Abstract
Leaf senescence and abscission in autumn are critical phenological events in deciduous woody perennials. After leaf fall, dormant buds remain on deciduous woody perennials, which then enter a winter dormancy phase. Thus, leaf fall is widely believed to be linked to the onset of dormancy. In Rosaceae fruit trees, DORMANCY-ASSOCIATED MADS-box (DAM) transcription factors control bud dormancy. However, apart from their regulatory effects on bud dormancy, the biological functions of DAMs have not been thoroughly characterized. In this study, we revealed a novel DAM function influencing leaf senescence and abscission in autumn. In Prunus mume, PmDAM6 expression was gradually up-regulated in leaves during autumn toward leaf fall. Our comparative transcriptome analysis using two RNA-seq datasets for the leaves of transgenic plants overexpressing PmDAM6 and peach (Prunus persica) DAM6 (PpeDAM6) indicated Prunus DAM6 may up-regulate the expression of genes involved in ethylene biosynthesis and signaling as well as leaf abscission. Significant increases in 1-aminocyclopropane-1-carboxylate accumulation and ethylene emission in DEX-treated 35S:PmDAM6-GR leaves reflect the inductive effect of PmDAM6 on ethylene biosynthesis. Additionally, ethephon treatments promoted autumn leaf senescence and abscission in apple and P. mume, mirroring the changes due to PmDAM6 overexpression. Collectively, these findings suggest that PmDAM6 may induce ethylene emission from leaves, thereby promoting leaf senescence and abscission. This study clarified the effects of Prunus DAM6 on autumn leaf fall, which is associated with bud dormancy onset. Accordingly, in Rosaceae, DAMs may play multiple important roles affecting whole plant growth during the tree dormancy induction phase.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Temperate woody perennials enter dormancy in autumn, during which external growth is halted, with bud break subsequently initiated after an exposure to a specific period of chilling conditions. Dormancy prevents premature sprouting under unfavorable environmental conditions, thereby protecting the plant from potential damage (Faust et al. 1997). In addition to bud dormancy, several phenological events typically occur during the tree dormancy induction phase. Saure (1985) reported that shoot growth cessation and leaf fall are closely linked to dormancy and cold acclimation in temperate deciduous woody perennials. To date, several studies generated evidence of the connection between leaf senescence/abscission and bud dormancy. For example, temperature increases in autumn delay leaf senescence and abscission, while also significantly affecting bud dormancy, resulting in delayed spring bud burst (Beil et al. 2021; Wang et al. 2022; Malyshev et al. 2024). An exogenous abscisic acid treatment significantly enhances shoot growth cessation and induces early dormancy, but also alters leaf senescence and cold acclimation in apple (Malus spp.) plants (Guak and Fuchigami 2001). However, the regulatory mechanisms underlying the relationship between leaf senescence/abscission and bud dormancy remain largely unclear.
Rosaceae DORMANCY-ASSOCIATED MADS-box (DAM) genes encode the primary regulators of tree dormancy; their expression patterns in dormant buds are positively correlated with the timing of the dormancy phase transition (i.e., up-regulated expression during dormancy induction and down-regulated expression during dormancy release) (Bielenberg et al. 2008; Jiménez et al. 2009; Sasaki et al. 2011; Zhu et al. 2015; Tuan et al. 2017; Falavigna et al. 2019; Yamane et al. 2019; Lloret et al. 2021; Hsiang et al. 2024). DAM genes, which are phylogenetically closely related to SHORT VEGETATIVE PHASE (SVP) and AGAMOUS-LIKE 24 (AGL24) in Arabidopsis, were initially identified as the genes associated with the failure of the peach (Prunus persica) evergrowing (evg) mutant to enter dormancy (Bielenberg et al. 2008; Li et al. 2009). MdDAM1 RNA interference (RNAi) transgenic apple plants are reportedly unable to enter dormancy in winter, displaying evergrowing traits similar to those of the dormancy-insensitive peach evg mutant (Bielenberg et al. 2008; Moser et al. 2020). Additionally, simultaneously silencing several DAM and SVP genes in apple results in an evergrowing phenotype (Wu et al. 2021). A recent study showed that the Prunus mume gene DAM6 (PmDAM6) encodes a transcription factor that modulates the expression of genes related to phytohormone and lipid body metabolism as well as the cell cycle in the dormant vegetative meristem, thereby controlling dormancy (Hsiang et al. 2024). However, other studies suggest Rosaceae DAMs affect tree growth in addition to dormancy. The expression of PmDAM6 in leaves is up-regulated toward leaf fall (Sasaki et al. 2011). The overexpression of peach PpeDAM6 and P. mume PmDAM6 promotes bud set, while also repressing growth (Lloret et al. 2021; Yamane et al. 2019). Additionally, the overexpression of PmDAM6 in poplar (Populus tremula × Populus tremuloides) and apple promotes growth cessation, whereas the RNAi-based silencing of DAM expression delays leaf shedding in transgenic plants (Bielenberg et al. 2008; Moser et al. 2020; Wu et al. 2021; Sasaki et al. 2011; Yamane et al. 2019). These results reflect the importance of DAMs on physiological processes other than bud dormancy.
