Abstract
Aims
Root system architecture traits (RSAT) are crucial for crop productivity, especially under drought and low soil fertility. The “shovelomics” method of field excavation of mature root crowns followed by manual phenotyping enables a relatively high throughput as needed for breeding and quantitative genetics. We aimed to develop a new sampling protocol in combination with digital imaging and new software.
Methods
Sampled rootstocks were split lengthwise, photographed under controlled illumination in an imaging tent and analysed using Root Estimator for Shovelomics Traits (REST). A set of 33 diverse maize hybrids, grown at 46 and 192 kg N ha−1, was used to evaluate the method and software.
Results
Splitting of the crowns enhanced soil removal and enabled access to occluded traits: REST-derived median gap size correlated negatively (r = −0.62) with lateral root density based on counting. The manually measured root angle correlated with the image-derived root angle (r = 0.89) and the horizontal extension of the root system (r = 0.91). The heritabilities of RSAT ranged from 0.45 to 0.81, comparable to heritabilities of plant height and leaf biomass.
Conclusion
The combination of the novel crown splitting method, combined with imaging under controlled illumination followed by automatic analysis with REST, allowed for higher throughput while maintaining precision. The REST Software is available as supplement.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Enormous breeding efforts and the establishment of modern high-yielding hybrid cultivars contributed substantially to increased maize (Zea mays L.) yields during the last decades (Hammer et al. 2009; Abendroth et al. 2011). Generally, shoot traits like short anthesis silking intervals, increased starch contents in the kernels and tolerance of high stand densities were of interest (Abendroth et al. 2011). Due to the lack of high-throughput phenotyping systems and ignorance of trait utility, root traits have not been directly selected so far (Campos et al. 2004; Zhu et al. 2011; Passioura 2012). Based on computer simulations, Hammer et al. (2009) proposed that the root systems of maize have become steeper and deeper due to the selection towards higher stand densities. This greater rooting depth may have contributed to increased water uptake and yield in the U.S. Corn Belt during the last decades. There is some experimental evidence that a more vertical orientation of the roots leads to increased rooting depth and is an adaptation to low nitrogen conditions (Trachsel et al. 2013).
The increasing number of scientific publications dealing with crop root systems indicates the growing awareness regarding their importance for crop productivity (Zhu et al. 2011; Lynch 2013). Root system architecture traits (RSAT), which determine the spatial configuration of coarse root structures in the soil, plays a crucial role (Lynch 1995). To allow for comparison between species, root traits are distributed into distinct classes following the terminology proposed by the international society of root research (Zobel and Waisel 2010) and further divided into subclasses by Zobel (2011). In the case of mature root stocks of maize, mainly the subclass of stem nodal roots within the class of shoot-born roots can be observed. In case of maize, these nodal roots can be further divided into below ground nodal roots (crown roots) and above ground nodal roots (prop or brace root). The terms crown and prop roots are widely used in the maize literature (Hund et al. 2011) and, therefore, also adopted here. Major traits include nodal root angles, which correlate with rooting depth (Araki et al. 2000; Trachsel et al. 2013; Grieder et al. 2014), the number of nodal roots (Giuliani et al. 2005; Lynch 2013; Saengwilai et al. 2014) and the density and length of lateral roots (Wiesler and Horst 1994; Wu et al. 2005; Yu et al. 2007).
Remarkable genotypic diversity of root system architecture has been reported in artificial systems and in the field for young and fully developed plants. Information about root system architecture is mainly derived from plants growing under controlled conditions: Significantly different nodal root angles at the two-leaf stage were observed among nine inbred lines (Hund 2010) and two hybrids (Singh et al. 2010). Hund (2010) also confirmed the positive effect of steeper roots on rooting depth during early growth. A study with 74 maize inbred lines showed differences in root numbers 14 days after planting: nodal root numbers ranged from 1.5 to 5.8 per plant; lateral root numbers ranged from 40.8 to 235.7 per plant (Kumar et al. 2012). Liu et al. (2008) observed significant differences in terms of lateral root length. Since the root system architecture can change tremendously during growth, it is important to monitor the entire growth period under field conditions (Hund et al. 2011; Kumar et al. 2012; Passioura 2012). A number of studies tackled this challenge: at the changeover of vegetative to reproductive growth, where the root system reaches its maximum extent (Hochholdinger 2009), highly diverse nodal root angles were reported among genotypes. Panel screenings in the field with more than 200 inbred lines showed angles of ten to almost ninety degrees from horizontal (Giuliani et al. 2005; Grift et al. 2011; Trachsel et al. 2011). Also the rooting depth (Worku et al. 2012; Trachsel et al. 2013) and the number of nodal roots of mature plants varied strongly among different accessions (Trachsel et al. 2011; Burton et al. 2013). Furthermore, branching densities of lateral roots ranging from five to more than 30 lateral roots per centimeter have been reported (Giuliani et al. 2005; Zhu et al. 2005a; Trachsel et al. 2011; Worku et al. 2012; Burton et al. 2013). Studying the relationship between all these traits and their utilities for soil resource acquisition will be important for understanding how root traits integrate to give rise to whole root system functioning (York et al. 2013).
Pioneering work about crop root systems in the field showed basic characteristics of root system architecture and its responses to edaphic factors (Weaver 1925; Kutscherea and Lichtenegger 1960). However, for the incorporation of RSAT into breeding programs, phenotyping approaches are needed that enable high-throughput evaluation of mature root systems in the field (Campos et al. 2004; Zhu et al. 2011; Passioura 2012). Such methods were used to characterize smaller samples of field-grown varieties (Stoffella et al. 1979; Kahn and Stoffella 1991), but still lacked the required number of genotypes needed for genetic mapping. The need of higher throughput for the characterization of root traits of mapping populations led to new concepts of root phenotyping. Zhong et al. (2009) used a support vector machine to analyse the complexity of maize root stocks by means of image analysis. Grift et al. (2011) developed an imaging methodology in combination with the digita l measurement of the outermost root angle and the fractal dimensions of excavates root stocks of maize. Trachsel et al. (2011) coined the term “shovelomics,” for the process of excavating and evaluating large numbers of field-grown plants for basic root characteristics. Though the initial “shovelomics” method was based on simple rating (Trachsel et al. 2011), it was subsequently updated to use manual measurements (Trachsel et al. 2013). The time needed to assess the RSAT by scoring was 2 min per rootstock (Trachsel et al. 2011) while no time requirements for the manual measurement were reported. To complement shovelomics with image processing the software Digital imaging of root traits (DIRT) was developed, allowing to measure about 30 root traits of dicots and monocots with an online interface (Bucksch et al. 2014). However, whereas DIRT focused on image analysis capable of handling a variety of image qualities, progress would likely be made by improving image quality and sample preparation. For example, current manual shovelomics and the DIRT-based approach only phenotype the outer brace and crown roots which occlude the internal root system and ignore the root traits of the younger nodes even though these roots contribute greatly to maize growth.
Successful incorporation of RSAT into modern breeding schemes is highly dependent on the heritability of the respective traits. The correlation between trait expression in hybrids and their parental lines is still unclear or weak (Chun et al. 2005; Gallais and Coque 2005; Coque et al. 2008; Hochholdinger and Tuberosa 2009). In early vegetative growth stages broad sense heritabilities of the number of nodal roots of 0.34 (Hund et al. 2004) and 0.65 (Kumar et al. 2012) were observed. Kumar et al. (2012) estimated the heritability for the number of lateral roots as 0.81. The estimations for the length of lateral and axial roots were in the same range as for the branching densities (Hund et al. 2004; Ruta et al. 2010; Kumar et al. 2012). Furthermore, that these heritabilities tend to decrease under abiotic stress due to an increasing environmental variance has been observed (Gallais and Hirel 2004; Ruta et al. 2010).
