Abstract
Background and aims
Dieback is pervasive in many populations of invasive woody weeds globally. Previous studies on dieback have focused on specific potential causative biotic agents, but most cases remain unexplained. The potential role of endophytic microbial communities in dieback, including the relative importance of endophytes with pathogenic or protective capabilities, remains poorly studied. We tested whether changes in archaeal, bacterial and fungal endophyte community structure is associated with dieback occurrence in the invasive, leguminous tree, Parkinsonia aculeata L. (parkinsonia).
Methods
We sampled roots, stems and stem tips from healthy and dieback-affected parkinsonia and conducted terminal restriction fragment length polymorphism (T-RFLP) analysis on DNA extracted from these samples using domain-specific primers for archaea, bacteria and higher fungi.
Results
Microbial community composition strongly differed with parkinsonia disease status (archaea, bacteria and fungi) and plant part (archaea and fungi). Plant part and disease status effects were strongest in archaea. We also found evidence implicating both pathogenic and potentially protective endophytes in the onset of dieback.
Conclusions
This is the first study that has shown significant associations between changes in endophyte community composition and dieback presence. Our results highlight the complexity of those changes and provide support for the hypothesis that diverse pathogenic and protective endophytes may be implicated in dieback.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
We define dieback in perennial plants as a progressive reduction in health resulting in the death of plant parts and often outright death of the whole plant that is not age-related or explained by a specific stress such as fire or prolonged flooding. It can result in local extinctions, either as a gradual process over many years or as a more sudden occurrence. Reports of dieback are increasingly common in native and invasive plant species globally (La Porta et al. 2008). Microorganisms are often implicated (e.g., Mueller-Dombois 1987; Kowalski and Holdenrieder 2009; Herrero et al. 2011; Ismail et al. 2012; Mehl et al. 2013), and dieback in some tree and shrub species has been found to result from single pathogens, sometimes in the presence of stressors (Rice et al. 2004; La Porta et al. 2008; Pautasso et al. 2013; Scarlett et al. 2013; Aghighi et al. 2014). More often, causal explanations remain elusive (Houston 1992; Slippers and Wingfield 2007; Sakalidis et al. 2011; Jami et al. 2013). This might be due, in part, to complex interactions between pathogens, the host and the environment as described in dieback models by Houston (1992) and Manion (1991). To add a further layer of complexity, the absence of potentially protective endophytes, which normally improve the host’s resistance to pathogen infection (Arnold et al. 2003; Newcombe et al. 2009), might cause the host to become more susceptible to infection by generalist pathogens, resulting in dieback (Evans 2008). Explaining dieback is therefore difficult for several reasons. First, many organisms are resistant to culture (Hawksworth and Rossman 1997) and cultivation is often biased towards ubiquitous, fast-growing organisms which outcompete more specialised species with specific requirements (Aly et al. 2011). Second, dieback in individual plants may be caused by multiple pathogens. Third, expression of dieback is often mediated by complex interactions between these pathogen(s), the host and potential dieback suppressing organisms (Mendes et al. 2011). Improving our understanding of the role of all endophytes associated with dieback, including those not easily cultured, may therefore provide new insights into this phenomenon.
Over the past two decades there have been an increasing number of reports of dieback in invasive woody plants in Australia, including many of the 20 taxa ranked as weeds of national significance (Wilson and Pitkethley 1992; van Klinken et al. 2009; Bissett et al. 2012). These dieback-affected invasive plants include Mimosa pigra (Wilson and Pitkethley 1992; Sacdalan et al. 2012), Rubus anglocandicans (Aghighi et al. 2012), Acacia nilotica ssp. indica (Haque et al. 2012) and Chrysanthemoides monilifera ssp. rotundata (Morin et al. 2014). Reported symptoms include loss of leaves, browning of the stem tips that gradually moves down the plant, loss of vigour and death of plant parts and finally whole plant death. Population decline can be local or extensive and in some cases results in populations dying out (Manion 1991). Since there is potential for dieback to be an effective method of natural weed control, identification of the cause would be beneficial. Invasive tree decline has been observed in other countries too, for example in South Africa with Acacia cyclops (Impson et al. 2011) and Hakea sericea (Cronk and Fuller 1995), and in the USA with Melaleuca quinquenervia (Rayachhetry et al. 1997). These are all woody plant species native to Australia that have become serious invaders elsewhere. Dieback in invasive plants is at odds with the enemy-release hypothesis (Keane and Crawley 2002; Mitchell and Power 2003) which suggests that successful invasive plants often leave their natural, native enemies behind. In these cases however, there is no evidence that dieback occurs in the native ranges, suggesting that the cause of dieback is encountered upon introduction to the invasive range.
