Abstract
We present a method to visually score 10 root architectural traits of the root crown of an adult maize plant in the field in a few minutes. Phenotypic profiling of three recombinant inbred line (RIL) populations of maize (Zea mays L.; B73xMo17, Oh43xW64a, Ny821xH99) was conducted in 2008 in a silt loam soil in Pennsylvania and in a sandy soil in Wisconsin, and again in 2009 in Pennsylvania. Numbers, angles and branching pattern of crown and brace roots were assessed visually at flowering. Depending on the soil type in which plants were grown, sample processing took from three (sand) to 8 min (silt-loam). Visual measurement of the root crown required 2 min per sample irrespective of the environment. Visual scoring of root crowns gave a reliable estimation of values for root architectural traits as indicated by high correlations between measured and visually scored trait values for numbers (r 2 = 0.46–0.97), angles (r 2 = 0.66–0.76), and branching (r 2 = 0.54–0.88) of brace and crown roots. Based on the visual evaluation of root crown traits it was possible to discriminate between populations. RILs derived from the cross NY821 x H99 generally had the greatest number of roots, the highest branching density and the most shallow root angles, while inbred lines from the cross between OH43 x W64a generally had the steepest root angles. The ranking of genotypes remained the same across environments, emphasizing the suitability of the method to evaluate genotypes across environments. Scoring of brace roots was better correlated with the actual measurements compared to crown roots. The visual evaluation of root architecture will be a valuable tool in tailoring crop root systems to specific environments.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Root system architecture is important for plant productivity under edaphic stress (Lynch 1995). The root system of maize (Zea mays L.) consists of an embryonic root system comprised of a single primary root and a variable number of seminal roots (Abbe and Stein 1954), and a post-embryonic root system of shoot-borne roots. Shoot-borne roots formed at underground nodes are called crown roots, while those formed at above-ground nodes of the shoot are called brace roots (Hochholdinger and Tuberosa 2009).
A root system architecture specifically adapted to the prevailing soil conditions might be advantageous. After the onset of drought, water is often found in deeper soil layers. Deeper soil layers are predominantly reached by maize genotypes forming a sparsely branched axile root system (Cahn et al. 1989; Hund et al. 2009b). In contrast, phosphorus availability is typically greatest in the topsoil (Lynch 1995). When phosphorus was evenly distributed in the soil, root shallowness did not confer any competitive advantage in common beans (Rubio et al. 2003). However, when phosphorus availability was restricted to the topsoil, common bean (Lynch and Brown 2001) and maize genotypes (Zhu et al. 2005a) having root architectural traits enhancing topsoil foraging grew best. In common bean, trade-offs for soil resource acquisition were incurred when roots were not deployed where the limiting resource was in greatest availability (Lynch and Ho 2005; Ho et al. 2005). In order to improve plant performance breeders need to select genotypes with a root architecture adapted to the conditions of the target environment.
Quantitative genetic studies require phenotyping protocols that are rapid, accurate, and robust. Root architecture is difficult to evaluate directly in soil. Several high-throughput procedures to measure root systems have been reported. Sanguineti et al. (2006) investigated morphophysiological characteristics of root traits in hydroponics. Paper rolls have been used to investigate the genetic basis of lateral root (Zhu et al. 2005b), seminal root (Zhu and Lynch 2004) and root hair (Zhu et al. 2005a) responses to phosphorus availability. Growth pouches, consisting of germination paper covered by plastic (Hund et al. 2009b), have been used to investigate the root angle of common bean in response to high and low P (Bonser et al. 1996), to investigate the stress tolerance of tropical maize, and to map quantitative trait loci (QTL) for different root traits of tropical maize inbred lines (Trachsel et al. 2009). In all these systems, maize can only be grown for a limited duration. Detection of genetic differences among genotypes might therefore be biased by effects of the seed on germination and initial growth as described by various authors (e.g. Pommel 1990; Manga and Yadav 1995; Smith et al. 2003).