The onset of leaf abscission and senescence in temperate woody perennials during autumn is a crucial indicator of water and nutrient remobilization within plants (Koyama 2014; Walde et al. 2024). Leaf senescence and abscission are influenced by auxin, ethylene, and jasmonic acid (JA) levels (He et al. 2002; Ferrante and Francini 2006; Koyama 2014; Hu et al. 2017; Li et al. 2018; Zhang et al. 2020). Specifically, leaf abscission is regulated by both auxin and ethylene (Ferrante and Francini 2006; Iqbal et al. 2017), with ethylene initiating senescence, especially in ethylene-sensitive species (Iqbal et al. 2017). In tomato and Arabidopsis, a prolonged exposure to ethylene results in starch and chlorophyll degradation, further suppressing photosynthesis and causing early leaf senescence (Zhang et al. 2022; Mohorović et al. 2023b). Furthermore, JA-induced leaf senescence depends on components of the ethylene signaling pathway, particularly the key components EIN2 and EIN3; EIN3 overexpression and silencing accelerates and delays leaf senescence, respectively (Li et al. 2013). Therefore, ethylene regulates leaf senescence more than JA.
The objective of this study was to determine whether PmDAM6 affects leaf senescence and abscission. We used two mRNA-seq datasets to examine the leaves of 35S:PmDAM6-GR transgenic apple (this study) and 35S:PpeDAM6 transgenic European plum (Prunus domestica) (Lloret et al. 2021). A comparative transcriptome analysis revealed the conserved mechanisms underlying the functions of DAM6 in P. mume and P. persica. Notably, this study provides the first evidence of the possible conserved regulatory effect of Prunus DAM6 on leaf senescence and abscission, suggesting it contributes to the regulation of bud dormancy as well as the seasonal tree growth/dormancy cycle during the dormancy induction phase.
Materials and methods
Plant materials
This study was completed using previously reported transgenic apple lines (35S:PmDAM6-GR and 35S:PmDAM6) (Yamane et al. 2019). Specifically, two 35S:PmDAM6-GR (chemical-inducible overexpression) transgenic apple lines (GR21 and GR22) were examined in the 2015–2016 (4-year-old) and the 2022–2023 (11-year-old) seasons, whereas two 12-year-old 35S:PmDAM6 (stable overexpression) transgenic apple ‘JM2’ lines (35S-2 and 35S-4) and WT plants were included for detecting ethylene and analyses of leaf abscission and senescence in the 2023–2024 season. All transgenic lines were cultivated under natural photoperiodic conditions in a sealed greenhouse in Kyoto, Japan. The greenhouse was cooled when temperatures exceeded 25 °C (May–September) or 15 °C (October–April), but was not heated during the experimental period.
Adult ‘Nanko’ (early leaf shedding, high-chill) and ‘Ellching’ (late leaf shedding, low-chill) P. mume trees (> 15 years old) from the Kyoto experimental farm of Kyoto University as well as adult ‘Nanko’ and ‘SC’ (late leaf shedding, low-chill) trees from the Kizu experimental farm of Kyoto University were also used. For seasonal analyses, leaves were collected from ‘Nanko’ and ‘Ellching’ trees twice per month from September to December 2023. They were immediately frozen in liquid nitrogen and stored at − 80 °C prior to extracting RNA. Leaf ethylene emission was measured using ‘Nanko’ and ‘Ellching’ leaves collected on November 23, 2023 at the Kyoto experimental farm as well as ‘Nanko’ and ‘SC’ leaves collected on November 27, 2023 at the Kizu experimental farm. Treatment analysis was performed using ‘Nanko’ and ‘JM2’ trees. The analyzed plant materials are listed in Supplementary Table S3.
DEX treatment of 35S:PmDAM6-GR apple plants
A solution consisting of 100 μM DEX (Wako, Osaka, Japan) and 0.1% Tween 20 was thoroughly mixed and uniformly sprayed onto the leaf surface of 35S:PmDAM6-GR transgenic apple lines and WT plants in October 2015 as well as on June 28 and October 31, 2023. Leaf samples collected at 0, 8, and 16 h after the DEX treatment were rapidly frozen in liquid nitrogen and stored at − 80 °C prior to extracting RNA. The leaf samples from the 2015 season were used for the mRNA-seq analysis, ACC measurement, and hormone (IAA and JA) extraction, whereas the leaf samples from the 2023 season were used for analyses of gene expression (via qRT-PCR) and ethylene emission.
