments or replicates were separated by a blank space with 1 m in width and the core experimental area was surrounded by blank space greater than 1 m. In this study, soils were collected at 10–20 cm in depth from each replicate after rice was harvested but before A. sinicus was seeded or chemical fertilizer was applied on August 15, 2016. Five randomly selected sample repeats were equally mixed in situ to constitute the composite sample for each replicate and then kept in surface-sterilized ice box for transporting to the laboratory. For rhizobial trapping, the soil samples were mixed with sterilized vermiculite (1/5, v/v) and the mixtures were immediately used to fill surface-sterilized plastic pots (15 cm height × 10 cm diameter). Four surface-sterilized and pre-germinated seeds of the same A. sinicus variety used in Xinyang were sown in each pot and grown for 45 days under the previously described greenhouse conditions (Zhang et al. 2014). Then six plants were randomly sampled for each replicate of the soil treatment (in total 18 plants/treatment), and two root nodules per plant were randomly selected for rhizobial isolation (in total 12 root nodules per treatment). In parallel, physicochemical characteristics of the composite soil samples were analyzed as reported previously (Zhang et al. 2012). Statistical analysis was subsequently performed on soil characteristics by using SPSS version 22. ANOVA method and Duncan test were applied to analyze the average of the data and the significance.
Isolation, storage, and nodulation tests of rhizobia
Nodules were surface-sterilized by immersion in NaClO solution (2.5%, 5 min) and rhizobia were isolated from the nodules on yeast extract-mannitol agar (YMA) by a standard procedure (Vincent 1970; Zhang et al. 2014). After incubated for 3 days at 28 °C, single colony was picked and purified by repeatedly striking on the same medium and incubating at the same conditions. All purified isolates were subcultured on YMA slants at 28 °C and maintained at 4 °C for temporary storage and in YM broth supplied with 20% (w/v) glycerol at − 80 °C for long-term storage (Zhang et al. 2012). All isolates were checked for their capacity to induce root nodules on A. sinicus with the standard procedures (Tena et al. 2017; Zhang et al. 2017).
PCR-based restriction fragment length polymorphism (RFLP) of 16S rRNA and 16S-23S rRNA intergenic spacer (IGS)
Genomic DNA of each rhizobial isolate was extracted according to Terefework et al. (2001) and used as template for 16S rRNA gene and intergenic spacer (IGS) PCR amplifications with primers P1 and P6, corresponding to the position of 8–37 bases and 1479–1506 bases respectively on the 16S rRNA sequence of Escherichia coli (Tan et al. 1997; Zhang et al. 2017) and IGS1490 (forward) and IGS132′ (reverse) (Zhang et al. 2012), respectively. PCR products (1600 bp for 16S rRNA gene and 900 bp for IGS) were digested separately with each of the following restriction endonucleases: MspI, HaeIII, and AluI for 16S rRNA gene; HaeIII, NdeI, and XhoI for IGS (Zhang et al. 2012). Separation and visualization of restriction fragments were done by agarose gel electrophoresis (2.5%, w/v) (Wang et al. 1999) containing GoldView type I nucleotide dye (Solarbio, Lot No. 20140820) (Zhang et al. 2017). The results were analyzed by DNR Bio-Imaging System (MiniBIS Pro, Israel). Then, the rRNA gene type of each strain was designated according to its restriction patterns, and a grouping analysis was performed (Wang et al. 1999) with the UPGMA method of Vauterin (1992).
Sequencing and phylogenetic analysis of 16S rRNA
Isolates representing different IGS types were used for 16S rRNA gene amplifications with the forward primer P1 and reverse primer P6 (Zhang et al. 2012). All the PCR products were checked by electrophoresis in agarose gel (1.0%, w/v) containing Goldview type I nucleotide dye and then sent for sequencing. The acquired sequences were deposited in the NCBI database and were used for alignment and reconstruction of the phylogenetic tree by using the NJ method (Neighbor-Joining) with 1000 bootstrap replications of each sequence.