At later growth stages pots can be used to create more natural conditions. For instance Liao et al. (2001) investigated the gravitropic response of root angles in response to phosphorous availability in sand-filled 20 l pots. Although pot experiments are more representative of natural conditions than the seedling assays described above, plant growth can be restrained by soil volume and nutrient availability. At the flowering stage, roots have been measured in the field (Laboski et al. 1998; Kato et al. 2006), in soil boxes (Araki et al. 2000) and in soil columns (Hund et al. 2009a; Araki and Iijima 1998; Zhu et al. 2010). Growing plants in columns or boxes, filled with soil or artificial substrate, can help to reduce sampling efforts compared to field studies and allows growth under controlled conditions. However, the excavation of roots and measurement of root traits in these systems remains labor-intensive and does not allow for high throughput. Moreover artificial systems fail to mimic the complex interaction between the plant, intrinsic abiotic and biotic soil parameters and prevailing environmental conditions as suggested by Walter et al. (2009). In the field, roots and shoots are exposed to very different environmental conditions, especially with regard to temperature, which is an important regulator of root development (Hund 2010). In controlled conditions, field environments to which the shoot is exposed are typically simulated, leading to highly artificial conditions for the root system. The root system is buffered from the atmospheric environment in a completely different way when grown in a small container compared to the field. Hence, there is a high risk for artifacts of root growth or of root-shoot-interaction in such investigations, when aiming to simulate field-situations. Overall, information about root architecture in the field and information about the genetic control of root architecture remains scarce.
Visual scoring using a defined rating system has been employed for high throughput phenotyping of shoot traits. For example, visual scoring has been used to monitor stay-green in sorghum (Xu et al. 2000), onset of senescence (Thomas and Howarth 2000), disease monitoring in barley (Hill et al. 2008), for the quantification of leaf retention in cassava to select towards desiccation tolerance (Lenis et al. 2006) and for root length of cucumbers (Walters and Wehner 1994). To our knowledge, a high throughput method which utilizes visual scoring of the numbers, angles and branching density of brace and crown roots has not yet been used for the investigation of root architecture. The objectives of the present study were to: i) develop a high throughput method to evaluate root architecture, and ii) assess its suitability to phenotype large sets of genotypes.
Material and methods
Experimental site
Experiments were carried out in 2008 and 2009 at the Russell Larson Research and Education Center of the Pennsylvania State University in Rock Springs, PA, USA (40°42′37″.52 N, 77°57′07″.54 W, 366 masl), in 2008 at the Hancock Agricultural research station of the University of Wisconsin in Hancock, WI, USA (44°07′56″.74 N, 89°30′43″.96 W, 331 masl) and in the 2009/2010 season in Alma, LP, ZA (24°33′00.12 S, 28° 07′25.84 E, 1235 masl). The experiment was conducted on a Hagerstown silt loam (fine, mixed, semiactive, mesic Typic Hapludalf) in Rock Springs, a Plainfield loamy sand (mixed, mesic Typic Udipsamment) in Hancock and a loamy sand (Clovelly Plinthic soil) in Alma.
Field management
In 2008, genotypes were planted on May 28 in Rock Springs and on May 21 in Hancock. Plants were evaluated at flowering (Table 1). In 2008, plants were sampled from August 11–14 in Rock Springs and August 25–27 in Hancock. In 2009 in Rock Springs, genotypes were planted on June 2 and harvested on August 25. In 2009 in Alma genotypes were planted on December 14 and harvested in 2010 on March 1 and 2. At sampling, plants had accumulated 717 growing degree days (GDD) in Rock Springs in 2008, 961 GDD in Hancock in 2008, 774 GDD in Rock Springs in 2009 and 1182 in 2010 in Alma. GDD were calculated from air temperature with a base temperature of 10°C. The mean temperature during the 2008 season was 21.5°C in Rock Springs and 19.9°C in Hancock, 20.3°C during the 2009 in Rock Springs, and 23.9°C during the 2009/2010 season in Alma. Precipitation within the experimental period was 173 mm in Rock Springs in 2008, 381 mm in Hancock in 2008, 254 mm in Rock Springs in 2009 and 299 mm in 2010 in Alma.
Row width was 75 cm, and distance between plants within a row was 23 cm, resulting in an overall planting density of 6 plants m−2. Based on soil analysis at the beginning of the cropping season the plots were not fertilized in Rock Springs. The field in Hancock was amended with 66 kg K2CO3 ha−1. The field in Alma was amended with 110 kg NH4NO3 ha−1, 25 kg K2SO4 ha−1 and 22 kg KH2PO4 ha−1. In all environments pest control was carried out as needed. Water was applied by sprinkler irrigation as needed. Two days prior to sampling the fields were irrigated using an irrigation cannon with 13 mm of water to soften the soil in order to facilitate excavation of root crowns.
Plant material
Two hundred and eighteen randomly selected Recombinant Inbred Lines (RILs) from the crosses between the parental lines B73 x Mo17 (IBM, 98 RILs), OH43 x W64a (OhW, 61 RILs) and NY821 x H99 (NyH, 59 RILs) were evaluated in the present study. These populations were chosen because our previous research indicated that they were segregating for root morphological traits and abiotic stress tolerance.