Recent studies show the important role endophytic communities play in plant health, and that interactions between these communities are complex, with members from at least three microbial domains (archaea, bacteria and fungi) potentially contributing to plant performance (Ruppel et al. 2013). Since all three groups may be involved in disease expression (as pathogens, mutualists or commensals), it is possible that interactions between these communities contribute to dieback occurrence or absence. Invasive plant species are not only likely to be exposed to a novel endophytic community (Mitchell and Power 2003) but may also have different suite of endophytes (Evans 2008). The enemy release hypothesis (Keane and Crawley 2002) suggests that upon introduction, some invasive plant species may leave behind specialist pathogens normally encountered in the native environment but maintain endophytes that confer potentially protective or beneficial effects. This gives the invasive species an advantage in the new range. Evans (2008) describes the endophyte-enemy release hypothesis where both beneficial endophytes and pathogens are left in the native range. Leaving native-range endophytes behind that normally protect the plant from pathogens could benefit the plant since they usually require resource allocation, but this is only advantageous if the host is not re-united with its natural enemies, as in the case of classical biocontrol. This is of particular concern since plants are constantly challenged with infection by potential pathogens from multiple microbial consortia. Over 33,000 archaeal and bacterial operational taxonomic units (OTUs) were detected in disease-suppressive soils, and certain soil-borne bacterial groups were consistently associated with disease suppression of the fungal plant pathogen Rhizoctonia solani (Mendes et al. 2013). Metabolites produced by endophytic bacteria (Brader et al. 2014) or fungi (Aly et al. 2011) may also play a role in disease suppression in their host. Since the presence of beneficial endophytes may prevent disease onset or progression, it is possible that dieback occurrence is a result of the lack of these endophytes, and not simply the presence of certain pathogens.
Classical pathology research on dieback has usually focused on either root-associated pathogens, or on endophytic pathogens from stems of affected plants. Nonetheless, microbial community structure has been shown to vary across different parts of plants, which provide unique habitats for these microorganisms (Rudgers and Orr 2009; Thongsandee et al. 2012). The cultivation of endophytic streptomycetes from above- and below-ground parts of Quercus serrata, for example, concluded that some species may be organ specific (Thongsandee et al. 2012) and a wider range of actinomycetes is thought to be associated with roots rather than with stems in rice (Tian et al. 2007). Archaeal, bacterial and fungal endophytes respond to changes in external environmental stress (Ruppel et al. 2013), which may affect different plant parts in different ways. These microorganisms can fulfil a range of functions, including carbon conversion throughout the plant, transport and stress response processes, methanogenesis in the rhizosphere (Knief et al. 2012) or via the production of secondary metabolites that alter host physiology and increase resistance to abiotic or biotic stressors (van Loon et al. 1998). Since microbial associations with host plants might differ depending on their location within the plant (e.g., arbuscular mycorrhizal fungi vs. other endophytes), both above- and below-ground endophytic communities should be considered when investigating the cause of dieback.
Parkinsonia aculeata L. (Leguminosae: Caesalpinioideae; commonly referred to as parkinsonia) is a tree invasive to northern Australian rangelands (Pichancourt and van Klinken 2012) originally introduced from South America (Hawkins et al. 2007). Parkinsonia now occurs across northern Australia, with its dense, thorny thickets impacting the pastoral industry and the environment (van Klinken and Heard 2012). Dieback has been observed across most of its Australian distribution (Diplock et al. 2008) and may be an important factor limiting its environmental and economic impacts (van Klinken et al. 2009). Although the effects are often spectacular, the cause of dieback in parkinsonia remains unknown. Parkinsonia occurs in diverse climates and habitats across Australia (Pichancourt and van Klinken 2012), so dieback is not obviously associated with specific stressors such as drought or flood. Extensive studies using classical pathology techniques have isolated generalist pathogens from parkinsonia stems (Diplock et al. 2006, 2008; Toh et al. 2008; Toh 2009), but have as yet failed to identify any causal link with the observed dieback phenomenon (van Klinken et al. 2009). A range of native herbivores are present on parkinsonia, and three biocontrol agents from the native range have been introduced, but insect herbivory on parkinsonia is generally negligible in Australia (van Klinken et al. 2009; van Klinken and Heard 2012). Extensive surveys of natural enemies have been conducted across the native range of parkinsonia (Bell et al. 2013) but dieback has never been observed there (van Klinken et al. 2009; van Klinken and Heard 2012).
We hypothesise that the occurrence of parkinsonia dieback is associated with a change in endophytic microbial community structure, and that this change might vary between different parts of the plant. Most dieback studies focus on pathogens from specific taxa – higher fungi in the case of parkinsonia. In this study we test whether multiple endophytic organisms are responsible for dieback, possibly as a disease complex with multiple taxa causing an infection or as part of a community involving both pathogenic and endophytic taxa. We expect pathogenic endophytes to be more closely associated with dieback-affected parkinsonia and protective endophytes with healthy parkinsonia.
Materials and methods
Tree sampling
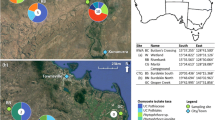
We sampled five trees from each of three replicate dieback-affected and three healthy populations of parkinsonia on pastoral properties between 20 and 40 km north of Charters Towers, Queensland (Fig. 1, Appendix Table 5). For each tree we sampled three roots, stems and stem tips and pooled the samples by plant part. Sites were paired into one healthy and one dieback-affected population such that, where possible, pairs were closer to each other than to other replicates (Fig. 1). Parkinsonia was categorised as healthy if plants were unambiguously healthy with 70–100 % foliage cover, all major and minor stems healthy, and no consistent vascular staining or browning of branches (Fig. 2a). Dieback-affected parkinsonia trees (Fig. 2b) were classified as live specimens that showed evidence of dieback including defoliation (1–40 % foliage cover), dead or dying stems (Fig. 2c), vascular staining when sampled (Fig. 2d) and evidence of tissue death in the stem tips. All trees sampled were mature: >25 cm trunk circumference at 30 cm above ground level and >2 m tall.