In this study, the approaches described by Trachsel et al. (2011) and Grift et al. (2011) were combined and partially modified. By using a set of test hybrids, with a common father line, heritabilities of RSAT and shoot traits could be compared. To obtain information about the outer shape and inner complexity of the root systems at once, root crowns were split lengthwise. The aim was to facilitate washing and enable an insight into the root crowns without the need of cutting off single roots. Manual phenotyping of the RSAT was done similar to Trachsel et al. (2011) and image derived parameters were obtained from standardized images photographed under controlled illumination in an imaging tent. The tent is constructed in a way to place the root in the imaging plane of the camera, while the black, light absorbing background is 0.58 m further away. This diffuse background differs strongly in its pixel values compared to the root object. The resulting images were processed with the newly developed software Root Estimator for Shovelomics Traits (REST), which is available as supplement or from SorceForge (rest4roots).
The objectives of the current study were (i) to develop a new sampling and imaging strategy in combination with a custom image analysis software, (ii) to evaluate the power of this approach to describe the root system architecture by comparing image-derived traits with manually assessed traits and (iii) to obtain a general idea about the degree of heritability of RSAT and its robustness under nitrogen deficiency. The utility of the approach was tested using data from a field experiment in South Africa with 33 test hybrids and two levels of nitrogen fertilization (46 and 192 kg N ha−1).
Material and methods
Experimental site
Experiments were carried out in 2013 near Alma, Limpopo province, South Africa (24°33′00.12 S, 28°07′25.84 E, 1235 m. a.s.l.). The site was characterized by a loamy sand (clovelly plinthic in South African nomenclature: Typic Ustipsamment according to USDA Soil Taxonomy). Average day and night temperatures were 25.0 and 18.5 °C respectively. Growing degree days (GDD) were calculated according to Abendroth et al. (2011) as:
where Tmin and Tmax is the daily minimum and maximum temperature in °C and i the number of days after sowing (DAS) (n total = 97 DAS). Irrigation was provided from the on-site centre pivot irrigation system.
Plant material
The plant material consisted of the core EURoot maize panel (www.euroot.eu) with 33 dent inbred lines (A310, A347, B100, B73, B89, B97, EC169, EP52, EZ11A, EZ37, F1808, F7028, F894, F912, F98902, FC1890, FV353, LAN496, LH38, Mo17, MS153, MS71, N25, N6, Oh33, Oh43, Pa405, PB40R, PH207, PHK76, RootABA1-, RootABA1+, W64A) crossed to UH007 as common flint tester. The EURoot panel is part of the larger DROPs (www.dropsproject.eu) panel and consists of temperate germplasm from the United States and Europe. Three other genotypes were added for spatial adjustment and were not evaluated.
Experimental design and agronomic treatments
The design was a split-plot design with four replications, two nitrogen levels (LN = 46, HN = 192 kg N ha−1) as the whole-plot factor and the 33 genotypes as the split-plot factor. The split-plots were arranged in three rows by 12 columns. The replications were independently distributed in the pivot and surrounded by border plants. To control for spatial variability, six incomplete blocks were arranged within the whole-plots. Each plot consisted of three rows of 20 individuals planted by hand with a planting distance of 15 cm and a row spacing of 75 cm. Genotypes were sown December 5th 2012 and 50 % silking was reached 52 DAS or at 615 GDD.
Before planting both fertilizer treatments received 46 kg N ha−1 in the form of urea. The following urea applications in the HN treatment were split over the whole vegetative growth phase due to the heavy leaching in the sandy soil. Other nutrients, irrigation and pesticides were applied as needed.
Sampling
After flowering (BBCH: 67–73 from 61 to 73 DAS), three representative, non-adjacent plants were sampled in the middle row of each plot. About 80 rootstocks were sampled each morning following the order of incomplete blocks. Stalks were cut exactly 25 cm above the soil surface and the shoots collected. Afterwards the remaining stalk was split lengthwise with a knife either along or across the row’s orientation. After cutting, both halves were dug out in a cylinder of around thirty centimeter diameter and depth (Fig. 1) using a spade. The excavated root crowns were shaken briefly to remove a large fraction of the soil adhering to the root crown. Afterwards, the root crowns were washed under low pressure using a water hose and nozzle, then kept in water until RSAT analysis.
Sampling of root stocks in the field: a removal of shoots 25 cm above the soil surface, b lengthwise splitting of the stalk with a knife, c cutting of entire root crown in the soil with a sharpened and flat spade, d and excavation of the root stock as a cylinder of 30 cm depth and diameter e excavated root stocks
Analysis of RSAT
The larger half of each rootstock, in which the full root top angle and stem diameter were visible, was phenotyped. Angles were measured using a scoreboard and numbers of nodal roots were counted before the rootstock was photographed in a custom-made imaging tent, both as described below. Representative nodal roots for manually determining branching density were excised from the other half of the rootstock before that half was discarded. Time requirements for both manual and digital evaluation were recorded.
Trait abbreviations
Hund et al. (2011) proposed a system of trait abbreviations used for a QTL meta-analysis. Here we aim to use the same system but adapt it to the situation of mature root system. As the root system matures, it develops 5 to 6 underground whorls bearing crown root, eventually followed by several above ground whorls bearing prop roots. The system proposed by Hund et al. (2011) starts counting from old to young roots starting with the seminal root emerging from node 0, followed by the crown roots emerging from internode 1 and so on. When assessing root systems with high-throughput, it is usually too time consuming to count the exact number of whorls upwards. Accordingly the system was adapted to a downwards count, where the youngest whorl of a mature root system is noted as No-0 (i.e., the youngest whorl of nodal roots minus 0) indicating the last whorl of prop roots. Increasingly older whorls would then be noted as No-1, No-2 … No-n, where the counting includes first the prop and then the crown root whorls. Note that both prop and crown roots are nodal roots. As in the system of Hund et al. (2011) the trait abbreviation is followed by the root index in lower case letters, e.g., AngNo-2 for the angle of the third youngest whorl of nodal roots, which were usually below ground crown roots.
Score board method
The manual scoring was based on the published “shovelomics” approach (Trachsel et al. 2011) but with some modifications. Measured traits were the number of above ground whorls occupied with nodal roots (#WhPp) and the number of nodal roots on the youngest whorl (#NoNo-0). Root angles from horizontal were determined on an angle scoreboard where the vertex of the angle was set to the middle of the stalk at soil surface. The nodal root angles were measured along an arc with a 10 cm radius from the vertex. The angles to the left and the right of the youngest (AngNo-0), the second (AngNo-1) and the third (AngNo-2) youngest whorl were recorded on a 10° scale. Additionally, the root top angle to the left and the right (AngRt) was also determined irrespectively of the whorls. For further analyses the average of right and left angles were used (AngRt, AngNo-0, AngNo-1, AngNo-2).
Branching density was measured on the second half of the root stock on excised representative roots from the youngest (BDNo-0) and the third youngest whorl (BDNo-2). First order lateral roots (Lat) were counted in a 2 cm segment starting at 5 cm from the basal zone of the respective nodal root. Branching density was expressed as lateral roots per cm of axile root.