Experimental design
In 2008, all 218 genotypes were grown in Rock Springs and Hancock. Genotypes were not replicated. Additionally the IBM RILs were grown in Alma in 2009/2010. In 2009, 10 genotypes (IBM3, IBM79, IBM368, NyH180, NyH227, NyH272, OhW21, OhW48, OhW163 and OhW206) were grown in Rock Springs with four replicates. Genotypes were randomly assigned to plots in each location and in both years using a randomized complete block design. One plot consisted of one 4.6 m long row containing 20 plants. Three representative plants for each plot were selected for excavation and visual scoring in all three season-by-environment combinations. Selection was carried out based on plant height and general appearance. Only fully bordered plants were selected. As selected root crowns within a plot were homogeneous root crowns of the three plants per plot were bulked for visual scoring and only a single rating was recorded. Root crowns were stored and preserved in their three-dimensional structure at 4°C for 3 days. Subsequently traits were measured and compared to the trait values obtained by scoring.
Data was normalized prior to the analysis of variance. Data was fitted using linear mixed effect model nlme() in R (Pinheiro et al., 2004). The linear mixed effect model was
where Yij is the trait value of the ith genotype within the jth environment (j = 1, 2), α is the main effect of the genotype, β is the main effect of the environment and ε ij is the random error term composed of interaction and true random error. Environment was treated as random while the genotype was treated as fixed. Genotypes were not replicated within environments, which did not allow us to estimate the genotype-by-environment interaction. Comparisons among populations and environments were carried out using the Tukey-Kramer multiple comparison test. Comparisons between populations and environments for the number of whorls occupied by brace roots (BW: either one or two) were carried out using a Chi-square test. The trait repeatability of 98 IBM lines grown across environments and years (Hancock and Rock Springs in 2008 and Alma in 2009/2010) was calculated according to (Falconer and Mackay 1996):
where σ 2G and σ2 GE are the ANOVA estimates of the variance for genotype and the error composed of genotype-by-environment interaction and true random error. σ 2G has two components: VG and VEG. VG is the genetic variance and VEG the general environmental variance associated with the permanent differences between individuals. e is the number of environments.
The Wilcoxon Rank Sum test was used to test for the ranking of 98 IBM RILs across environments, using the wilcox.test() function in R.
Analyses at sampling
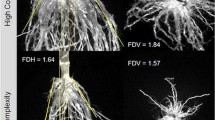
At harvest roots were excavated by removing a soil cylinder of 40 cm diameter and a depth of 25 cm with the plant base as the horizontal center of the soil cylinder. Excavation was carried out using standard shovels. The excavated root crowns were shaken briefly to remove a large fraction of the soil adhering to the root crown. Most of the remaining soil was then removed by soaking the root crown in mild detergent at a concentration of 0.5% (only in Rock Springs, PA; containing sodium laureth sulphate, cocamidophorol betaine, cocamide DEA, Styrene acrylate copolymer, chlorhexidine gluconate and sodium chloride). In a third step remaining soil particles were removed from the root crown by vigorous rinsing at low pressure. Soaking of the root crowns was not necessary in the sandy soil in Hancock. The clean roots were visually scored for the following traits (Fig. 1): Numbers of above-ground whorls occupied with brace roots (BW); brace root number (BO); angle of the 1st and 2nd arm of brace roots originating from the first and second whorl in relation to horizontal (BA1a, BA1b, BA2a, BA2b; The first arm of brace roots represents the basal part of the brace root growing on an initial trajectory, and the second arm represents the second part of the brace root growing at a trajectory angled in relation to the trajectory of the first arm); the branching of brace roots (BB); and the numbers (CN), angles (CA) and branching (CB) of crown roots. Traits were assigned values from one to nine where one indicates shallow root angles (10°), low root numbers and a low branching density (0.5 lateral root cm−1). Nine indicates steep root angles (90°), high numbers and a high branching density (7 lateral roots cm−1). Representative images depicting contrasts for the various traits are given in Fig. 2. Scoring in 2008 was carried out by a different researcher than in 2009 and 2010. Correlations between traits were established using the Spearman-Rank correlation. Significant correlations between traits with r < 0.5 were considered weak, 0.5 < r < 0.8 moderate and r > 0.8 strong.