Tools for collecting samples were sterilized between sites by immersing in 50 % NaClO for 5 min, and dried with sterile paper towel before next use. Where possible, samples were collected first from healthy parkinsonia, followed by dieback-affected trees. Three roots, stems and stem tips (each 5–8 cm long) were sampled from each individual tree. Stems were sourced from secondary branches of at least 1 cm diameter. Stem tips were defined as the terminal points of the tertiary stems from where only rachi grow. Roots were primary or secondary roots which we could see as connected to the sampled tree and were at least 1 cm diameter. These were rinsed free of excess soil immediately using tap water. Samples from the same plant part of the same plant were stored together in sealed plastic bags with silicon packets. All samples were maintained at 4 °C until processed in the lab within 48–72 h.
Sample processing was conducted in a UV-sterilized laminar flow cabinet and all instruments were surface sterilised between samples using 95 % ethanol and a flame. Plant parts were vigorously pre-washed in sterile, distilled H2O for 20 s. For stems and stem tips a three stage ethanol-bleach-ethanol surface sterilization method was then used as recommended by Bills (1996). Roots were washed for 30 s in sterile, distilled H2O containing 0.1 % Tween-20™ since harsher sterilization techniques are not recommended for roots (Thorn et al. 2007). All samples were blotted dry with sterile filter paper and surface sterilization was checked by sliding sterilized tissue over the surface of media as an imprint, and incubating at 30 °C for a week or longer (Bacon and Hinton 2007). The epidermis of stem tips and stems, and a small portion of root cortex was then removed using a sterile scalpel. Samples were stored in sealed, individual sterile plastic containers at −20 °C until surface sterilisation was established. Once surface sterilized, the inner tissue was shaved and placed in sterile paper envelopes for DNA extraction.
Nucleic acid analysis
Sample shavings were freeze-dried for 48 h and then ground to a fine powder using a beadbeater for 2–3 min in 2 mL Eppendorf tubes® containing two sterile 2 mm steel ball bearings. The MO BIO PowerSoil® DNA Isolation Kit was then used to extract total DNA from approximately 20 mg of each powdered sample, according to manufacturer’s instructions, and DNA presence was confirmed on an agarose gel.
For T-RFLP, extracted DNA was amplified using taxon-specific primers (Table 1) as per Singh et al. (2006), except that amplification for each taxon was performed individually, rather than in a multiplex reaction. PCR reaction contained: MyTaq DNA polymerase (Bioline), 4 μl 5× MyTaq Reaction Buffer, 0.5 μL MyTaq polymerase, 5 μL extracted DNA, and varying concentrations of primer depending on target DNA (0.4 μM for fungi, 0.2 μM for bacteria and 1.0 μM for fungi). PCR conditions for archaea and bacteria comprised 1 cycle of 95 °C for 2 min; 30 cycles of 95 °C for 15 s, 55 °C for 15 s and 72 °C for 50 s, and a final extension step of 72 °C for 10 min. PCR conditions for fungi were optimised as 1 cycle of 94 °C for 3 min; 30 cycles of 95 °C for 15 s, annealing at 56–50 °C, dropping 1 °C each cycle then maintained at 50 °C for subsequent cycles, 72 °C for 1 min, and a final extension step of 72 °C for 10 min. PCRs were purified using the Wizard®SV Gel and PCR Clean-Up System (Promega), quantified spectrophotometrically (NanodropTM ND-1000, Thermoscientific) and 30 ng PCR product digested with the restriction endonuclease HindII. Digests were isopropanol precipitated, resuspended in 10 μl formamide containing LIZ600 size standard (Applied Biosystems), denatured at 95 °C (3 min) and separated by size using capillary electrophoresis (ABI PRISM 3130xl Genetic 110 Analyzer, Applied Biosystems).
Statistical data analysis
True peaks were selected in R (R Core Team 2014) using the method by Abdo et al. (2006) with minimum peak size 40, maximum at 600 by “Area” with 1 standard deviation and minimum threshold 20. Peaks were then ‘binned’ using Interactive Binner v1.4 (Ramette 2009) with the following constraints: min = 40, max = 600, MinRFI = 0.09, window size = 1, shift size = 0.9. The resulting file contained the raw relative abundance data, which were then analysed in Primer v6 (Clarke and Gorley 2006). Both presence/absence data and relative abundance data were analysed, but since the analyses showed similar results, only relative abundance data are reported here. OTU accumulation plots for dieback-affected and healthy samples were then constructed for each of the three taxa using the Chao1 index and 9999 permutations.
To estimate the influence of dieback occurrence and plant part on microbial community composition, we constructed Bray-Curtis dissimilarity matrices (Legendre and Legendre 1998) after square-root transformation of the archaeal, bacterial and fungal OTU abundance data. The transformed data were subjected to permutational multivariate analyses of variance (PERMANOVA) using type III sum of squares, permutation of residuals under a reduced model with 9999 permutations. This was conducted for interactions between plant part and disease status. Since the samples were gathered from multiple sites (Fig. 1), we also tested for geographic location effects on OTU community. We constructed canonical analysis of principle coordinates (CAP) graphs on the Bray-Curtis resemblance matrices using combined plant part and disease status factors as groups. Finally, we conducted indicator species analysis by plant part to test whether there were OTUs significantly associated with either dieback parkinsonia (potentially pathogenic endophytes) or healthy parkinsonia (potentially protective endophytes). This was done using the statistical package IndicSpecies v1.7.4 (De Cáceres and Legendre 2009) available through R (R Core Team 2014), using 999 permutations and a 0.01 alpha significance level.