Imaging tent and camera for automated method
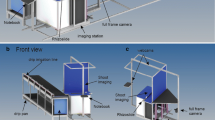
The imaging tent was constructed using aluminium bars connected with plastic joints (Phoenix Mecano Komponenten AG, Stein am Rhein, Switzerland) and covered with a black velvet casing. The tent consisted of a cuboid-shaped backside (70*58*100 cm) and a prism-shaped front side (70*94*100 cm). A ten-megapixel digital camera (Canon EOS 400D, Canon, Tokyo, Japan) with a 35 mm fixed focal length objective (Canon EF 35 mm f/2.0, Canon, Tokyo, Japan) was placed on the top of the prism at 50 cm above the ground. The imaging plane was at the conjunction of the cuboid and the prism. There the rootstocks were clamped with the cut plane facing the camera. Two flashlights (Mecablitz 44 AF1-digital, Metz-Werke GmbH & Co KG, Zirndorf, Germany), covered with a diffusor, illuminated the root systems at a 90° angle from the side and the bottom. Aluminium foil was placed as a reflector at the opposite side of the flashlights to minimize shadows. The whole set up was controlled by a notebook computer using the EOS utility software (EOS Utility version 2.12.0, Canon, Tokyo, Japan). The picture size at the focal plane was 35*52.5 cm resulting in a pixel size of 0.13 mm. Images were recorded and stored as JPEG (Fig. 2). An aperture value of 9.0 and an exposure time of 1 sec allowed for optimal image quality with minimized background noise. A label was placed in the field of view of the camera for sample identification.
Root imaging tent with cuboid-shape backside (70*58*100 cm) and prism-shape frontside (70*94*100 cm); root crown (a) with 25 cm stalk hanged on clap; uniform illumination was achieved with two flashlights (b) and reflector foil on the opposites of the flashlights(c); pictures taken with 10 megapixel camera (d) as JPEG
Image processing with REST
For image analyses, we developed a software called Root Estimator for Shovelomics Traits (REST), in MatLab 7.12 (The Mathworks, Natick Massachusetts, United States). It is available as supplement or from SorceForge (rest4roots). For the identification of the root crown samples, an optical character recognition (OCR) algorithm was programmed to detect the labels, read their content and rename the images accordingly. Rootstocks were segmented from the background and converted into binary images using Otsu’s method (Otsu 1979). Due to the optimized image quality, uniform illumination in the imaging tent and contrast between object and background, this could be done in a batch processing routine with a minimal loss of details.
Due to the standardized removal of the shoots in the field at 25 cm, the soil surface was on a fixed level in the images. An arc with a 10 cm radius was virtually drawn from the middle of the stalk. The angles of the left and right uppermost root were determined and their average was recorded as the image derived root top angle (IAngRt) (Fig. 3). To reduce errors due to single roots standing out of the bulk root stock, the evaluated area was reduced to 95 % in width and depth for further analyses (Fig. 3). On the vertical axis, 5 % of root pixels from the lower edge were discarded; on the horizontal axis 2.5 % of root pixels on each side were discarded. All following analysis are based on these images comprising 90 % of pixel classified as roots.
Image processing with RootEstimatorforShovelomicsTraits: a original RGB picture taken with a 10 megapixel and segmented into binary images; b I) automated determination of image derived root top angle (IAngRt), II) correction for outstanding roots by subtracting 2.5 % of the pixels at the left and the right side and 5 % at the bottom of the root stock, III) determination of Filling factor along the arc; c I) area of the convex hull (AcH) and II) maximum width (mWRt); Within AcH: determination of the projected total structure length (PLSt), number of holes and the filling factor (Ff)
The filling factor along the arc was measured as the rate of root derived pixels along the arc of the opening angle (Fig. 3). The size of the entire root system was estimated as the area of convex hull (AcH), starting at the soil surface. In addition, the projected total structure length, calculated as the sum of the weighted length of root derived structures and the number of background patches within the AcH were determined. Further information about the spatial extension and the shape of the root system in the soil were obtained from the maximum width (mWRt) of the root system.
Further parameters were measured to generate more detailed information about the internal structure of the root system. By dividing the number of root pixels within the convex hull by the pixel number of the AcH the root system filling factor (Ff) was determined. The size of all the background gaps within the convex hull was measured as a measure for the root system complexity (Fig. 4). Furthermore, a distance map within the same area provided information about the diameters of the root clusters in the binary images. Therefore, distances of every root derived pixel to the closest background pixel were measured (Fig. 5). According to Grift et al. (2011), fractal dimensions (FD) of the root systems were obtained as an additional indicator for the internal complexity of the root crown. By using a standard “box-counting” algorithm (Moisy 2006), a grid was applied on the picture and its fineness was reduced stepwise by the factor two. Grid mashes, which coincidence with pixels, were counted in each step. Logarithms of these counts were plotted against the logarithm of the respective reciprocal mash size values. Finally FD was calculated as the slope of the linear regression in these plots (Supplemental Fig. S1).
Distribution of gap sizes measured as size of holes enclosed by roots in the test crosses a) FV353 × UH007 under high nitrogen (HN; 192 kg N ha−1), b) B97 × UH007 under high nitrogen (HN; 192 kg N ha−1) and c) B97 × UH007 under low nitrogen (LN; 46 kg N ha−1); Lateral branching density at youngest whorl a) 9 lateral roots cm−1, b) 39 lateral roots cm−1 and c) 20 lateral roots cm−1; Median gap size a) 0.00383 cm2, b) 0.00274 cm2 and c) 0.00374 cm2
Distribution of cluster diameters measured as distances from root crown derived structures to the background in the test crosses a) W64A × UH007 and b) F1808 × UH007 under high nitrogen (HN; 192 kg N ha−1); Number of nodal roots at youngest whorl a) 10, b) 14; Filling factor a) 0.550, b) 0.699 and median cluster diameter a) 0.171 cm, b) 0.324
Shoot measurements
Flowering data
Silking data were recorded in the middle row of each plot. Ten plants were randomly selected and silking was recorded 47, 50, 52 and 58 DAS. Linear interpolation allowed estimating the date of 50 % silking (SD) in each plot.
Plant vigor
Vigor traits were recorded from the same plants as those investigated for RSAT. Plant height from the soil surface to the collar of the flag leaf was measured after tasseling at 58 DAS. At the same day SPAD values (SPAD-502, Konica Minolta, Tokyo, Japan) were recorded by taking three random measurement points on the second leaf above the ear.
After the shoots were cut from the rootstocks in the field, they were stored in a cool room at 4 °C to avoid water loss. The leaf fresh weight (FWLf) was assessed with precision spring scales (Medio-Line 40600 and 41002, Pesola AG, Switzerland) on a five gram scale.
Data analysis and statistics
All statistics were computed in R version 3.0.1 (R Core Team 2013) and data were fitted by using linear mixed models in the ASReml package (Guilmour et al. 2009) for R.
Diversity of traits in the set
According to the randomization in the field design, the following linear mixed model was used to control for spatial variability:
where Y is the plot-mean trait value of the ith (i = 1, 2,…, 32, 33) genotype within the jth nitrogen treatment (j=HN, LN), within the kth replicate (k = 1, 2, 3, 4) and the lth incomplete block (l = 1, 2,…, 5, 6); α is the main genotype effect, β the main nitrogen level effect, αβ the interaction effect between genotype and nitrogen level, r the replicate effect, βr the effect of the interaction between nitrogen level and replicate, βrb the effect of the incomplete block and ε the residual error. For phenotypic investigations, genotype, nitrogen level, genotype-nitrogen level interaction and the replicate were set as fixed factors while the rest was set as random factors. Analyses of variances (ANOVA) and Wald F-statistic tests were used to identify significant effects of fixed effects.
Best linear unbiased estimators (BLUEs) were taken to investigate the phenotypic diversity in the set (Piepho and Möhring 2007). Tukey’s honestly significant differences (Tukey HSD) were used as post-hoc test and calculated as:
where q is the critical value according to the chosen significance level and degree of freedom, MSE is the mean square error calculated from the average standard error of the difference (avsed) supplied by the predict function of ASReml-R and n is the number of treatment levels.