Sequencing and phylogenetic analyses of housekeeping and symbiotic genes
All PCR amplifications were performed in 30 μl PCR mixtures containing dNTPs, buffer, primer, TaqE, and template DNA. The housekeeping genes atpD (encoding for ATP synthase beta chain), rpoB (RNA polymerase beta subunit), and glnII (glutamine synthetase II) that have been used to differentiate rhizobial species (Ji et al. 2015; Martens et al. 2008; Vinuesa et al. 2005a, b) were independently amplified by using primer pairs atpD255F/atpD782R, rpoB83F/rpoB1061R, and glnII12F/glnII689R, respectively (Vinuesa et al. 2005b). While the symbiotic genes of nodC and nifH were amplified by using primers pairs nodC540F/nodC1160R and nifH F/nifH R (Terefework et al. 2001), respectively, under the PCR conditions reported previously (Zhang et al. 2012), all purified amplicons were sequenced using the same primers for PCR in the Sangon Biotech company (Shanghai, China). The obtained sequences were 465 bp for atpD, 800 bp for rpoB, 600 bp for glnII, 336 bp for nodC, and 390 bp for nifH and deposited in the NCBI database. Phylogenetic analyses of the acquired sequences were performed using the procedure described above for 16S rRNA.
Multilocus sequence analysis (MLSA) of housekeeping genes
The acquired sequences of atpD, glnII, and rpoB were concatenated and aligned using ClustalW with the manually concatenated sequences of the corresponding genes of type strains for defined Mesorhizobium and Bradyrhizobium species, which were extracted from NCBI database (Zhang et al. 2017). Distance calculation and construction of the concatenated gene tree were performed with both the maximum likelihood (ML) and neighbor-joining (NJ) method and bootstrapping algorithms contained in the MEGA 6.0 (Tamura et al. 2013).
Correlation between soil characteristics, treatments, and rhizobial genotypes
The canonical version 4.54 of redundancy analysis (RDA) (Braak and Smilauer 2002) was used to examine the relationship among the genotypes of rhizobia and the soil parameters, totalnitrogen (TN), available phosphate (AP), available potassium (AK), organic matter (OM), total salt (TS), and pH value. Before the RDA, a linear or unimodal ordination model was determined by DCA (detrended canonical analysis) (Leps and Smilauer 2003). The maximal value of the lengths of the gradient in four ordination axes was below 3, suggesting that the linear gradient analysis model was more suitable, but the unimodal also could be used. The average values of the three replicates in each treatment (Table 2) and the total number of IGS types in a certain treatment (Table 1) were used for the correlation analysis.
Results
Bacterial isolation and nodulation tests
In this study, a total of 257 pure isolates were obtained and all of them could form single colonies ranging from 2 to 4 mm in diameter after 3–4 days of incubation on YMA at 28 °C. All isolates induced effective nodules on A. sinicus in subsequent inoculation tests, as evidenced by the red color inside the nodules and dark-green leaves of the plants. The negative controls had no nodule and grew poorly with short shoots and yellow-green leaves.
Soil characteristics and statistical analysis
The statistical analysis of physicochemical characteristics of soil samples corresponding to different treatments is available in Table 2. In general, the soil AK contents (93.00–99.00 mg kg−1) and pH values (5.63–5.90) for F1 to F6 treatments were lower than those (AK, 109–110 mg kg−1; pH, 6.53–6.70) in CF or CK treatments. Except the soil AP, most of the parameters did not differ significantly between the CF and CK treatments. Furthermore, the soil OM and TN increased while AP and TS decreased in the F1 to F6 treatments compared to CK and CF. Overall, OM and TN increased while the others decreased after the long-term input of green manure (F1 to F6) compared to those in CK and CF.
RFLP analysis
All isolates shared the same 16S rRNA RFLP pattern but were distinguished into 12 IGS types (Table 1). IGS types 1 and 2 had a prevalent distribution in all the treatments and type 1 is dominant. Furthermore, more IGS types (3–8 types) were found in F1 to F6 treatments than in CK or CF (only 2 types). Except types 1 and 2, the other 10 IGS types were only found in F1 to F6 at a frequency of one to four isolates in each IGS type per treatment.
Phylogenetic analyses of 16S rRNA and housekeeping genes
Sixteen strains representing the 12 IGS types from different treatments (CK, CF, F1 to F6) were chosen for further sequencing and phylogenetic analysis (Table 1). As shown in the 16S rRNA gene-based phylogeny (Suppl. Fig. S1, Table 1), all representative isolates were found in a single clade with sequence similarities ranging from 99.9 to 100% between them and from 99.1 to 100% with the type strains of 16 defined species, including Mesorhizobium jarvisii ATCC 33669T (100%) and Mesorhizobium huakuii IFO 15243T (99.6%).