Results
Sampling effort
The number of bacterial OTUs recovered from the T-RFLP analysis was far fewer than the number recovered from the archaeal and fungal analyses (Fig. 3), and for some samples there was no bacterial amplification (data not shown). With the exception of archaeal diversity in dieback plants, there was indications of OTU richness plateauing as the number of samples increased (Fig. 3). OTU richness in dieback samples was double that observed in healthy samples for archaea and fungi, but was unaffected by plant disease status for bacteria.
Plant part and disease status effects
The effects of plant part, disease status and the interaction between the two, on endophyte community composition were significant for each microbial group (Table 2). There were no effects on community composition due to the geographic location of sites (data not shown). Archaeal community composition was highly structured within healthy plants, being distinctive in each plant part (Fig. 4a). Archaeal communities in stem tips and roots was significantly different in dieback-affected plants compared to healthy plants (Table 3), with stem tip communities showing similarities with stem communities (Fig. 4a). Archaeal community composition in stems was similar for healthy and dieback plants (Table 3). Bacterial community composition differed significantly with health status for stems and tips but not roots (Table 3). However, it was not as structured as archaeal communities (Fig. 4a), with considerable unexplained variation apparent between samples (Fig. 4b). Explanations for this pattern are limited since Fig. 4 only describes the first two axes of ordination in the CAP analysis. The archaeal data was also plotted along the other axes (data not shown) but we did not find any evidence that archaeal community structure in dieback-affected stem tips differed to that in healthy or dieback stems. Bacterial community composition showed diversity within both stem and stem tip samples for healthy plants (Table 3), but this is greatly reduced in dieback plants with communities becoming more homogenous throughout the plant (and possibly more like that in healthy/dieback roots; Fig. 4b). Although fungal community composition was affected by disease status in all three plant parts (Table 3), it was not strongly structured (Fig. 4c). The effect of disease status on fungal communities was most apparent in roots (along CAP 1 axis in Fig. 4c) and stem tips (along CAP 2 axes in Fig. 4c).
Canonical analysis of principle coordinates (CAP) constrained by the interaction between Parkinsonia aculeata disease status and plant part for a archaeal, b bacterial and c fungal OTU community data. Axis label percentages indicate amount of variance explained. Shaded symbols indicate samples from dieback-affected trees and open symbols from healthy trees
Indicator species analysis
The indicator species analysis (Table 4) supports the patterns observed in the canonical analysis (Fig. 4). There were 12 archaeal OTUs significantly associated with specific plant parts within healthy plants, and a further two associated with stem tips and roots (Table 4). Most notably there were 18 archaeal OTUs associated with roots of dieback plants, representing 8 % of the archaeal community in dieback plants (Table 4). There were only four bacterial OTUs associated with healthy plants and one with dieback plants (Table 4), although this also reflected low OTU diversity (Fig. 2). OTUs associated with health status were proportionally lowest for fungi, of which most were in stem tips (Table 4).
Discussion
Dieback in plant populations is widely observed globally but in most cases remains difficult to explain, and its presence in invasive species is counter to general expectations (Callaway and Ridenour 2004). Our study is the first to show significant difference in microbial community composition between healthy and dieback-affected plants of an invasive species. Differences were evident in all studied microbial groups (archaea, bacteria and fungi), but were strongest for archaea, which is also the least understood group of microorganisms. Community composition was structured across plant parts, most strongly in archaea, which also showed interaction with health status. Differences in community structure was partially explained by OTUs that were affiliated either with healthy or dieback-affected plants. This provides some support for our hypothesis that both pathogenic and potentially protective endophytes are involved in parkinsonia dieback. These results extend current thinking, that multiple organisms within multiple taxonomic domains may be involved in the onset of disease or disease suppression in plants (Coats and Rumpho 2014).
Most studies on microbial endophyte communities and dieback investigate one specific microbial taxon, typically fungi (Wilson and Pitkethley 1992; Diplock et al. 2006; Kowalski and Holdenrieder 2009; Haque et al. 2012; Ismail et al. 2012; Sacdalan et al. 2012), although there are exceptions (Mendes et al. 2011; Knief et al. 2012; Ma et al. 2013; Oliveira et al. 2013; Ruppel et al. 2013). Our study found a very diverse microbial community (as assessed using OTUs) whose structure differed with plant health status. OTU diversity was high for archaea and fungi, and low for bacteria. However, the lack of resolution in the bacterial OTU data might be due to the use of only one restriction enzyme, since one bacterial OTU may represent more than one bacterial species (Avaniss-Aghajani et al. 1996). At least for fungi, high endophytic fungal biodiversity has previously been observed in plants, which can host hundreds of fungal species or genotypes (Hawksworth 2001). High diversity of bacterial and archaeal endophytes is also expected (Ma et al. 2013). Archaeal and fungal OTUs were twice as numerous per sample in dieback-affected plants compared to those in healthy plants. This may be due to the subsequent colonisation of dieback-affected tissue by opportunists or saprotrophs (Arnold 2007), although the indicator species analysis did not show a consistent pattern to support this. Alternatively, we suggest that the observed OTUs may have to co-occur to trigger dieback symptoms.