Heritability estimations
Variance components were used to estimate the heritability of selected traits at the hybrid level (Vargas et al. 2013). To check for the stability of trait inheritance under stress, estimations were done separately for HN and LN using the following linear mixed model:
with Y as the trait value of the ith (i = 1, 2,…, 32, 33) genotype within the kth replicate (k = 1, 2, 3, 4) and within the lth incomplete block (l = 1, 2,…, 5, 6); α as the main genotype effect, r the replicate effect, rb the effect of the incomplete block and ε the residual error. The replicate was set as a fixed factor, the rest as random.
Broad-sense heritability based on the mean of the four replications was calculated according to Falconer and Mackay (1996) as:
where σ 2 g and σ 2 e are the genotypic and residual error variance respectively obtained from the linear mixed model fitting (Eq. 4) and r as the number of replications.
Correlations among RSAT
Relationships between genotype means of manually assessed RSAT and image derived traits were assessed with linear correlations. This allowed identifying possible proxy parameters for manually measured RSAT. Significant correlations based on Pearson correlation coefficients were considered to be weak, moderate, reasonably strong and strong if r < 0.5, 0.5 < r < 0.7, 0.7 < r < 0.85 and r > 0.85 respectively.
Rotational symmetry of maize root crowns
The response of the rotational symmetry of the maize root crowns with respect to the row orientation was obtained from the BLUEs of a slightly modified linear mixed model as shown in Eq. (2):
where Y is the plant trait value of the ith (i = 1, 2,…, 32, 33) genotype and the nth split direction (n=along the row orientation, against the row orientation), within the kth replicate (k = 1, 2, 3, 4), within the lth incomplete block (l = 1, 2,…, 11, 12) and within the mth plot (m = 1, 2,…,5, 6). The parameters α, r and ε describe the same effects as in Eq. (3). The effects of the cut, cut-genotype interaction, the incomplete block and the plot respectively are described by c, αc, rb and rbp respectively. Fixed effects were α, c, αc and r while the other effects were set as random.
Results
The time needed to determine the RSAT by rating followed by counting of lateral roots was around 4.3 min per root crown. Imaging was 6.5 times faster, requiring only 40 sec per root stock for image capture, followed by 6 sec per image for the image analysis with REST.
Different RSAT among genotypes and comparably high heritability for root angles, branching densities and gap size distribution
The evaluated test crosses covered a remarkable range of phenotypic diversity. All measured RSAT except of FD showed significant (p < 0.05) genotype responses (Table 1). The differences between minimum and maximum genotype mean values of these RSAT exceeded the respective Tukey HSD (Table 1). Significant responses to the nitrogen treatment could be observed for all the traits except AngNo-2, BDNo-0, BDNo-2, Ff and the 3rd quartile of the gap size (Table 1). Most manual scores and counts showed significant interactions between genotype and nitrogen fertilization, whereas image-derived traits except of the IAngRt did not show interactions (Table 1).
The date of 50 % silking varied among genotypes between 49 and 56 DAS and was not affected by the level of nitrogen fertilization (Table 1). Plant height, SPAD values and leaf fresh weight differed among genotypes and the latter two responded significantly to the nitrogen level as well (Table 1). Average SPAD values were reduced from 51.4 in the HN treatment to 40.4 in the LN treatment while FWLf dropped from 89.9 to 67.5 g per plant under nitrogen deficiency.
The estimated heritabilities of the assessed RSAT ranged from almost 0 to values higher than 0.8 and were generally reduced in the LN treatment. For the nodal root angles, H2 ranged from 0.54 to 0.77 under high nitrogen and from 0.48 to 0.63 under low nitrogen. Thereby, the manual AngRt and the digital IAngRt showed very similar heritability estimations (Table 1). The heritabilities of the image-derived traits Ff, AcH and mWRt were in the same range as the estimations for the nodal root angles. A very high H2 could be observed for the branching density of lateral roots at the youngest whorl under well fertilized conditions. The heritability of BDNo-0 was 0.81 under HN and 0.54 under LN; heritability of BDNo-2 was 0.67 under HN and 0.67 under LN (Table 1). Under both nitrogen levels, median gap size and median cluster diameters showed heritabilities between 0.45 and 0.64 (Table 1). Relatively low H2 values were observed of #WhPp, #NoNo-0 and FD. The heritabilities of these traits covered a range between 0.00 and 0.41 over both nitrogen treatments (Table 1). By comparison, heritability of most shoot traits ranged between 0.44 and 0.61 with the exception of flowering time, being well above 0.8. Thus, we could identify root traits, such as root angles and branching density that had an even higher heritability than most shoot traits (Table 1).
REST parameters explained focal RSAT
Nodal root angles correlated with image derived root top angles
A common pattern of nodal root angels of different whorls was observed under both nitrogen levels for all evaluated genotypes. Nodal root angles of older whorls were steeper compared to angles of the later emerged whorls (Supplemental Fig. S6). The correlation between genotype means of the AngRt and the IAngRt was strong (r = 0.89) (Fig. 6). Furthermore, the AngRt was correlated with the AngNo-0 and AngNo-1 (r = 0.98 and 0.64, respectively). The correlation of AngRt and AngNo-2 was not significant (Supplemental Fig. S2) yet AngNo-1 and AngNo-2 were weakly (r = 0.38) correlated with each other. With the applied methodology AngRt or IAngRt were reliable measurements to describe the nodal root angles of at least the two latest emerged whorls within a maize root crown.
Nodal root angles defined the size and lateral expansion of the root system
Lower and, therefore, shallower AngRt caused a more horizontal expansion of the root system. The root top angles correlated reasonably strong with the area of the convex hull (r = −0.82 Fig. 7). Since the root crowns were always excavated at the same depth, the genotypic differences in the size of the root systems were mainly measurable by the lateral expansion. The strong correlation between AcH and the maximum width of the root system (r = 0.91) confirmed this suggestion (Fig. 7). A further strong positive correlation among size describing traits was observed between AcH and the projected total structure length (r = 0.89). In contrast, the number of nodal roots on the youngest whorl was not significantly correlated to the nodal root angles or the AcH (Supplemental Fig. S3).
As shown in table 1, AngRt, AcH and mWRt responded to the level of nitrogen fertilization. Thereby root top angles became steeper and AcH and mWRt became smaller under low nitrogen (data not shown).
Weak relationships of RSAT and plant vigor were observed. Increasing leaf fresh weight were related to an increased #NoNo-0 (r = 0.49), increased FD (r = 0.42) but not to AcH (Supplemental Fig. S4) or AngRt. Furthermore, the leaf fresh weight was related to the projected total structure length (r = 0.44) (Fig. 8).
Branching density was closely related to gap size distribution
The two lateral branching densities of nodal lateral roots were assessed to describe the inner architecture of the root crowns. Similar to the pattern of nodal root angles described above, the branching density of lateral roots was lower at older whorls than at the youngest (data not shown). Thereby BDNo-0 and BDNo-2 were correlated reasonably well (r = 0.77). Altered branching intensities of lateral roots due to different genotypes or nitrogen levels changed the size distribution of the background gaps in the images (Fig. 4). Increased BDNo-0 and BDNo-2 caused lower median gap sizes as indicated by the negative correlations of −0.62 (Fig. 9) and −0.44, respectively (Supplemental Fig. S5).