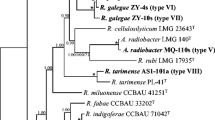
In the phylogeny (ML tree) of the concatenated sequences of atpD, glnII, and rpoB, the 16 representative strains were divided into two clusters (genospecies) within the genus Mesorhizobium (Fig. 1), and similar topology was observed in the phylogenies of the individual housekeeping genes (Suppl. Figs. S2, S3, and S4). Cluster 1 (C1) includes the 10 representative strains of IGS types 4, 9, 10, and 11 and Cluster 2 (C2) includes the representatives for the other 8 IGS types consisting of 247 strains (Fig. 1). The MLSA similarities among the strains were 99.3–100% intra-cluster and 97.4–98.2% inter clusters. Similarities of 99.6–99.9% between C1 members and M. huakuii USDA 4779T, and 99.2–99.7% between C2 members and M. jarvisii ATCC 33669T, while similarities less than 98.2% among the two clusters and the other references (Table 1) were observed. So, cluster 1 and cluster 2 were affiliated to M. huakuii and M. jarvisii, respectively. The topology in the ML tree was very similar with that of the NJ tree (Suppl. Fig. S6).
Maximum likelihood phylogenetic tree of MLSA based on concatenated sequences of atpD-glnII-rpoB, showing the relationships of the rhizobia isolated from Xinyang, China. The tree was constructed by using a distance matrix (Kimura 2-parameter model). Bootstrap confidence levels > 50% are indicated at the internodes. Bar = 2% nucleotide divergence. All the acquired sequences were used to reconstruct the phylogenic tree
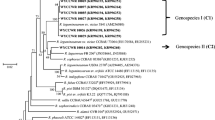
Phylogenies of nodC and nifH genes
In the nodC phylogenetic tree (Fig. 2), the symbionts of A. sinicus, including the 16 representative strains, M. huakuii 7653R, and M. qingshengii CCBAU 33460T formed a unique branch with 100% internal similarity. The nifH gene phylogeny (Suppl. Fig. S5) showed similar topology with internal branch similarities of 99.5–100% between strains.
Rhizobial distribution in the treatments
The MLSA cluster 2 (C2) was the main group in all treatments, representing 11.3–13.6% of the total isolates in different treatments, while cluster 1 (C1) was the minor group representing 0.4–1.6% of the total isolates in the green fertilizer treatments F2, F4, F5, and F6 (Table 1). At the level of IGS types, IGS type 1 (C2) was evenly distributed in all treatments with 20 to 33 isolates, ranging from 7.8 to 12.8% of the total isolates in each treatment, while the other prevalent IGS type (type 2 in C2) in all the treatments represented only 0.8 to 3.5%. The other IGS types in C1 or C2 were found in one to four treatments, such as type 4 in four different treatments (F2, F4, F5, and F6), while types 7 to 12 only in one of the F2, F3, or F6 treatments.
According to the RDA results (Fig. 3), soil pH, AP, TN, AK, OM, and TS had different effects on the distribution of the rhizobial groups (MLSA cluster and IGS types). The high values of pH, TS, AK, and AP strongly and positively selected the IGS type 1 (C2) and negatively selected the other 11 types except TS. In addition, IGS types 2, 3, 5, 7, and 8 in C2 and 4 and 9 in C1 were comprehensively and positively selected by high levels of TN and OM; and IGS types 6 and 12 in C2 and 10 and 11 in C1 were positively selected by both TS and OM. Furthermore, IGS type 1 (C2), together with the soil characters, TS, pH, AP, and AK, strongly correlated with CK and CF, two treatments without green fertilizer input. Soil OM and TN strongly correlated with treatments F3 and F5, respectively. Positive selection was observed for IGS types 2, 5, 7, 8, and 9 by treatments F3, F4, and F6; IGS type 3 and 4 by F5; and IGS types 6 and 10 to 12 by F1 and F2.