Microbial communities can differ by plant part due to their role as endophytes (e.g., production of alkaloids as herbivory inhibitors in above ground parts; Alexopoulos et al. 1996), the way in which these microorganisms are transmitted between hosts (via wind dispersal to aerial tissues or animal dispersal to roots or flowers), or whether infection by these microorganisms is systemic or local (Rudgers and Orr 2009; Knief et al. 2012; Thongsandee et al. 2012). Expectations of within-plant structuring were supported in our study irrespective of health status. Changes in community composition with plant health status differed according to plant part and endophyte taxon. Structuring was strongest for archaea and not as obvious for fungi, a comparison which has not been previously tested in the literature. Structuring of fungal community composition across plant parts may be the result of the function of fungal species differing with plant part (Rudgers and Orr 2009). However, since the majority of OTUs were not restricted to specific plant parts, we suggest that community structure across plant parts was driven by the presence or absence of the indicator OTUs. Most previous studies on dieback in parkinsonia have taken fungal isolations from stems (Diplock et al. 2006; Toh 2009). These and many other dieback studies could therefore be biased by limiting sampling to a particular plant part. Archaeal communities in stems were similar, regardless of plant disease status, but changed substantially in roots and stem tips of dieback-affected trees, with community structure in stem tips converging with that in stems. The functional significance of this, if any, is yet to be determined.
Endophytes in invasive plants can enhance their competitiveness and invasiveness in non-native ranges (Callaway et al. 2008; Mangla and Callaway 2008; Rudgers and Orr 2009; Rout et al. 2013) and the protective role of fungi and bacteria have been observed in other systems (Newcombe et al. 2009; Rout et al. 2013). The apparent absence of dieback in parkinsonia from the native range suggests that any potential dieback-causing pathogen(s) would have been acquired in Australia, contrary to the enemy-release hypothesis (Keane and Crawley 2002), or that endophytes protecting parkinsonia from pathogens in the native range are absent in Australia, which aligns more with the endophyte-enemy release hypothesis (Evans 2008). The association in our study of archaea, bacteria and fungi with both healthy and dieback-affected parkinsonia provides support that both protective and pathogenic endophytes could be involved in dieback in Australia, potentially as new associations. Most notable was a diverse archaeal community associated with dieback-affected roots. The OTUs associated with dieback-affected plants may be opportunists that take over the niche left by those endophytes which have been displaced as a result of dieback-causing organisms. Conversely, these OTUs may be pathogens in their own right and infected parkinsonia upon introduction to Australia, causing dieback either before or after the loss of OTUs unique to healthy parkinsonia.
We found preliminary support for the presence of potentially protective archaeal endophytes in healthy stem tips. We also found evidence for potential protective endophytes from the fungi and bacteria, as expected (although it should be noted that these OTUs might also represent endophytes which are neutral and are displaced by opportunists with the onset of dieback). Previous experimental studies have shown that some fungal endophyte species do limit damage to their hosts by pathogens (Arnold et al. 2003) and some bacterial endophytes have a positive effect on disease-affected hosts (Bacon and Hinton 2007). However, the potential pathogenic and protective organisms identified in our study need to be identified, and whether they are causally related to dieback (or confer protection) is yet to be ascertained. To determine this, a sequencing-based approach is required, followed by isolation and pathogenicity trials. Since multiple taxa were identified as potential causal agents of dieback or to confer protection, the next step might be to utilise innovative inoculation strategies to test entire communities. This could be done in a minimally-invasive inoculation trial by inoculating healthy plants using ground-up material from different healthy or dieback-affected plant parts. Furthermore, it is important to compare endophyte community composition of invasive Australian parkinsonia in this study, to that of parkinsonia from native populations in meso-America. We also note that due to the specificity of the primers selected for this study, communities other than those characterised might also contribute to dieback occurrence.
Historically, the focus in dieback studies has been on potential fungal pathogens, but our results have shown that a diverse community of archaea are present in both healthy and dieback-affected parkinsonia. Despite archaeal communities showing the most significant difference between healthy and dieback-affected parkinsonia, very few archaeal species have been identified in the literature – e.g., methanogens associated with human or animal health – and even fewer have been cultured (Schleper et al. 2005). Without representative cultures of these species, it is difficult to observe or analyse their ecosystem function, physiology or pathogenic potential (Killham and Prosser 2007). Additionally, no known archaeal pathogen has been identified or shown to be the primary cause of disease in any plant or animal (Cavicchioli 2011), so we can only make assumptions about the significance of the results for archaea presented in this study. At most we can conclude that archaea are more involved in dieback than previously thought, and should not be ignored in future studies. From this perspective, the next step might be to evaluate gene expression associated with archaea in healthy and dieback-affected tissue in order to infer ecological functions, and sequencing to identify known species.
Conclusion
Our study demonstrated the value of taking a community-level analysis approach to investigating the complex phenomenon that is dieback, which has allowed us to analyse the composition and structure of these communities. Using this approach, we have shown that there are strong correlations between changes in endophytic archaeal and fungal community structure, and the occurrence of dieback in parkinsonia, and that the degree of structuring also varies by plant part. Nevertheless, one difficulty is that very little is known about one of these domains: archaea, but we now know that archaea should be considered an important part of the dieback phenomenon. Despite this, a number of OTUs have been implicated as potentially pathogenic or protective endophytes. These results open new avenues for research into understanding the dieback phenomenon. For invasive species this may also lead to novel management solutions.