Ten traits were assessed visually on the excavated root crowns: number of whorls occupied by brace roots (BW), number of brace roots (BO), 1st (BA1a, BA2a) and 2nd (BA1b, BA2b) arm of the brace roots originating from whorl 1, whorl 2, respectively, the branching density of brace roots (BB), the number (CN), angles (CA) and branching density (CB) of crown roots
Spearman-Rank correlation coefficients between traits obtained by measuring and scoring and representative images showing roots contrasting for the evaluated traits. Images displayed were scored with 1, 3, 5, 7 and 9. As no crown root angles were scored with 1 no image is displayed for this trait-by-score combination
Correlation between measured and scored traits
In 2009, ten genotypes were grown in Rock Springs to compare traits values scored visually with trait values measured and counted, and to quantify trait variability within specific genotypes. The genotypes chosen were: IBM3, IBM79, IBM368, NyH180, NyH227, NyH272, OhW21, OhW48, OhW163 and OhW206. Genotypes were replicated four times. In 2009, excavated root crowns were visually scored and subsequently used to measure root angles and to count numbers of brace roots, crown roots and lateral roots originating from brace and crown roots. Angles of roots were measured using a large protractor. The number of lateral roots was counted on a 4 cm root segment obtained from 5 cm below the soil surface. On the same 4 cm segment the mean length of 3 randomly selected lateral roots was measured. Linear density was calculated as the number of lateral roots per cm. Root angles for crown and brace roots were averaged after measuring the angles of three typical roots for each root class. In 2010 30 IBM RILs were randomly selected to evaluate how the accuracy of scoring for CN and CA would be affected by removing the brace roots on one side of the root crown. Prior to scoring of CN and CA brace roots were removed from one side of the root crown. CN and CA were thereafter scored as described above. Subsequently CN were counted and CA measured with a large protractor. Trait values obtained by scoring and counting (CN)/measuring (CA) were thereafter correlated. Significant correlations between traits with r < 0.5 were considered weak, 0.5 < r < 0.8 moderate and r > 0.8 strong. Variability among plants was assessed by comparing the values measured on the three root crowns excavated for one genotype. Variability is described using the coefficient of variation (C.V.).
Results
Large variability was observed among genotypes
Phenotypes varied widely in both years and environments within and between populations (Figs. 2 and 3). Scores for the number of brace roots (BO), the angles (BA) and branching (BB) of brace roots ranged from 1 to 9, numbers (CN) and angles (CA) of crown roots from 3 to 9 and branching (CB) of crown roots from 2 to 9. As brace roots had only formed at two (above-ground) whorls, scores for BW were either 1 or 2. To assess variability within a plot for 10 selected genotypes, four root crowns were measured. The largest C.V.s were observed for BB (14–38%), BW (14–34%), BA1, (11–30%; the distinction between the first and the second arm of brace roots is not made since too few plants showed two distinct arms). BO (9–27%) and CB (11–22%) had intermediate variability, while BA2 (9–17%), CA (6–10%) and CN (6–9%) had little variability.
Frequency distribution of 218 recombinant maize inbred lines replicated once in Rock Springs, PA and Hancock, WI based on visual scoring for the following traits: number of whorls occupied by brace roots (BW), number of brace roots (BO), 1st (BA1a, BA2a) and 2nd (BA1b, BA2b) arm of the brace roots originating from whorl 1, whorl 2, respectively, the branching density of brace roots (BB), the number (CN), angles (CA) and branching density (CB) of crown roots
Scored and measured trait values are highly correlated
The measurement of root crowns for ten randomly selected genotypes was used to calibrate the values obtained by scoring. Moderate-strong correlations between scored and measured trait values were obtained for five brace root traits BW (Fig. 2): (r = 1, either one or two whorls occupied with brace roots), BO (0.97) and BA2 (0.71), BA1 (0.66) and BB (0.54). Removal of brace roots from the root crown resulted in similarly strong correlations between measured and scored trait values for crown roots: CB (0.88) and CA (0.76). A weak correlation between scored and measured trait values was only found for CN (0.46). Correlations between scored and measured trait values for the 1st and 2nd arm of the brace roots were not established since too few genotypes in the subset of ten genotypes showed two distinctive brace root arms. Branching of brace roots occurred in patches. Measurement of the linear density over a given segment did not necessarily reflect the same branching pattern as visual scoring, resulting in a moderate correlation between scored and measured trait values (0.54). Visual scoring, therefore, gives a better estimate of overall brace root branching than does the measurement of linear density at a given position along an axial root.
Excavation and evaluation of one root crown required 5–10 min
The soil type in which plants had been grown influenced the time needed to uproot and evaluate the root crowns (Table 2). Visual evaluation was not affected by the soil type and averaged 2 min per plot irrespective of the environment. Excavation (1 min in WI vs. 3 min in PA), soaking (1 min in WI vs. 3 min in PA) of the root and time needed to rinse (1 min in WI vs. 2 min in PA) of soil residues from the root crowns took more than twice as much time in the silt loam soil (8 min) in PA compared to the sandy soil in WI (3 min).