Biplot of the redundancy analysis (RDA) on the 12 IGS types of the isolates and the soil factors from soil samples collected from different treatments by CANOCO software v4.5. The gray arrows represent different soil properties and the black arrows show different IGS types. The longer the gray arrow, the greater the influence of that specific soil property on the distribution of the IGS types; the smaller the angle between the gray and black arrows, the closer the relationship between the soil factor and the rhizobial IGS types
Discussion
Due to the major importance and environmental value of A. sinicus, its nitrogen-fixing microsymbionts have been investigated in China since 1944, while M. huakuii (Chen et al. 1991) and M. qingshengii (Zheng et al. 2013) have been described previously for the rhizobia associated to A. sinicus in Zhejiang, Hubei, and Jiangxi provinces. In addition, diverse genotypes have been detected among the rhizobia of this plant by sequence analyses of 16S rRNA and 16S-23S IGS (Guo et al. 1999). In the present study, a collection of 257 rhizobial isolates trapped by A. sinicus from soil samples were characterized by genotypic analysis and some important findings were generated from the results.
Firstly, our study enlarged the diversity of A. sinicus rhizobia by detecting 12 IGS types within M. javisii and M. huakuii in the studied field. The identification of M. javisii with eight genotypes added an additional species for the A. sinicus-nodulating rhizobia, and the detection of M. huakuii demonstrated its universal distribution in the A. sinicus-cultivated area in China. In addition, our results demonstrated the inability of 16S rRNA gene sequence analyses (PCR-RFLP and phylogeny) in differentiation of M. huakuii and M. jarvisii (Supp. Fig. S1).
Secondly, our results evidenced a novel symbiovar in M. jarvisii that was originally isolated from Lotus corniculatus (Martínez-Hidalgo et al. (2015). The present study reports for the first time that M. jarvisii is the symbiont of A. sinicus dominant (96.1%) in soils at Xinyang City. Based upon phylogenies of symbiotic genes (nodC and nifH) (Fig. 2 and Suppl. Fig. S5) and housekeeping genes (atpD-glnII-rpoB) (Fig. 1) and results of nodulation tests, we propose a novel symbiovar, sv. astragali, in the species M. jarvisii.
Although conserved nodulation gene (nodC) in A. sinicus rhizobia was previously reported (Guo et al. 1999; Murooka et al. 1993; Zhang et al. 2000), the conservation in the nitrogen fixation gene nifH among these symbionts (Suppl. Fig. S5) is here reported for the first time. The identical or very similar symbiotic genes (Fig. 2 and Suppl. Fig. S5) in all M. jarvisii sv. astragali and M. huakuii isolates in this study and in the M. huakuii and M. qingshengii reference strains isolated from A. sinicus in other regions in China (Chen et al. 1991; Guo et al. 1999; Murooka et al. 1993; Zhang et al. 2000; Zheng et al. 2013) and in Japan (Murooka et al. 1993) demonstrated that this host legume exerts a stringent selection for symbiotic genes. Symbiotic genes may have been transferred horizontally among different Mesorhizobium species under the stringent selection by A. sinicus, with similar mechanisms as reported for rhizobia nodulating chickpea in China (Zhang et al. 2012; Zhang et al. 2017) and Lotus in New Zealand (Sullivan et al. 1996), which may have helped this plant to get the most adapted symbionts in different regions.
Thirdly, our results in the present study, consistent with previous studies (Chen et al. 1991; Zheng et al. 2013), demonstrated the biogeographic distribution pattern of the A. sinicus-nodulating rhizobial species and revealed the effects of fertilization practice on rhizobial distribution. It is clear that M. huakuii is dominant in Hunan and Anhui, M. jarvisii is the predominant symbiont in Xinyang, while M. qingshengii was found in Jiangxi (Zheng et al. 2013). This biogeographic pattern might be related to the soil characteristics, as reported for the soybean rhizobia in China (Han et al. 2009; Zhang et al. 2011).