RNA extraction
Total RNA was isolated from the collected leaves using a modified CTAB method (Yamane et al. 2008). Briefly, the frozen leaf tissue was ground to a powder, which was resuspended in 6 mL pre-heated extraction buffer containing 3% (v/v) 2-mercaptoethanol (65 °C). The sample was gently mixed, incubated at 65 °C for 30 min, and centrifuged (8000 rpm for 5 min). The supernatant was transferred to a tube and combined with an equivalent amount of chloroform:isoamyl alcohol [24:1 (v/v)]. The solution was gently shaken and subsequently centrifuged (8000 rpm for 5 min). These steps were repeated, after which the supernatant was transferred to a new tube. Nucleic acids were precipitated by adding the same volume of isopropanol and then incubating the solution at − 20 °C for 2 h. After centrifuging the solution (14,000 rpm for 30 min), the pellet was resuspended in 500 μL DEPC-treated H2O and LiCl (one-third volume) was added. The solution was incubated overnight at − 20 °C and centrifuged (14,000 rpm for 30 min). The pellet was washed using 500 μL 70% ethanol and then centrifuged (14,000 rpm for 5 min). The pellet was air-dried and then resuspended in 30 μL DEPC-treated water.
mRNA-seq library preparation
To construct the mRNA-seq library, mRNA was isolated from the extracted total RNA (10–25 µg) using the Dynabeads® mRNA Purification Kit (Thermofisher scientific, MA, USA). Next, cDNA was synthesized from the isolated mRNA using Superscript III (Thermofisher scientific), with a double-stranded cDNA formed via nick translation. The KAPA HyperPlus Library Preparation Kit (NIPPON Genetics Co. Ltd., Tokyo, Japan) was employed to repair the cDNA ends, add adenine residues, and ligate indexing adapters for identifying samples. The cDNA fragments longer than 100 bp were purified after each step using AMPure XP beads (Beckman Coulter Inc., Brea, CA, USA). A small amount of purified cDNA solution was used as a template for a PCR amplification using Bio-amp primers to determine the optimal number of amplification cycles. The remaining cDNA solution was used for the PCR amplification with the optimal cycle number. The concentration of the generated mRNA-seq library was measured using a Qubit® 2.0 Fluorometer (Thermofisher scientific). After adjusting the library amount (at least 20 ng), the mRNA-seq analysis was performed on the Illumina HiSeq 2000 platform (Illumina, San Diego, CA, USA) to generate 100 bp paired-end reads. The sequencing analysis was conducted using two or three biological replicates per experimental time point.
Comparative transcriptome analysis
The following mRNA-seq datasets were analyzed: (1) mRNA-seq datasets for the DEX-treated and control leaves of 35S:PmDAM6-GR transgenic apple (current study) and (2) mRNA-seq datasets for the leaves of 35S:PpeDAM6 transgenic European plum and WT available in the National Center for Biotechnology Information Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo) (accession number PRJNA630876) (Lloret et al. 2021). For the analysis of apple mRNA-seq datasets, the GDDH13 genome (Daccord et al. 2017) served as the reference genome. Adapters and low-quality reads were removed using the Fastp software (Chen et al. 2018), after which clean reads were mapped to the reference genome using the STAR aligner (Dobin and Gingeras 2015). The European plum (P. domestica) mRNA-seq datasets were analyzed using the Prunus domestica Draft Genome Assembly v1.0.a1 (Callahan et al. 2021) as the reference genome. Because P. domestica is a hexaploid (2n = 6x = 48), the default settings of the cd-hit program (Li and Godzik 2006) were applied to exclude highly similar transcripts, while retaining longer and specific transcripts as references. Subsequently, Fastp (Chen et al. 2018) was used to eliminate adapters and low-quality reads. The clean reads were then mapped to unique transcripts in the Prunus domestica Draft Genome Assembly v1.0.a1 using the Salmon software (Patro et al. 2017).
Mapped reads were counted using featureCounts (Liao et al. 2014). Raw read counts were normalized to transcripts per million (TPM) values. The DESeq2 package in R (Love et al. 2014) and the following criteria were used to identify DEGs: |log2(fold-change)|> 0.5 and adjusted p < 0.05. Orthologs across species were classified using the OrthoFinder software (Emms and Kelly 2019). On the basis of orthogroups, conserved differentially expressed orthologs were identified and filtered out. A GO enrichment analysis was conducted using the default parameters of the ClusterProfiler package v3.16.1 (Yu et al. 2012). The criteria for identifying significantly enriched GO terms were as follows: pvalueCutoff = 0.01, pAdjustMethod = “BH”, and qvalueCutoff = 0.05. The obtained data were visualized using the Revigo platform (http://revigo.irb.hr/).