Angles and branching are consistent between brace and crown roots
Irrespective of root class, root angles were similar as indicated by the obtained correlations (Table 3): CA was strongly correlated with BA1a (0.53), BA1b (0.51), BA2a (0.53), and was moderately correlated with BA2b (0.29). Brace roots originating from the first whorl showed angles similar to roots originating at a later developmental stage from the second whorl as illustrated by strong correlations between BA1a and BA2a as well as between BA1b and BA2b. Within the first whorl, the first and second arm of brace roots were moderately correlated (0.49).
The populations are different for root angles and branching
Based on the visual evaluation of root crown traits, it was possible to discriminate between populations. All traits except BW were significantly affected by the genotype (Table 4). The NyH population had the greatest average number of brace and crown roots, greatest average branching density of crown and brace roots and the shallowest average angle for both crown and brace roots. The OhW population had the steepest angles for both crown and brace roots (Table 4). Compared to the IBM population, the OhW population had the steepest angles for brace roots (BA1b, BA2a) and crown roots (CA). The OhW population also had more brace roots than the other two populations. The greatest variability within a population was observed for IBM.
Trait values for root architecture were consistent across years
Highest repeatability for 98 IBM RILs across all three environments was obtained for CA (67%). Repeatability for BA1, BB and CB ranged from 44 to 49%. Lowest repeatability was obtained for BW (29.6%), BO (38.6%) and CN (31.8%) (Table 5). A Wilcoxon Rank Sum Test showed that genotypes changed their ranking based on the initial scoring data for most traits across environments. The ranking did not change across environments for CB and not for BB between PA and WI. When the Wilcoxon Rank Sum Test was performed on the quintiles the ranking of genotypes did not change across environments for any trait (Table 5). These findings indicate that the method presented discerns rather large differences (i.e. quintiles) while smaller trait differences based on the scoring scheme with 9 distinct categories were difficult to ascertain statistically. Moreover within an environment (PA) trait values obtained in 2008 were reproducible in 2009 as indicated by moderate (BA1b: 0.68, CB: 0.68, BB: 0.68, BW: 0.56, BO: 0.54) or weak (BA1a: 0.35) correlations among trait values between 2008 and 2009. Differences in angles of the first arm (BA1a) and brace root numbers (BO) were possibly related to differences in precipitation. As a result of fewer precipitation events in 2008 the soil surface was harder resulting in greater aboveground growth of overall fewer brace roots than in 2009 (data not shown).
Discussion
We present a method to rapidly evaluate 10 architectural traits of maize root systems in the field. We measured the number, angles and branching density of crown and brace roots. The measurement of these traits permits the description of the basic architecture of the root crown of a plant. Depending on the soil type in which plants were grown, excavation and cleaning of root crowns required from three (sand) to 8 min (silt loam). Plants grown in a silt-loam needed more time for soaking and removing the soil surrounding the root crowns. Removal of soil residues in the sandy soil was carried out by vigorous rinsing with water at low pressure. Subsequently visual measurement of the root crown required 2 min per sample irrespective of the environment.
The accuracy of the scoring method is highlighted by high correlations found between scored and measured values for BW (1), BO (0.97) and moderate correlations for BA1 (0.66), BA2 (0.76) and BB (0.54). Since BA has a broad range (20–90°) it should be possible to accurately identify differences in brace root angles by scoring. The accuracy of the measurements could be increased by means of a protractor for angles, while only minimally increasing the time needed for the evaluation per sample.
Removal of brace roots on one side of the root crown provides a good view on the crown roots which can thereafter be reliably scored as indicated by the correlation coefficient between measured and scored trait values for CN (0.46), CA (0.76) and CB (0.88). Araki and Iijima (1998) reported that the deepest roots originated from the oldest crown root tier. Crown roots supply the plant with water when water availability is limited to deeper soil layers (Araki et al. 2000). With respect to the ecophysiological impact of the crown root system at later developmental stages it may be justifiable to further increase the reliability of the trait values by counting the number of crown roots.
Architectural analysis of root traits revealed that root angles were similar between the different root types as indicated by correlations between CA, BA1 and BA2; similarly the branching pattern was similar irrespective of root type as indicated by correlations between CB and BB. These results are consistent with evidence presented previously (Firn and Digby 1997) suggesting that plants possess a mechanism that allows them to attain a stable growth angle, characteristic for each organ.