It has been reported that long-term soil management modifies the soil characteristics which in turn regulate the soil microbial communities, including the rhizobial populations (Yan et al. 2014). The fact that only M. jarvisii was detected in the soils of CK and CF treatments (Table 1) demonstrated its greater adaption to the local soil in Xinyang than that of M. huakuii, while the occurrence of M. huakuii and more IGS types in the soils of treatments F1 through F6 implied that long-term application of A. sinicus biomass as green manure increased the biodiversity of rhizobia and favored M. huakuii for nodulation. In addition, the relative occupancy decrease for M. jarvisii IGS type 1 and increase for M. jarvisii IGS type 2 in the green manure fertilized soils (Table 1) were consistent with the decrease of soil pH, AP, AK, and TS, and the increase of OM and TN, implying that the higher OM and TN contents and lower pH, AP, AK, and TS stimulated the diversification of A. sinicus-nodulating rhizobia in the tested soil and selected/inhibited some genotypes for nodulation (Fig. 3 and Table 2). Therefore, the supplement of A. sinicus biomass increased soil OM and decreased AP, AK, TS, and pH values, which in turn modified the rhizobial community composition as reported previously (Chen et al. 2009). Furthermore, the findings of 8 genotypes in the F2 treatment and of 3 to 6 in the F1 and F3–F6 treatments (Table 1) might reflect the effects of dose of A. sinicus fertilization on the diversity of rhizobia, via modification of the soil characteristics.
In this study, all the rhizobial strains were isolated from root nodules of trapping plants. Previous several studies evidenced that rhizobial strains isolated from soils by trapping plants may differ from those isolated from nodules directly collected in fields (Duodu et al. 2006, 2007; Duodu et al. 2006; Odair et al. 2006); however, the other studies (Van Cauwenberghe et al. 2016) revealed that these two strategies were comparable and only few differences in the minor genotypes or species occurred, or showed no any difference (Harrison et al. 1987). However, the arguments about the bias in rhizobial isolates between the plant trapping and direct field-sampled nodules should not affect the conclusions in this study since no A. sinicus plant was cultivated in CK and CF treatments, which made the comparative study on rhizobial isolates from the field nodules impossible.
In conclusion, M. jarvisii was identified for the first time as the main rhizobial group associated with A. sinicus in the acidic soils at Xinyang. The A. sinicus-nodulating M. jarvisii strains were proposed as a novel symbiovar, sv. astragali. The long-term application of green manure may modify the soil characteristics, and in turn, increase the genetic diversity and favored M. huakuii to nodulate with Chinese milk vetch.
References
Braak C, Smilauer P (2002) CANOCO reference manual and CanoDraw for Windows user’s guide: sofware for canonical community ordination (version 4.5). Section on Permutation Methods Microcomputer Power, Ithaca, Newyork
Chen DM, Yuan L, Liu YR, Ji JH, Hou HQ (2017) Long-term application of manures plus chemical fertilizers sustained high rice yield and improved soil chemical and bacterial properties. Eur J Agron 90:34–42. https://doi.org/10.1016/j.eja.2017.07.007
Chen WC, Wang KR, Xie XL (2009) Effects on distributions of carbon and nitrogen in a reddish paddy soil under long-term different fertilization treatments. Chin J Soil Sci
Chen WX, Li GS, QI YL, Wang ET, Yuan HL, Li JL (1991) Rhizobium huakuii sp. nov. isolated from the root nodules of Astragalus sinicus. Int J Syst Bacteriol 41:275–280
Duodu S, Carlsson G, Huss-Danell K, Svenning MM (2007) Large genotypic variation but small variation in N2 fixation among rhizobia nodulating red clover in soils of northern Scandinavia. J Appl Microbiol 102:1625–1635