Abbreviations
- T-RFLP:
-
Terminal restriction fragment length polymorphism
- OTU:
-
Operational taxonomic unit
- PERMANOVA:
-
Permutational multivariate analysis of variance
- CAP:
-
Canonical analysis of principle coordinates
References
Abdo Z, Schuette UME, Bent SJ, Williams CJ, Forney LJ, Joyce P (2006) Statistical methods for characterizing diversity of microbial communities by analysis of terminal restriction fragment length polymorphisms of 16S rRNA genes. Environ Microbiol 8:929–938
Aghighi S, Hardy GEJ, Scott JK, Burgess TI (2012) Phytophthora bilorbang sp nov., a new species associated with the decline of Rubus anglocandicans (European blackberry) in Western Australia. Eur J Plant Pathol 133:841–855
Aghighi S, Fontanini L, Yeoh PB, Hardy GES, Burgess TI, Scott JK (2014) A conceptual model to describe the decline of European blackberry (Rubus anglocandicans), a weed of national significance in Australia. Plant Dis 98(5):580–589
Alexopoulos CJ, Mims CW, Blackwell M (1996) Introductory mycology, 4th edn. WIley, NY
Aly AH, Debbab A, Proksch P (2011) Fungal endophytes: unique plant inhabitants with great promises. Appl Environ Microbiol 90:1829–1845
Arnold AE (2007) Understanding the diversity of foliar endophytic fungi: progress, challenges, and frontiers. Fungal Biol Rev 21:51–66
Arnold AE, Mejia LC, Kyllo D, Rojas EI, Maynard Z, Robbins N, Herre EA (2003) Fungal endophytes limit pathogen damage in a tropical tree. Proc Natl Acad Sci U S A 100:15649–15654
Avaniss-Aghajani E et al (1996) Molecular technique for rapid identification of mycobacteria. J Clin Microbiol 34:98–102
Bacon CW, Hinton DM (2007) Isolation, in planta detection, and uses of endophytic bacteria. In: Hurst C, Crawford RL, Mills AL, Garland JL, Stetzenbach LD, Lipson DA (eds) Manual of environmental microbiology, 3rd edn. ASM Press, Washington, DC, pp 638–651
Bell KL, Heard TA, Manion G, Ferrier S, van Klinken RD (2013) The role of geography and environment in species turnover: phytophagous arthropods on a Neotropical legume. J Biogeogr 40:1755–1766
Bills GF (1996) Isolation and analysis of endophytic fungal communities from woody plants. In: Redlin SC, Carris LM (eds) American phytopathological society. St. Paul, Minnesota, pp 31–65
Bissett A, Morin L, Scott J, Galea V, Goulter K, van Klinken RD (2012) Dieback of WoNS: their cause and prospects for biocontrol. In: National Weeds Research. Rural Industries Research and Development Corporation, Canberra, ACT, pp 116–117 https://rirdc.infoservices.com.au/downloads/12-079 Accessed 12 January 2015
Brader G, Compant S, Mitter B, Trognitz F, Sessitsch A (2014) Metabolic potential of endophytic bacteria. Curr Opin Biotechnol 27:30–37
Callaway RM, Ridenour WM (2004) Novel weapons: invasive success and the evolution of increased competitive ability. Front Ecol Environ 2:436–443
Callaway RM et al (2008) Novel weapons: invasive plant supresses fungal mutualists in America but not in its native Europe. Ecology 89:1043–1055
Cavicchioli R (2011) Archaea - timeline of the third domain Nat. Rev Microbiol 9:51–61
Clarke KR, Gorley RN (2006) Primer v6. PRIMER-E, Plymouth
Coats V, Rumpho ME (2014) The rhizosphere microbiota of plant invaders: an overview of recent advances in the microbiomics of invasive plants. Front Microbiol 5:1–10
Cronk QCB, Fuller JF (1995) Plant invaders vol 2. ‘People and Plants’ conservation manuals. Chapman & Hall, London
De Cáceres M, Legendre P (2009) Associations between species and groups of sites: indices and statistical inference. Ecology 90:3566–3574
Diplock N, Galea V, van Klinken RD, Wearing A (2006) A preliminary investigation of dieback on Parkinsonia aculeata. Paper presented at the 15th Australian Weeds Conference, Adelaide
Diplock N, Galea V, van Klinken RD (2008) Movement of dieback through a stand of Parkinsonia - a time series study. Paper presented at the 16th Australian Weeds Conference, Cairns
Evans HC (2008) The endophyte-enemy release hypothesis: implications for classical biological control and plant invasions. In: Julien MH, Sforza R, Bon MC, Evans HC, Hatcher PE, Hinz HL, Rector BG (eds) Proceedings of the XII International Symposium on Biological Control of Weeds, La Grande Motte, France, 2008. CAB International
Gardes M, Bruns TD (1993) ITS primers with enhanced specificity for basidiomycetes–application to the identification of mycorrhizae and rusts. Microb Ecol 2:113–118
Giovannoni SJ, Delong EF, Olsen GJ, Pace NR (1988) Phylogenetic group-specific oligodeoxynucleotide probes for identification of single microbial cells. J Bacteriol 170:720–726
Haque A, Galea V, Goulter K (2012) A preliminary investigation of prickly acacia dieback (Acacia nilotica ssp. indica). Paper presented at the 18th Australian Weeds Conference, Melbourne