Based on these results it appears feasible to reduce the number of traits to be evaluated in order to increase the speed of the method. It seems possible to measure CA and CB at an earlier stage, when brace roots have not yet developed, and subsequently extrapolate trait values for BA and BB. This would both reduce the time required for the evaluation of one sample as well as shorten the growth period in the field. From an ecophysiological point of view, it would be interesting to investigate genotypes that do not follow the typical pattern (high CA/high BA or high CB/high BB and vice versa). Roots with shallow angles will predominantly explore the topsoil (Lynch and Brown 2001), while roots with steep angles will predominantly explore deeper soil layers (Singh et al. 2010).
Roots typically show large variability, especially when grown under natural environmental conditions. Genotypic variability observed here was consistent with previous studies which found variation in the range from 15–33% in nodal root angles in rice (Abe and Morita 1994) and angles of seminal and nodal roots in maize (Araki et al. 2000). Variability observed in the present study for the branching of crown roots (11–22%) was smaller than variability observed for the number (Hund et al. 2007) and the linear density of seminal roots (50%; Ito et al. 2006; 25%; Zhu et al. 2005b). Preselection of plants prior to excavation (based on plant height, overall impression, only fully bordered plants) and general visual information obtained from the three excavated root crowns helped reducing trait variability to a level typical for root traits. We therefore conclude that the variability observed for root traits was in a range typically observed and that the method used to evaluate root architecture did not introduce additional variability. The accuracy of measured trait values could be further increased by replicating the genotypes within environments, however this would entail increases in labor and land requirements.
A Wilcoxon Rank Sum test revealed that based on the scoring scheme with 9 categories ranks of the genotypes did change across environments for most traits (except CB and for BB between PA and ZA), while performing the same test on the quintiles for each trait did not show a change in category. These findings indicate that the scoring method differentiated between genotypes with rather large phenotypic differences, while it was not possible to ascertain smaller phenotypic differences. This postulate is supported by the moderate to high repeatability obtained for trait values measured in the three environments. Repeatability obtained across years, environments and researcher carrying out the scoring, indicate that traits are genetically determined. QTLs identified for most root traits (notably for BA1 and CA) in 26 RIL populations (unpublished, 2009/2010 season in Alma, ZA) further support this claim. The cause of the changes in genotype ranking is not clear. Possible explanations are that the visual/mental gauge applied by the scorer in 2008 was slightly altered between the evaluations in the two environments, as well as differences in perception between the two scorers carrying out the evaluation in 2008 and the other years. Moreover differences in environment may have led to a genotype-by-environment interaction. Since genotypes were not replicated in each environment it was not possible to calculate the genotype-by-environment interaction. Overall these results indicate that the scoring method can be used to ascertain differences between genotypes across environments irrespective of the possible subjective bias of the scorer.
Despite differences in temperature and precipitation the scoring method yielded similar results for randomly selected genotypes grown in Rock Springs in 2008 and 2009. Different mean values observed among years can be explained by the precipitation pattern (BW, BA1a) and inaccuracies in the scoring method (CA). In 2008 we measured more shallow angles of the first arm of the brace roots originating from the first whorl and more aboveground growth of brace roots (data not shown). We speculate that these differences were caused by the lower precipitation in 2008; as a result of lower precipitation soil penetration resistance was greater in 2008 than in 2009.
Moderate to high correlations obtained across two experimental years and high repeatability obtained across different environments, years and scorers indicate that the scoring method can be used under various environmental conditions without modifications for specific environments. Results obtained with the scoring method could therefore be compared across years and locations.
We found that the NyH population generally had the greatest number of roots, the greatest branching density of brace and crown roots, and the most shallow root angles, while the OhW population generally had the steepest root angles. The OhW population also had more brace roots than the other two populations, while the greatest variability within a population was observed for IBM.
The method presented in this paper is high throughput, low cost, and easy to learn. The scoring method would also be suitable for the evaluation of root architecture for other crop species. It should be directly applicable to other graminaceous crops such as sorghum or millet, while several adaptations would need to be made for the evaluation of small grain cereals. For the evaluation of small grain cereals (e.g. wheat) it would be necessary to consider how to take into account tillering of the mother plant. The scoring scheme for non- graminaceous crops would have to account for a root architecture that is completely different than the one observed in maize. For example, we have developed and used a comparable system for common bean that takes into account the features of dicotyledonous root systems such as basal roots, hypocotyl-borne roots, etc. (C. Jochua, J.P. Lynch, unpublished).