Duodu S, Nsiah EK, Bhuvaneswari TV, Svenning MM (2006) Genetic diversity of a natural population of Rhizobium leguminosarum biovar trifolii analysed from field nodules and by a plant infection technique. Soil Biol Biochem 38:1162–1165. https://doi.org/10.1016/j.soilbio.2005.07.015
Fang Y, Yan ZL, Chen JC, Wang F, Wang MK, Lin XJ (2015) Effect of chemical fertilization and green manure on the abundance and community structure of ammonia oxidizers in a paddy soil. Chil J Agric Res 75:488–496. https://doi.org/10.4067/s0718-58392015000500015
Gao SJ, Cao WD, Gao JS, Huang J, Bai JS, Zeng NH, Chang DN, Shimizu K (2017) Effects of long-term application of different green manures on ferric iron reduction in a red paddy soil in Southern China. J Integr Agric 16:959–966. https://doi.org/10.1016/s2095-3119(16)61509-5
Guo XW, Zhang XX, Zhang ZM, Li FD (1999) Characterization of Astragalus sinicus rhizobia by restriction fragment length polymorphism analysis of chromosomal and nodulation genes regions. Curr Microbiol 39:358–0364
Han L, Wang E, Han T, Liu J, Sui X, Chen W, Chen W (2009) Unique community structure and biogeography of soybean rhizobia in the saline-alkaline soils of Xinjiang, China. Plant Soil 324:291–305
Harrison S, Young JPW, Jones DG (1987) Rhizobium population genetics: effect of clover variety and inoculum dilution on the genetic diversity sampled from natural populations. Plant Soil 103:147–150
Ji ZJ, Yan H, Cui QG, Wang ET, Chen WX, Chen WF (2015) Genetic divergence and gene flow among Mesorhizobium strains nodulating the shrub legume Caragana. Syst Appl Microbiol 38:176–183. https://doi.org/10.1016/j.syapm.2015.02.007
Leps J, Smilauer P (2003) Multivariate analysis of ecological data using CANOCO. Cambridge University Press
Lin DH, Gu RS (1998) Chinese milk vetch. Fujian Science and Technology Publishing House
Martens M, Dawyndt P, Coopman R, Gillis M, De Vos P, Willems A (2008) Advantages of multilocus sequence analysis for taxonomic studies: a case study using 10 housekeeping genes in the genus Ensifer (including former Sinorhizobium). Int J Syst Evol Microbiol 58:200–214
Martinez-Hidalgo P, Ramirez-Bahena MH, Flores-Felix JD, Rivas R, Igual JM, Mateos PF, Martinez-Molina E, Leon-Barrios M, Peix A, Velazquez E (2015) Revision of the taxonomic status of type strains of Mesorhizobium loti and reclassification of strain USDA 3471T as the type strain of Mesorhizobium erdmanii sp. nov. and ATCC 33669T as the type strain of Mesorhizobium jarvisii sp. nov. Int J Syst Evol Microbiol 65:1703–1708. https://doi.org/10.1099/ijs.0.000164
Murooka Y, Xu Y, Sanada K, Araki M, Morinaga T, Yokota A (1993) Formation of root nodules by Rhizobium huakuii biovar renge, bv. nov. on Astragalus sinicus cv. Japan. J Ferment Bioeng 76:38–44
Nuswantara S, Fujie M, Yamada T, Malek W, Inaba M, Kaneko Y, Murooka Y (1999) Phylogenetic position of Mesorhizobium huakuii subsp. rengei, a symbiont of Astragalus sinicus cv. Japan. J Biosci Bioeng 87:49–55
Odair A, Glaciela K, Mariangela H (2006) Sampling effects on the assessment of genetic diversity of rhizobia associated with soybean and common bean. Soil Biol Biochem 38:1298–1307
Sullivan JT, Eardly BD, van Berkum P, Ronson CW (1996) Four unnamed species of nonsymbiotic rhizobia isolated from the rhizosphere of Lotus corniculatus. Appl Environ Microbiol 62:2818–2825
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729. https://doi.org/10.1093/molbev/msr121
Tan Z, Xu X, Wang E, Gao J, Martinez-Romero E, Chen W (1997) Phylogenetic and genetic relationships of Mesorhizobium tianshanense and related rhizobia. Int J Syst Evol Microbiol 47:874
Tena W, Wolde-Meskel E, Degefu T, Walley F (2017) Genetic and phenotypic diversity of rhizobia nodulating chickpea (Cicer arietinum L.) in soils from southern and central Ethiopia. Can J Microbiol 63:690–707. https://doi.org/10.1139/cjm-2016-0776
Terefework Z, Kaijalainen S, Lindstrom K (2001) AFLP fingerprinting as a tool to study the genetic diversity of Rhizobium galegae isolated from Galega orientalis and Galega officinalis. J Biotechnol 91:169–180