Hauben L, Vauterin L, Swings J, Moore ERB (1997) Comparison of 16S ribosomal DNA sequences of all Xanthomonas species. Int J Syst Bacteriol 47:328–335
Hawkins JA, Boutaoui N, Cheung KY, van Klinken RD, Hughes CE (2007) Intercontinental dispersal prior to human translocation revealed in a cryptogenic invasive tree. New Phytol 175:575–587
Hawksworth DL (2001) The magnitude of fungal diversity: the 1.5 million species estimate revisited. Mycol Res 105:1422–1432
Hawksworth DL, Rossman AY (1997) Where are all the undescribed fungi? Phytopatholgy 87:888–891
Herrero ML, Toppe B, Brurberg MB (2011) First report of Phytophthora ramorum causing shoot dieback on bilberry (Vaccinium myrtillus) in Norway. Plant Dis 95:355
Houston DR (1992) A host-stress-saprogen model for forest dieback-decline diseases. In: Manion PD, Lachance D (eds) Forest decline concepts. The American Phytopathological Society, St. Paul, pp 3–25
Impson FAC, Kleinjan CA, Hoffmann JH, Post JA, Wood AR (2011) Biological control of Australian Acacia species and Paraserianthes lophantha (Willd.) Nielsen (Mimosaceae) in South Africa. Afr Entomol 19:186–207
Ismail AM, Cirvilleri G, Polizzi G, Crous PW, Groenewald JZ, Lombard L (2012) Lasiodiplodia species associated with dieback disease of mango (Mangifera indica) in Egypt. Australas Plant Pathol 41:649–660
Jami F, Slippers B, Wingfield MJ, Gryzenhout M (2013) Greater Botryosphaeriaceae diversity in healthy than associated diseased Acacia karroo tree tissues. Australas Plant Pathol 42:421–430
Jurgens G, Lindstrom K, Saano A (1997) Novel group within the kingdom Crenarchaeota from boreal forest soil. Appl Environ Microbiol 63:803–805
Keane RM, Crawley MJ (2002) Exotic plant invasions and the enemy release hypothesis. Trends Ecol Evol 7:164–170
Killham K, Prosser JI (2007) The prokaryotes. In: Eldor AP (ed) Soil microbiology, ecology, and biochemistry. Elsevier, Oxford, pp 119–144
Knief C et al (2012) Metaproteogenomic analysis of microbial communities in the phyllosphere and rhizosphere of rice. ISME J 6:1378–1390
Kowalski T, Holdenrieder O (2009) The teleomorph of Chalara fraxinea, the causal agent of ash dieback. For Pathol 39:304–308
La Porta N, Capretti P, Thomsen IM, Kasanen AM, Hietala AM, Von Weissenberg K (2008) Forest pathogens with higher damage potential due to climate change in Europe. Can J Plant Pathol 30(2):177–195
Legendre P, Legendre L (1998) Numerical ecology. Elsevier Science, Amsterdam
Ma B, Lv XF, Warren A, Gong J (2013) Shifts in diversity and community structure of endophytic bacteria and archaea across root, stem and leaf tissues in the common reed, Phragmites australis, along a salinity gradient in a marine tidal wetland of northern China. Antonie Van Leeuwenhoek 104:759–768
Mangla S, Callaway RM (2008) Exotic invasive plant accumulates native soil pathogens which inhibit native plants. J Ecol 96(1):58–67
Manion PD (1991) Tree disease concepts. Prentice-Hall, Englewood Cliffs
Marchesi JR, Sato T, Weightman AJ, Martin TA, Fry JC, Hiom SJ, Wade WG (1998) Design and evaluation of useful bacterium-specific PCR primers that amplify genes coding for bacterial 16S rRNA. Appl Environ Microbiol 64:795–799
Mehl JWM, Slippers B, Roux J, Wingfield MJ (2013) Cankers and other diseases cause by the botryosphaeriaceae. CABI, Wallingford
Mendes R et al (2011) Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science 332:1097–1100
Mendes R, Garbeva P, Raaijmakers JM (2013) The rhizosphere microbiome: significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol Rev 37:634–663
Mitchell CE, Power AG (2003) Release of invasive plants from fungal and viral pathogens. Nature 421:625–627
Morin L, Bissett A, van Klinken RD (2014) Do fungal endophytes play a role in bitou bush ‘Sudden Death Syndrome’ in Australia? In: Impson FAC, Kleinjan CA, Hoffmann JH (eds) XIV International Symposium on the Biological Control of Weeds, Cape Town, South Africa, 2014. University of Cape Town, p 97
Mueller-Dombois D (1987) Natural dieback in forests. Bioscience 37:575–583
Newcombe G et al (2009) Endophytes influence protection and growth of an invasive plant. Commun Integr Biol 2:29–31
Oliveira MNV et al (2013) Endophytic microbial diversity in coffee cherries of Coffea arabica from southeastern Brazil. Can J Plant Pathol 59:221–230
Pautasso M, Aas G, Queloz V, Holdenrieder O (2013) European ash (Fraxinus excelsior) dieback - A conservation biology challenge. Biol Conserv 158:37–49
Pichancourt JB, van Klinken RD (2012) Phenotypic Plasticity Influences the Size, Shape and Dynamics of the Geographic Distribution of an Invasive Plant. PLoS One 7:12
R Core Team (2014) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna
Ramette A (2009) Quantitative community fingerprinting methods for estimating abundance of operational taxanomic units in natural microbial communities. Appl Environ Microbiol 75:2495–2505