Scatter plots and Pearson correlation coefficients between genotype means of the a) branching density of lateral roots at youngest whorl (BDNo-0) and the median gap size and b) the filling factor (Ff) in the convex hull and median cluster diameter; (**), (*) denote significant correlations on p-level 0.01 and 0.05 respectively
The other image derived parameters describing the inner structure of the root system (Ff, fill factor along the arc and cluster diameters) were not correlated with the manually assessed branching densities. However, higher #NoNo-0 caused larger median cluster diameters (r = 0.44). Furthermore, #NoNo-0 was positively correlated with Ff (r = 0.46). In Fig. 5 the relationship between #NoNo-0, median cluster diameter and Ff is shown on exemplary contrasting plants of two genotypes.
The filling factor itself was related to other image derived traits. Increasing filling factors were related to smaller median gap sizes and larger median cluster diameters. The filling factor showed relatively high correlations with the median cluster diameter (r = 0.81) (Fig. 9) and median gap size (r = −0.59) (Supplemental Fig. S5).
Rotational symmetry of maize root crowns
Due to the two different cut directions of the root crowns it was possible to check their rotational symmetry. Thereby the lateral extension of roots into the intra- and inter-row space was of interest. Significant effects to the split direction could be observed for the AngRt and AngNo-0. The root top angle showed an average angle of 56.6° when the root crown was split along the direction of the rows in the field. When the root stocks were split at 90° orientation to the row, the average AngRt was slightly shallower and decreased by 1.9°. A similar 1.8° difference was observed for AngNo-0. The other nodal root angles, mWRt (p = 0.06) and AcH (p = 0.25) did not show significant responses to the cut direction. Thus, the deviation from rotational symmetry where statistically significant but too small that they would be of major relevance for root foraging within the inter rows.
Discussion
The current study aimed for the development of a high-throughput phenotyping method for mature maize root systems in the field. This was achieved with advancements of the approaches presented by Trachsel et al. (2011) and Grift et al. (2011). We combined an improved sampling method (half-cut) and an improved imaging acquisition in an imaging tent with custom software (REST). The software was programmed to supply summary parameters describing basic characteristics of root stocks of maize based on standardized images. Hand measured traits were correlated with digital parameters obtained from REST. Cutting the root crowns lengthwise (half-cut) aimed at improving handling related to transport and cleaning and the accessibility of the inner structures, such as branching densities, without the need of cutting off single roots. Furthermore, due to this splitting the root crowns were exactly at the focal plane of the camera, facilitating the discrimination of object and background during image processing. Furthermore, a diverse set of 33 test hybrids allowed getting a general idea of the phenotypic diversity at the hybrid level and the heritability of focal traits. The panel was selected to represent genotypes with a comparably narrow window of flowering time to allow for field trials within the DROPS consortium (Welcker, personal communication). The narrow flowering, with differences of 7 days between the earliest and latest genotype confirmed this selection.
Standardized acquisition of high quality images enabled detailed and fast image processing
Phenotyping based on scoring of certain traits, such as number and angles of nodal roots and lateral branching densities required 2 min per root stock (Trachsel et al. 2011). Time expenditures for manual measurements have not been reported (Trachsel et al. 2011; Trachsel et al. 2013). In the current study the required time for manual measurements was more than twice as much as presented by Trachsel et al. (2011), in which visual scoring was used. Image capturing with a standardized procedure as presented here or by Grift et al. (2011), reduced the duration of the whole procedure tremendously while maintaining an optimal image quality. Pictures of high quality and with a good contrast between object and background were the basis for a fast and precise image processing (Zhong et al. 2009; Grift et al. 2011). Furthermore, the standardization of the imaging allowed detecting the soil level in the images which is crucial when quantifying the position of the roots in the soil (Bucksch et al. 2014). The readout of the label and the processing with REST of a single image was completely automatized and took just a few seconds. Hence, automated phenotyping required less than 1 min per root crown, while the manual measurements took more than 6 min per rootstock. Importantly, automated phenotyping requires no oversight from the researcher so trait measurement time is effectively close to zero. The REST software is specifically adapted to measure root traits in maize. With some modifications on the image acquisition side, e.g., an increased resolution of the camera, it may be also used to measure root systems of small grain cereals. However, to measure root systems of dicots, one may refer to more appropriate software, such as DIRT (Bucksch et al. 2014).
Heritabilities of REST parameters were comparable with those of manually assessed RSAT and shoot traits
The phenotyping of root traits is useful to breeders only if the variability caused by genetic effects is much larger than the variance due to environmental influences. The study showed a remarkable genotypic variation among the hybrids (Table 1). Nevertheless, this variation is much narrower than the phenotypic diversity reported from large inbred panel screenings (Grift et al. 2011; Trachsel et al. 2011; Trachsel et al. 2013). This can possibly be explained by the overriding effect of the common father line. It may have masked many allelic effects of the mother inbred lines due to additive or dominant effects. However, dent-by-flint hybrids are typical for temperate regions. Accordingly, the observed variation at the hybrid level serves as better estimate about the genetic variability available to breeders. The degree of heritability for RSAT found in the literature cover a broad range from <0.4 to >0.8 (Hund et al. 2004; Ruta et al. 2010; Kumar et al. 2012; Grieder et al. 2014), what is in accordance with our results. Low H2 estimations were observed for #WhPp, #NoNo-0 and FD, while the heritabilities of AngRt, IAngRt,BDNo-0, size and diameter distributions of gaps and clusters respectively were comparable or even higher than the H2 of shoot traits (Table 1). Decreasing heritabilities under low nitrogen have been observed for most of the assessed RSAT. This can be explained by the increasing environmental error due a pronounced effect of soil inhomogeneity (Gallais and Hirel 2004) and fits in well with previous studies (Chun et al. 2005; Gallais and Coque 2005; Ruta et al. 2010; Zhu et al. 2011). Yet, heritability was so far mainly calculated based on replication within one experimental setup under similar environmental conditions. It remains to be elucidated, if heritabilities of root traits stay high, when compared across years and soil types (Liedgens et al. 2000; Cai et al. 2012). Furthermore, as mean-based heritability increases with the number of measurements on the same genotype, shoot traits that are simple to measure and related to root traits may be a better choice for selection as discussed by Grieder et al. 2014.
REST parameters explained focal RSAT and allowed observing genotype and nitrogen effects
Previous studies used traits such as FD (Grift et al. 2011) or support vector machine parameters (Zhong et al. 2009; Iyer-Pascuzzi et al. 2010) as proxy parameters for RSAT. In this study correlations between measured traits and a set of digitally assessed parameters could be shown. Furthermore, significant genotype and nitrogen effects were observed for those image-derived traits.
Image derived root top angle as reliable proxy value for nodal root angles
Genotype responses of nodal root angles were highly significant (Table 1). The difference in the used set between maximum and minimum values of AngRt, IAngRt, AngNo-0 and AngNo-1 were around 20°. Digitally assessed root top angles in a population of 200 inbred lines showed a similar degree of variation (Grift et al. 2011). In another study the observed variation of scored nodal root angles among 218 inbred lines was almost 90° (Trachsel et al. 2011). It has been suggested that AngRt is inherited additively (Grift et al. 2011). Since the current study just evaluated the test-crosses and not the parental lines it was not possible to verify this claim. Nevertheless, it could be shown that the broad sense heritability of nodal root angles is between 0.54 and 0.77 under well-fertilized conditions and decreased by around 20 % under low nitrogen (Table 1). These estimations are comparable to H2 estimations of other RSAT such as nodal root number or branching density (Kumar et al. 2012).