Van Cauwenberghe J, Lemaire B, Stefan A, Efrose R, Michiels J, Honnay O (2016) Symbiont abundance is more important than pre-infection partner choice in a Rhizobium-legume mutualism. Syst Appl Microbiol 39:345–349
Vauterin LaV P (1992) Computer-aided objective comparison of electrophoresis patterns for grouping and identification of microorganisms. Eur Microbiol 1:37–41
Vincent JM (1970) A manual for the practical study of rootnodule bacteria. International Biological Programme, Blackwell Scientific, Oxford
Vinuesa P, Silva C, Lorite MJ, Izaguirre-Mayoral ML, Bedmar EJ, Martinez-Romero E (2005a) Molecular systematics of rhizobia based on maximum likelihood and Bayesian phylogenies inferred from rrs, atpD, recA and nifH sequences, and their use in the classification of Sesbania microsymbionts from Venezuelan wetlands. Syst Appl Microbiol 28:702–716. https://doi.org/10.1016/j.syapm.2005.05.007
Vinuesa P, Silva C, Werner D, Martínez-Romero E (2005b) Population genetics and phylogenetic inference in bacterial molecular systematics: the roles of migration and recombination in Bradyrhizobium species cohesion and delineation. Mol Phylogenet Evol 34:29–54
Wang ET, Van Berkum P, Sui XH, Beyene D, Chen WX, Martínez-Romero E (1999) Diversity of rhizobia associated with Amorpha fruticosa isolated from Chinese soils and description of Mesorhizobium amorphae sp. nov. Int J Syst Bacteriol 49:51–65
Yan J, Han XZ, Ji ZJ, Li Y, Wang ET, Xie ZH, Chen WF (2014) Abundance and diversity of soybean-nodulating rhizobia in black soil are impacted by land use and crop management. Appl Environ Microbiol 80:5394–5402
Zhang JJ, Lou K, Jin X, Mao PH, Wang ET, Tian CF, Sui XH, Chen WF, Chen WX (2012) Distinctive Mesorhizobium populations associated with Cicer arietinum L. in alkaline soils of Xinjiang, China. Plant Soil 353:123–134. https://doi.org/10.1007/s11104-011-1014-5
Zhang JJ, Yang X, Guo C, de Lajudie P, Singh RP, Wang ET, Chen WF (2017) Mesorhizobium muleiense and Mesorhizobium gsp. nov. are symbionts of Cicer arietinum L. in alkaline soils of Gansu, Northwest China. Plant Soil 410:103–112. https://doi.org/10.1007/s11104-016-2987-x
Zhang JJ, Yu T, Lou K, Mao PH, Wang ET, Chen WF, Chen WX (2014) Genotypic alteration and competitive nodulation of Mesorhizobium muleiense against exotic chickpea rhizobia in alkaline soils. Syst Appl Microbiol 37:520–524. https://doi.org/10.1016/j.syapm.2014.07.004
Zhang XX, Turner SL, Guo XW, Yang HJ, Debelle F, Yang GP, Denarie J, Young JP, Li FD (2000) The common nodulation genes of Astragalus sinicus rhizobia are conserved despite chromosomal diversity. Appl Environ Microbiol 66:2988–2995
Zhang YM, Li Y, Chen WF, Wang ET, Tian CF, Li QQ, Zhang YZ, Sui XH, Chen WX (2011) Biodiversity and biogeography of rhizobia associated with soybean plants grown in the north China plain. Appl Environ Microbiol 77:6331–6342. https://doi.org/10.1128/aem.00542-11
Zheng WT, Li Y, Wang R, Sui XH, Zhang XX, Zhang JJ, Wang ET, Chen WX (2013) Mesorhizobium qingshengii sp nov., isolated from effective nodules of Astragalus sinicus. Int J Syst Evol Microbiol 63:2002–2007. https://doi.org/10.1099/ijs.0.044362-0
Funding
This work was financed by the China Agriculture Research System—Green Manure (Project No. CARS-22), the National Natural Science Foundation of China (Project No. 31400008 and 31571778), National Human and Social Security Ministry of Science and Technology for Overseas Returnees (Project No. 21104000005). ETW is financially supported by grants SIP20171259 authorized by Instituto Politécnico Nacional, Mexico.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Responsible Editor: Katharina Pawlowski.
Rights and permissions
About this article
Cite this article
Zhang, J., Shang, Y., Wang, E. et al. Mesorhizobium jarvisii sv. astragali as predominant microsymbiont for Astragalus sinicus L. in acidic soils, Xinyang, China. Plant Soil 433, 201–212 (2018). https://doi.org/10.1007/s11104-018-3830-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11104-018-3830-3