Rayachhetry MB, Elliott ML, Van TK (1997) Natural epiphytotic of the rust Puccinia psidii on Melaleuca quinquenervia in Florida. Plant Dis 81:831
Rice K, Matzner S, Byer W, Brown J (2004) Patterns of tree dieback in Queensland, Australia: the importance of drought stress and the role of resistance to cavitation. Oecologia 139:190–198
Rout ME, Chrzanowski TH, Westlie TK, DeLuca TH, Callaway RM, Holben WE (2013) Bacterial endophytes enhance competition by invasive plants. Am J Bot 100:1726–1737
Rudgers JA, Orr S (2009) Non-native grass alters growth of native tree species via leaf and soil microbes. J Ecol 97:247–255
Ruppel S, Franken P, Witzel K (2013) Properties of the halophyte microbiome and their implications for plant salt tolerance. Funct Plant Biol 40:940–951
Sacdalan A, Galea V, Goulter K, Elliot L, Van Klinken RD (2012) Preliminary investigations of the Mimosa pigra dieback phenomenon. Paper presented at the 18th Australian Weeds Conference, Melbourne
Sakalidis ML, Hardy GES, Burgess T (2011) Endophytes as potential pathogens of the baobab species Adansonia gregorii: a focus on the Botryosphaeriaceae. Fungal Ecol 4:1–14
Scarlett K, Guest D, Daniel R (2013) Elevated soil nitrogen increases the severity of dieback due to Phytophthora cinnamomi. Australas Plant Pathol 42:155–162
Schleper C, Jurgens G, Jonuscheit M (2005) Genomic studies of uncultivated archaea. Nat Rev Micro 3:479–488
Singh BK, Nazaries L, Munro S, Anderson IC, Campbell CD (2006) Use of multiplex terminal restriction fragment length polymorphism for rapid and simultaneous analysis of different components of the soil microbial community. Appl Environ Microbiology 72:7278–7285
Slippers B, Wingfield MJ (2007) Botryosphaeriaceae as endophytes and latent pathogens of woody plants: diversity, ecology and impact. Fungal Biol Rev 21:90–106
Thongsandee W, Matsuda Y, Shimizu M, Ehara H, Ito S (2012) Isolation of endophytic streptomycetes from above- and belowground organs of Quercus serrata. J For Res 18:179–189
Thorn RG, Scott J, Lachance MA (2007) Methods for studying terrestrial fungal ecology and diversity. In: Reddy CA (ed) Methods for general and molecular microbiology, 3rd edn. ASM Press, Washington, pp 929–950
Tian X, Cao L, Tan H, Han W, Chen M, Liu Y, Zhou S (2007) Diversity of cultivated and uncultivated actinobacterial endophytes in the stems and roots of rice. Microb Ecol 53:700–707
Toh R (2009) Investigation of fungi pathogenic towards seedlings of Parkinsonia aculeata – their potential for use as mycoherbicides. Dissertation, The University of Queensland
Toh R, Galea V, Diplock N, van Klinken RD (2008) Evaluation of fungal isolates for potential use as mycoherbicides for seed bank reduction of Parkinsonia aculeata. Paper presented at the 16th Australian Weeds Conference, Cairns
van Klinken RD, Heard TA (2012) Parkinsonia aculeata L. – parkinsonia. In: Julien MH, McFadyen R, Cullen JM (eds) Biological control of weeds in Australia. CSIRO Publishing, Canberra, pp 437–446
van Klinken RD, Campbell SD, Heard TA, McKenzie J, March N (2009) The biology of Australian weeds: Parkinsonia aculeata L. Plant Prot Quart 24:100–117
van Loon LC, Bakker P, Pieterse CMJ (1998) Systemic resistance induced by rhizosphere bacteria. Annu Rev Phytopathol 36:453–483
White TJ, Bruns T, Lee S, Taylor J (1990) Analysis of phylogenetic relationship by amplification and direct sequencing of ribosomal RNA genes. In: Innis MA, Gelfond DH, Sainsky JJ, White TJ (eds) PCR protocol: a guide to method and applications. Academic, New York, pp 315–322
Wilson CG, Pitkethley RN (1992) Botryodiplodia dieback of Mimosa pigra, a noxious weed in northern Australia. Plant Pathol 41:777–779
Acknowledgments
We would like to thank Kurt Rheinhart, an anonymous reviewer and our internal reviewers Dr Louise Morin and Dr Donald Gardiner (CSIRO). We also thank Kelli Pukallus (Biosecurity Queensland) for field work assistance; Dr Donald Gardiner and colleagues (CSIRO) for laboratory support with the extractions; and Dr Shamsul Hoque (CSIRO) for technical support with the T-RFLP lab work. This project was supported by Meat and Livestock Australia via a technical assistance grant (B.STU.0271) and the Australian Government via an Australian Postgraduate Award.
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible Editor: Birgit Mitter.
Appendix
Appendix
Rights and permissions
About this article
Cite this article
Steinrucken, T.V., Bissett, A., Powell, J.R. et al. Endophyte community composition is associated with dieback occurrence in an invasive tree. Plant Soil 405, 311–323 (2016). https://doi.org/10.1007/s11104-015-2529-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11104-015-2529-y