Previous studies reported moderate to strong correlations between nodal root angles of different whorls (Trachsel et al. 2013) which corresponds to our results (Supplemental Fig. S2). Furthermore, we could show that AngRt, and AngNo-0 can be replaced by the digitally assessed IAngRt (Fig. 6). Since the vertex of nodal root angles from different whorls was fixed at the soil surface, angle patterns were not comparable with other studies, where the vertex was set at the respective whorl. With the applied methodology roots at younger whorls seemed to be shallower than at older ones, that disagrees with previous results (Araki et al. 2000; Trachsel et al. 2011; Grieder et al. 2014).
Area of the convex hull as measure for root system size and lateral extension
Nodal root angles were the major trait measured manually defining the size of the excavated root crown, expressed as AcH. Beside AcH also mWRt and the projected total structure length described the size of the root system in the images (Fig. 7 and Supplemental Fig. S3). For all these RSAT genotype and nitrogen responses could be observed (Table 1). These findings correspond well with previous studies in the field and under controlled conditions, where root system size differed among genotypes (Kumar et al. 2012; Worku et al. 2012; Manavalan et al. 2012) and fertilization levels (Vamerali et al. 2003; Yu et al. 2007; Liu et al. 2008; Worku et al. 2012). As reported by Trachsel et al. (2013), root angles became steeper in the LN treatment. Furthermore, mWRt and AcH decreased under low nitrogen. Due to the excavation of the rootstock at a fixed depth, the AcH was mainly defined by its lateral extent (mWRt) and the steepness of the nodal root angles (Fig. 7). The vertical dimension was affected by the excavation depth, only. Links between the steepness of nodal root angles and the degree of topsoil root density have been shown in greenhouse and field experiments (Zhu et al. 2005b). Similar results were obtained in other studies with mature maize root systems in the field. Correlations between the steepness of nodal root angles and the maximum rooting depth could be shown (Araki et al. 2000; Hund 2010; Trachsel et al. 2013).
Further genotype and fertilization responses were observed for #WhPp and #NoNo-0 (Table 1). These findings have been reported in several studies, where #WhPp differed between nitrogen levels (Ku et al. 2012) and genotypes (Trachsel et al. 2011; Ku et al. 2012; Burton et al. 2013). The same effects have also been observed for the nodal root number (Trachsel et al. 2011; Burton et al. 2013; Trachsel et al. 2013). Similar to the findings of Trachsel et al. (2013), we did not find significant correlations (p < 0.05) between #NoNo-0 and nodal root angles, AcH (Supplemental Fig. S3) or mWRt. Compared to other studies, where the H2 of #WhPp and #NoNo-0 were estimated under controlled conditions (Kumar et al. 2012; Ku et al. 2012) our estimations were much lower (0.15 < H2 < 0.41). Hund et al. (2004) reported similar degrees of heritability for #NoNo-0. The heritabilities of AcH, mWRt and projected total structure length were between 0.33 and 0.71 and therefore comparable to H2 for RSAT reported by other authors (Hund et al. 2004; Ruta et al. 2010; Kumar et al. 2012).
Fractal dimensions have been suggested to be a proxy measurement for the complexity of the root system (Grift et al. 2011). Although fractal geometry of whole root systems differs from the fractal geometry of linear and planar root system transects (Nielsen et al. 1997), and FD alone is less informative than integrated analysis of FD, fractal abundance and lacunarity (Walk et al. 2004), most previous studies focus on planar FD computed just from a subsection of the root system. These authors observed significant differences between maize (Grift et al. 2011) and common bean genotypes (Nielsen et al. 1999; Walk et al. 2004). In REST planar FD were computed from 90 % of the pixels classified as roots, which resulted in non-significant differences of FD among genotypes (Table 1). This would also be an explanation for the low heritability estimations for FD. Unlike to Walk et al. (2004) we could observe fertilizer responses of FD (Table 1) and a correlation to FWLf (Supplemental Fig. S4). These findings suggest that with the applied methodology FD describes plant vigor and the size of the excavated root stock rather than the complexity or the inner structure of the root system per se. Burton et al. (2013) showed correlations of plant biomass and the diameter of the rootstock. This corresponds to our results, since planar FD, as computed with REST, is influenced to a certain extent by the diameter of the root system. Further relations of RSAT and FWLf have been observed for #NoNo-0 and the projected total structure length. This confirms results from other studies, where number and length of nodal roots (Burton et al. 2013) or root biomass (Manavalan et al. 2012) were positively correlated with shoot biomass.
Half-cut enabled insights into the root system and REST provided measures for inner complexity
While nodal roots define the very coarse structures and the outer shape of a root system, lateral roots are responsible for finer structures and the inner complexity of a root crown (Lynch 1995; Hochholdinger 2009; Gaudin et al. 2011). In this study different lateral branching densities were observed among the test-crosses (Table 1). The manual measurements ranged from 11.88 to 27.33 and from 7.76 to 13.48 lateral roots per centimeter of crown root for BDNo-0 and BDNo-2 respectively. In large panel screenings with maize inbred lines (Trachsel et al. 2011) and land races (Burton et al. 2013) genotype responses with a larger variation were reported. Branching densities of lateral roots at different whorls correlated well, what agrees with the results by Kumar et al. (2012) and Trachsel et al. (2013). Furthermore, the estimated heritabilities of BDNo-0 and BDNo-2 were comparably high (Table 1) and in the same range as reported previously (Kumar et al. 2012).
Planar fractal dimension as computed in this study could not explain the lateral branching patterns. Rather using the number of root tips (Bucksch et al. 2014), this study used gap sizes within the root crown to explain lateral branching patterns. The current results showed for the first time, that the median size of the gaps between root derived pixels in the images correlate moderately with BDNo-0 (Fig. 9) and to a smaller extent with BDNo-2 (Supplemental Fig. S5). Our findings suggested that Ff provides a more general idea of the inner structure and is a more robust measurement with respect to nitrogen deficiency while median gap size provided more detailed information about the inner structure and is clearly related to the apparent density of lateral roots. By increasing the resolution to 20 or more megapixels new insights into the lateral root system can be obtained such as diameters of lateral roots or even secondary lateral branching.
Half-cut enabled to study rotational symmetry of root crowns
Plant-to-plant competition might affect rotational symmetry of the root system. Leaf orientation is adjusted by modern varieties to point toward the inter row, closing the large gaps between rows (Maddonni et al. 2002). This might be also the case of roots. The approach of lengthwise splitting of the root crowns in the field either along or against the planting row, allowed investigating the rotational symmetry of the rootstocks without extra effort. Compared to gel-based systems (Iyer-Pascuzzi et al. 2010) or computed tomography (Zhu et al. 2011) this could be done much faster, at lower acquisition costs and with mature root systems. The effects of the split directions on the lateral extension were significant in the case of AngRt and AngNo-0, but with 2° steeper angles within the row the effect is very small. In the case of mWRt and AcH no significant responses to the split direction could be seen. This suggests that maize root crowns keep their rotational symmetry, even if more unoccupied soil volume is provided between the planting rows than within. However, we did not evaluate to what extent the excavation method might bias the results. In a more specific experiment the question of inter vs. intra row foraging may be addressed by extending the dimensions of the excavated root stock. For example, two adjacent neighboring plants might be harvested together with the target plant and carefully cleaned. Certainly, such a methodology would come at the costs of lower throughput.