In order to further increase the accuracy and reliability of the scoring method for maize, several adaptations should be made for future experiments: The number of crown roots should be counted and angles of brace and crown roots in relation to horizontal should be measured rather than scored. Increased standardization of the method for angles and branching would allow several persons to carry out the scoring while ruling out subjective bias. The involvement of multiple scorers would allow the evaluation of large sets of genotypes as required in breeding or in studies of quantitative genetics. The increase in precision should make it easier to detect small differences among genotypes. Time required for root excavation and processing would be reduced by sampling smaller plants, at an earlier phenological stage than the one at which this study was carried out, when they are easier to excavate and clean; concomitantly the duration of field occupancy would be reduced. It will therefore be important to identify the earliest phenological stage at which plants show root architecture relevant at later growth stages. Moreover it will be interesting to see how root angles measured on the root crown affect root distribution in the soil. We are currently investigating these topics.
In the breeding process, large sets of genotypes are usually evaluated over a wide range of environments for phenotypic selection and/or the identification of QTLs that are stable across environments. Although shovelomics is labor intensive, requires a large acreage and has a long field occupancy, the advantage of measuring root architecture in the field ultimately relevant for crop performance may justify the additional effort compared to screening at the seedling stage. Shovelomics will permit rapid visual selection of root architecture phenotypes that are well adapted for efficient water and nutrient acquisition in marginal environments, as are often encountered in developing countries. Since plant breeders will be able to use the scoring method to screen large populations in multiple environments, it will also become possible to identify the genetic basis of root architecture. As a consequence, marker assisted selection (MAS) will become possible. MAS in combination with phenotypic selection will in turn permit researchers and breeders to tailor root systems for adaptation to target environments.
References
Abbe EC, Stein OL (1954) The origin of the shoot apex in maize: embryogeny. Am J Bot 41:285–293
Abe J, Morita S (1994) Growth direction of nodal roots in rice: its variation and contribution to root system formation. Plant Soil 165:333–337
Araki H, Iijima M (1998) Rooting nodes of deep roots in rice and maize grown in a long tube. Plant Prod Sci 1:242–247
Araki H, Hirayama M, Hirasawa H, Iijima M (2000) Which roots penetrate the deepest in rice and maize root systems? Plant Prod Sci 3:281–288
Bonser AM, Lynch J, Snapp S (1996) Effect of phosphorus deficiency on growth angle of basal roots in Phaseolus vulgaris. New Phytol 132:281–288
Cahn MD, Zobel RW, Bouldin DR (1989) Relationship between root elongation rate and diameter and duration of growth of lateral roots of maize. Plant Soil 119:271–279
Falconer DS, Mackay TF (1996) Introduction to quantitative genetics, 4th edn. Longman, Harlow
Firn RD, Digby J (1997) Solving the puzzle of gravitropism—has a lost piece been found? Planta 2003:S159–S163
Hill NS, Neate SM, Cooper B, Horsley R, Schwarz P, Dahleen LS, Smith KP, O’Donnell K, Reeves J (2008) Comparison of ELISA for fusarium, visual screening, and deoxynivalenol analysis of fusarium head blight for barley field nurseries. Crop Sci 48:1389–1398
Ho MD, Rosas JC, Brown KM, Lynch JP (2005) Root architectural tradeoffs for water and phosphorus acquisition. Funct Plant Biol 32:737–748
Hochholdinger F, Tuberosa R (2009) Genetic and genomic dissection of maize root development and architecture. Curr Opin Plant Biol 12:172–177
Hund A (2010) Genetic variation in the gravitropic response of maize roots to low temperatures. Plant Root 4:22–30
Hund A, Richner W, Soldati A, Fracheboud Y, Stamp P (2007) Root morphology and photosynthetic performance of maize inbred lines at low temperature. Eur J Agron 27:52–61
Hund A, Ruta N, Liedgens M (2009a) Rooting depth and water use efficiency of tropical maize inbred lines, differing in drought tolerance. Plant Soil 318:311–325
Hund A, Trachsel S, Stamp P (2009b) Growth of axile and lateral roots of maize: I Development of a phenotyping platform. Plant Soil In Press. doi:10.1007/s11104-009-9984-2
Ito K, Tanakamaru K, Morita S, Abe J, Inanaga S (2006) Lateral root development, including responses to soil drying, of maize (Zea mays) and wheat (Triticum aestivum) seminal roots. Physiologia Plantarum 127:260–267
Kato Y, Abe J, Kamoshita A, Yamagishi J (2006) Genotypic variation in root growth angle in rice (Oryza sativa L.) and its association with deep root development in upland fields with different water regimes. Plant Soil 287:117–129
Laboski CAM, Dowdy RH, Allmaras RR, Lamb JA (1998) Soil strength and water content influences on corn root distribution in a sandy soil. Plant Soil 203:239–247
Lenis JI, Calle F, Jaramillo G, Perez JC, Ceballos H, Cock JH (2006) Leaf retention and cassava productivity. Field Crops Res 95:126–134
Liao H, Rubio G, Yan XL, Cao AQ, Brown KM, Lynch JP (2001) Effect of phosphorus availability on basal root shallowness in common bean. Plant Soil 232:69–79
Lynch JP (1995) Root architecture and plant productivity. Plant Physiol 109:7–13
Lynch JP, Brown KM (2001) Topsoil foraging—an architectural adaptation of plants to low phosphorous availability. Plant Soil 237:225–237
Lynch JP, Ho MD (2005) Rhizoeconomics: Carbon costs of phosphorus acquisition. Plant Soil 269:45–56
Manga K, Yadav OP (1995) Effect of seed size on development traits and ability to tolerate drought in pearl-millet. J Arid Env 29:169–172
Pinheiro J, Bates D, DebRoy S, Sarkar D (2004) nlme: linear and nonlinear mixed effects models. R package version 3.
Pommel B (1990) Effects of seed weight and sowing depth on growth and development of maize seedlings. Agr 10:699–708
Rubio R, Liao H, Yan X, Lynch JP (2003) Topsoil foraging and its role in plant competitiveness for phosphorus in common bean. Crop Sci 43:598–607
Sanguineti MC, Duvick DN, Smith S, Landi P, Tuberosa R (2006) Effects of long-term selection on seedling traits and ABA accumulation in commercial maize hybrids. Maydica 51:329–338
Singh V, van Oesterom EJ, Jordan DR, Messina CD, Cooper M, Hammer GL (2010) Morphological and architectural development of root systems in sorghum and maize. Plant Soil 333:287–299
Smith KF, McFarlane NM, Croft VM, Trigg PJ, Kearney GA (2003) The effects of ploidy and seed mass on the emergence and early vigour of perennial ryegrass (Lolium perenne L.) cultivars. Austr J Exp Agr 43:481–486
Thomas H, Howarth CJ (2000) Five ways to stay green. J Exp Bot 51:329–337
Trachsel S, Messmer R, Stamp P, Hund A (2009) Mapping of QTLs for lateral and axile root growth of tropical maize. Theor Appl Genet 119:1413–1424
Walter A, Silk WK, Schurr U (2009) Environmental effects on spatial and temporal patterns of leaf and root growth. Annu Rev Plant Biol 60:279–304
Walters SA, Wehner TC (1994) Evaluation of U.S. cucumber germplasm collection for root size using a subjective rating technique. Euphytica 79:39–43
Xu W, Rosenow DT, Nguyen HT (2000) Stay green trait in grain sorghum: relationship between visual rating and leaf chlorophyll concentration. Plant Breeding 119:365–367
Zhu JM, Lynch JP (2004) The contribution of lateral rooting to phosphorus acquisition efficiency in maize (Zea mays L.) seedlings. Funct Plant Biol 31:949–958
Zhu JM, Kaeppler SM, Lynch JP (2005a) Mapping of QTL controlling root hair length in maize (Zea mays L.) under phosphorus deficiency. Plant Soil 270:299–310
Zhu JM, Kaeppler SM, Lynch JP (2005b) Mapping of QTLs for lateral root branching and length in maize (Zea mays L.) under differential phosphorus supply. Theor Appl Genet 111:688–695
Zhu JM, Brown KM, Lynch JP (2010) Root cortical aerenchyma improves the drought tolerance of maize (Zea mays L.). Plant Cell Environ 33:740–749
Acknowledgements
This research was supported by the National Science Foundation PGRP grant DBI 0820624. We thank Bob Snyder for the management of the experiment in Rock Springs, as well as Amy Burton, Johanna Mirenda, Lauren Gelesh and the field crew at Hancock for their technical assistance with all the experiments. We would also like to thank two anonymous reviewers for their valuable comments improving the quality of the manuscript. We thank Thomas Mitchell-Olds for suggesting the name “shovelomics”.
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible Editor: Peter J. Gregory.
Rights and permissions
About this article
Cite this article
Trachsel, S., Kaeppler, S.M., Brown, K.M. et al. Shovelomics: high throughput phenotyping of maize (Zea mays L.) root architecture in the field. Plant Soil 341, 75–87 (2011). https://doi.org/10.1007/s11104-010-0623-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11104-010-0623-8