Conclusion
The results of this study illustrated clearly the potential and the power of image processing as a phenotyping approach of mature maize root crowns. A standardized acquisition of images of high resolution and contrast are crucial for fast and detailed image processing. Furthermore, the lengthwise split of the root crowns enabled to determine the outer shape and the inner structure of root systems at once without cutting of roots. The software REST provided a set of parameters, which allowed observing significant differences between genotypes and nitrogen levels at the periphery and the interior of the crowns. Moderate to strong correlations between focal RSAT measured by hand and parameters provided by REST were observed. The image derived root top angle showed a strong correlation with the root top angle determined manually. Further traits, such as the area of the convex hull and the maximum width of the root system measured the size, and even more importantly, the lateral extent of the excavated root system. These traits were also strongly related to nodal root angles. Based on the distribution of root clusters diameters and background gap sizes the inner structure of the crowns with respect to genotypes and the level of nitrogen fertilization could be quantified. The median gap size was correlated to the apparent lateral branching density and the filling factor within the root crown.
Furthermore, the results showed, that manually assessed RSAT and REST parameters have a similar degree of heritability like shoot traits such as plant height or leaf fresh weight. This indicated the opportunity to incorporate RSAT into hybrid breeding programs. However, to have a more evident idea about the heritability of RSAT, the study needs to be repeated at several locations and under various stress conditions.
Currently, the adaptation of sampling and imaging strategies to measure root systems of other crop species is in progress. As REST serves only a rough estimate of branching density, further image-based approaches to measure length, diameters and branching density of lateral roots on individual roots, are certainly desirable. This will further increase the power and utility of REST for large genotype screenings of various crops.
References
Abendroth LJ, Elmore RW, Boyer MJ, Marlay SK (2011) Corn growth and development. PMR 1009. Iowa State Univ. Extension, Ames
Araki H, Hirayama M, Hirasawa H, Iijima M (2000) Which roots penetrate the deepest in rice and maize root systems. Plant Prot Sci 3:281–288
Bucksch A, Burridge J, York LM et al (2014) Image-based high-throughput field phenotyping of crop roots. Plant Physiol 166:470–486. doi:10.1104/pp. 114.243519
Burton AL, Brown KM, Lynch JP (2013) Phenotypic diversity of root anatomical and architectural traits in Zea species. Crop Sci 53:1–15
Cai H, Chen F, Mi G et al (2012) Mapping QTLs for root system architecture of maize (Zea mays L.) in the field at different developmental stages. Theor Appl Genet 125:1313–24. doi:10.1007/s00122-012-1915-6
Campos H, Cooper M, Habben JE et al (2004) Improving drought tolerance in maize: a view from industry. Fields Crop Res 90:19–34
Chun L, Mi G, Li J et al (2005) Genetic analysis of maize root characteristics in response to low nitrogen stress. Plant Soil 276:369–382
Coque M, Martin A, Veyrieras JB et al (2008) Genetic variation for N-remobilization and postsilking N-uptake in a set of maize recombinant inbred lines. 3. QTL detection and coincidences. Theor Appl Genet 117:729–747
Falconer DS, Mackay TF (1996) Introduction to quantitative genetics, 4th edn. Longman, Harlow
Gallais A, Coque M (2005) Genetic variation and selection for nitrogen use efficiency in maize: a synthesis. Maydica 50:531–547
Gallais A, Hirel B (2004) An approach to the genetics of nitrogen use efficiency in maize. J Exp Bot 55:295–306
Gaudin ACM, McClymont SA, Holmes BM et al (2011) Novel temporal, fine-scale and growth variation phenotypes in roots of adult-stage maize (Zea mays L.) in response to low nitrogen stress. Plant Cell Environ 34:2122–2137
Giuliani S, Sanguineti MC, Tuberosa R et al (2005) Root-ABA1, a major constitutive QTL, affects maize root architecture and leaf ABA concentration at different water regimes. J Exp Bot 56:3061–3070
Grieder C, Trachsel S, Hund A (2014) Early vertical distribution of roots and its association with drought tolerance in tropical maize. Plant Soil 377:295–308. doi:10.1007/s11104-013-1997-1
Grift TE, Novais J, Bohn M (2011) High-throughput phenotyping technology for maize roots. Biosyst Eng 110:40–48
Guilmour AR, Gogel BJ, Cullis BR, Thompson R (2009) ASReml user guide release 3.0. VSN Int. Ltd, Hemel Hempstead, HP1 1ES, UK
Hammer G, Dong Z, McLean G et al (2009) Can changes in canopy and/or root system architecture explain historical maize yield trends in the US corn belt? Crop Sci 49:299–312
Hochholdinger F (2009) The maize root system: morphology, anatomy, and genetics. In: Hake SC, Bennetzen JL (eds) Handb. maize Its Biol. Springer New York, New York, pp 145–160
Hochholdinger F, Tuberosa R (2009) Genetic and genomic dissection of maize root development and architecture. Curr Opin Plant Biol 12:172–177
Hund A (2010) Genetic variation in the gravitropic response of maize roots to low temperatures. Plant Roots 4:22–30. doi:10.3117/plantroot.4.22
Hund A, Fracheboud Y, Soldati A et al (2004) QTL controlling root and shoot traits of maize seedlings under cold stress. Theor Appl Genet 109:618–29
Hund A, Reimer R, Messmer R (2011) A consensus map of QTLs controlling the root length of maize. Plant Soil 344:143–158
Iyer-Pascuzzi AS, Symonova O, Mileyko Y et al (2010) Imaging and analysis platform for automatic phenotyping and trait ranking of plant root systems. Plant Physiol 152:1148–1157
Kahn BA, Stoffella JP (1991) Nodule distribution among root morphological components of field-grown cowpeas. J Am Soc Hortic Sci 116:655–658
Ku LX, Sun ZH, Wang CL et al (2012) QTL mapping and epistasis analysis of brace root traits in maize. Mol Breed 30:697–708
Kumar B, Abdel Ghani AH, Reyes-Matamoros J et al (2012) Genotypic variation for root architecture traits in seedlings of maize (Zea mays L.) inbred lines. Plant Breed 131:465–478
Kutscherea L, Lichtenegger E (1960) Wurzelatlas mitteleuropäischer Ackerunkräuter und Kulturpflanzen. DLG-Verlag, Frankfurt am Main
Liedgens M, Soldati A, Stamp P, Richner W (2000) Root development of maize (Zea mays L.) as observed with minirhizotrons in lysimeters. Crop Sci 40:1665–1672
Liu J, Li J, Chen F (2008) Mapping QTLs for root traits under different nitrate levels at the seedling stage in maize (Zea mays L.). Plant Soil 305:253–265
Lynch JP (1995) Root architecture and plant productivity. Plant Physiol 109:7–13
Lynch JP (2013) Steep, cheap and deep: an ideotype to optimize water and N acquisition by maize root systems. Ann Bot. doi:10.1093/aob/mcs293
Maddonni GA, Otegui E, Andrieu B et al (2002) Maize leaves turn away from neighbors. Plant Physiol 130:1181–1189. doi:10.1104/pp. 009738.nated
Manavalan LP, Musket T, Nguyen HT (2012) Natural genetic variation for root traits among diversity lines of maize (Zea Mays L.). Maydica 56
Moisy F (2006) “Boxcount” (Matlab Central, 2006). http://www.mathworks.com/matlabcentral/fileexchange/13063-boxcount. Accessed 30 May 2013
Nielsen KL, Lynch JP, Weiss HN (1997) Fractal geometry of bean root systems: correlations between spatial and fractal dimension. Am J Bot 84:26–33
Nielsen KL, Miller CR, Beck D, Lynch JP (1999) Fractal geometry of root systems: field observations of contrasting genotypes of common bean (Phaseolus vulgaris L.) grown under different phosphorus regimes. Plant Soil 206:181–190
Otsu N (1979) A threshold selection method from gray-level histogrmas. IEEE Trans Syst Man Cybern 9:62–6
Passioura JB (2012) Phenotyping for drought tolerance in grain crops: when is it useful to breeders? Funct Plant Biol 39:851–859
Piepho H-P, Möhring J (2007) Computing heritability and selection response from unbalanced plant breeding trials. Genetics 177:1881–1888
R Core Team (2013) R: a language and environment for statistical computing
Ruta N, Liedgens M, Fracheboud Y (2010) QTLs for the elongation of axile and lateral roots of maize in response to low water potential. Theor Appl Genet 120:621–631
Saengwilai P, Tian X, Lynch JP (2014) Low crown root number enhances nitrogen acquisition from low-nitrogen soils in maize. Plant Physiol 166:581–9. doi:10.1104/pp. 113.232603
Singh V, Oosterom EJ, Jordan DR et al (2010) Morphological and architectural development of root systems in sorghum and maize. Plant Soil 333:287–299
Stoffella PJ, Sandsted RF, Zobel RW, Hymes WL (1979) Root characteristics of black beans. II. Morphological differences among genotypes. Crop Sci 19:826–830
Trachsel S, Kaeppler SM, Brown KM, Lynch JP (2011) Shovelomics: high throughput phenotyping of maize (Zea mays L.) root architecture in the field. Plant Soil 341:75–87
Trachsel S, Kaeppler SM, Brown KM, Lynch JP (2013) Maize root growth angles become steeper under low N conditions. Fields Crop Res 140:18–31
Vamerali T, Saccomani M, Bona S et al (2003) A comparison of root characteristics in relation to nutrient and water stress in two maize hybrids. Plant Soil 255:157–167
Vargas M, Combs E, Alvarado G et al (2013) META: a suite of SAS programs to analyze multienvironment breeding trials. Agron J 105:11–19
Walk TC, Van Erp E, Lynch JP (2004) Modelling applicability of fractal analysis to efficiency of soil exploration by roots. Ann Bot 94:119–28. doi:10.1093/aob/mch116
Weaver JE (1925) Investigations on the root habits of plants. Am J Bot 12:502–509
Wiesler F, Horst WJ (1994) Root growth and nitrate utilization of maize cultivars under field conditions. Plant Soil 163:267–277
Worku M, Bänziger M, Friesen D, Diallo AO, Horst WJ (2012) Nitrogen efficiency as related to dry matter partitioning and root system size in tropical mid-altitude maize hybrids under different levels of nitrogen stress. Fields Crop Res 130:57–67
Wu L, McGechan MB, Watson CA, Baddeley JA (2005) Develping existing plant root system architecture models to meet future agricultural challenges. Adv Agron 85:181–219
York LM, Nord EA, Lynch JP (2013) Integration of root phenes for soil resource acquisition. Front Plant Sci 4:1–15. doi:10.3389/fpls.2013.00355
Yu G-R, Zhuang J, Nakayama K, Jin Y (2007) Root water uptake and profile soil water as affected by vertical root distribution. Plant Ecol 189:15–30
Zhong D, Novais J, Grift TE et al (2009) Maize root complexity analysis using a support vector machine method. Comput Electron Agric 69:46–50
Zhu J, Kaeppler SM, Lynch JP (2005a) Mapping of QTLs for lateral root branching and length in maize (Zea mays L.) under differential phosphorus supply. Theor Appl Genet 111:688–95. doi:10.1007/s00122-005-2051-3
Zhu J, Kaeppler SM, Lynch JP (2005b) Topsoil foraging and phosphorus acquisition efficiency in maize (Zea mays). Funct Plant Biol 32:749. doi:10.1071/FP05005
Zhu J, Ingram PA, Benfey PN, Elich T (2011) From lab to field, new approaches to phenotyping root system architecture. Curr Opin Plant Biol 14:310–317
Zobel RW (2011) A developmental genetic basis for defining root classes. Crop Sci 51:1410. doi:10.2135/cropsci2010.11.0652
Zobel RW, Waisel Y (2010) A plant root system architectural taxonomy: a framework for root nomenclature. Plant Biosyst 144:507–512. doi:10.1080/11263501003764483
Acknowledgments
The authors would like to thank the anonymous reviewers for their helpful suggestions and Achim Walter for his support. Thanks for assistance in the field go to Johan Prinsloo and the farmworkers in South Africa. Thanks to Claude Welcker for his assistance in assembling the EURoot maize panel and to Delley Seeds and Plants Ltd. for hybrid production. We kindly thank the donors of the genetic material: Department of Agroenvironmental Science and Technologies (DiSTA), University of Bologna, Italy (RootABA lines); Misión Biológica de Galicia (CSIC), Spain (EP52); Estación Experimental de Aula Dei (CSIC), Spain (EZ47, EZ11A, EZ37); Centro Investigaciones Agrarias de Mabegondo (CIAM), Spain (EC169); Misión Biológica de Galicia (CSIC), Spain, (EP52); University of Hohenheim, Versuchsstation für Pflanzenzüchtung, Germany (UH007, UH250); and INRA CNRS UPS AgroParisTech, France (supply of the remaining, public lines). We thank the Forschungszentrum Jülich GmbH, Germany for the MatLib package and Oliver Dressler for the CAD illustrations. Support for field research in South Africa was provided to Jonathan Lynch by the Howard G. Buffett Foundation. This research received funding from the European Community Seventh Framework Programme FP7-KBBE-2011-5 under grant agreement no.289300 and the Walter Hochstrasser-Stiftung.
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible Editor: Peter J. Gregory.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Figure S1
Image processing with RootEstimatorForShovelomicsTraits from binary images: Determination of fractal dimensions by stepwise reduction of the grid fineness, here exemplary: a) 512*512 mashes, b) 256*256 mashes, c) 128*128 mashes and d) 64*64 mashes. (TIFF 248 kb)
Figure S2
Scatter plot and Pearson correlation coefficients between genotype means of root top angle (AngRt) and a) root angle of the youngest whorl (AngNo-0), b) the second youngset whorl (AngNo-1) and c) the third youngest whorl (AngNo-2); (**) denotes significant correlations on p-level 0.01, (n.s.) denotes non-siginificant correlations. (TIFF 86 kb)
Figure S3
Scatter plots and Pearson correlation coefficients between genotype means of the area of the convex hull (AcH) and a) nodal root number at the youngest whorl (#NoNo-0) and b) the projected total structure length; (**), (°) denote significant correlations on p-level 0.01 and 0.1 respectively. (TIFF 68 kb)
Figure S4
Scatter plots and Pearson correlation coefficients between genotype means of the leaf fresh weight (FWLf) and a) nodal root number at the youngest whorl (#NoNo-0), b) the area of the convex hull (AcH) and c) the fractal dimension (FD); (**), (*) denote significant correlations on p-level 0.01 and 0.05 respectively, (n.s.) denotes non-siginificant correlations. (TIFF 80 kb)
Figure S5
Scatter plots and Pearson correlation coefficients between genotype means of the a) branching density of lateral roots at 3rd youngest whorl (BDNo-2) and the median gap size and b) the filling factor (Ff) in the convex hull and the median gap size; (*) denote significant correlations on p-level 0.05. (TIFF 72 kb)
Figure S6
Mean root angles of the youngest (AngNo-0), second youngest (AngNo-1) and third youngest (AngNo-2) nodal root whorl of each genotype under high (HN; open circles) or low (LN; cross) nitrogen. (TIFF 136 kb)
ESM 7
(ZIP 26370 kb)
Rights and permissions
About this article
Cite this article
Colombi, T., Kirchgessner, N., Le Marié, C.A. et al. Next generation shovelomics: set up a tent and REST. Plant Soil 388, 1–20 (2015). https://doi.org/10.1007/s11104-015-2379-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11104-015-2379-7