Gene expression analysis via qRT-PCR
The cDNA synthesized from approximately 1 μg total RNA was used for the qRT-PCR analysis, which was performed using the SYBR Green Master mix (Roche), gene-specific primers, and the LightCycler 480 system (Roche, Basel, Switzerland). The apple SAND gene (MDP0000185470 and/or MDP0000202305) (Velasco et al. 2010) was selected as the reference control. The PCR program was as follows: 95 °C for 30 s and then 45 cycles of 95 °C for 15 s, 57 °C for 30 s, and 72 °C for 60 s. Gene-specific amplifications were confirmed by a dissociation curve analysis. Three biological replicates were analyzed, with three leaf samples per RNA extraction. Details regarding the qRT-PCR primers are provided in Supplementary Table S4.
Measurement of ethylene emission via gas chromatography
Ethylene emitted from leaf samples was detected according to an established protocol (Nakano et al. 2003) involving a gas chromatograph (GC8 CMPF, Shimadzu). Collected leaf samples were enclosed in 50 mL airtight tubes containing 20 mL water and kept at room temperature. Ethylene emission was quantified after 24 and 72 h for DEX-treated leaves or after 3, 5, and 7 days for P. mume leaves. More specifically, 1 mL headspace air was extracted from each tube; the minimum detectable ethylene concentration was 0.05 nL. At least three replicates were analyzed per time point.
Quantification of IAA, JA, and their derivatives in 35S:PmDAM6-GR transgenic apple and WT leaves
Fresh leaves were stored at − 80 °C until they were ground to a fine powder using the Multi-beads shocker (Yasui Kikai Co., Osaka, Japan). Published solid phase extraction procedures were used to extract IAA, JA, and JA-isoleucine (JA-Ile) (Yamane et al. 2019). The IAA, JA and JA-Ile contents were quantified using a liquid chromatography–triple quadrupole mass spectrometry system (6410-LC/MS QQQ, Agilent).
Quantitative analysis of ACC, a precursor of ethylene in the ethylene biosynthesis pathway
In November 2015, control and DEX-treated GR22 leaves were collected at 0, 24, and 72 h post-treatment. Leaf tissues (three replicates with 40 mg per replicate) were frozen in liquid nitrogen and stored at − 80 °C. The primary metabolite profiles of the leaves were analyzed using a CE-MS system (G2201AA, Agilent) as described previously (Oikawa et al. 2011).
Evaluation of ethylene- and JA-induced leaf abscission and senescence in apple and P. mume plants
Wild-type apple ‘JM2’ and P. mume ‘Nanko’ trees were selected for an analysis of the effects of ethephon and JA treatments on leaf abscission and senescence. For the apple trees, 100 or 500 ppm ethephon solutions containing 0.1% Tween 20 were applied to the leaf surface using a hand sprayer on December 2, 2022. The leaf abscission rate was determined 2 weeks later. On October 26, 2017, samples were treated with 1 or 5 mM JA solutions containing 0.1% Triton X-100. They were then photographed 3 weeks later. For the ‘Nanko’ trees, 100 or 500 ppm ethephon solutions containing 0.1% Tween 20 were applied to the leaf surface on October 16, 2023, after which the leaves were photographed at 10 days post-treatment, whereas the leaf abscission rate was determined at 3 weeks post-treatment. On October 21, 2023, samples were treated with 1 or 5 mM JA solutions containing 0.1% Tween 20. They were photographed 3 weeks later.
Statistical analyses
Data were analyzed using Student’s t-test, Fisher’s exact test, and the Tukey Honestly Significant Difference test in R Studio. The threshold for determining statistically significant differences was p < 0.05.
Results
Comparative transcriptome analysis of transgenic apple and European plum leaves overexpressing PmDAM6 and PpeDAM6, respectively
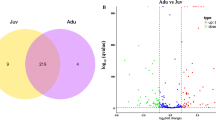
In 35S:PmDAM6-GR transgenic apple leaves, 2862 and 1922 genes had up- and down-regulated expression levels, respectively, following the dexamethasone (DEX) treatment. The 2,862 up-regulated genes included 2,079 orthogroups, whereas the 1,922 down-regulated genes included 1,465 orthogroups (Fig. 1a; Supplementary Fig. S1a). A total of 4,896 and 4,948 genes were expressed at significantly higher and lower levels, respectively, in the leaves of 35S:PpeDAM6 transgenic European plum lines TG1 and TG2 (Lloret et al. 2021) than in the wild-type (WT) leaves. The classification of these genes into orthogroups revealed that 3522 and 3702 orthogroups were expressed at higher and lower levels, respectively, in transgenic lines (Fig. 1a; Supplementary Fig. S1b). The expression levels of 713 and 491 orthogroups were higher and lower, respectively, in both DEX-treated 35S:PmDAM6-GR transgenic apple and 35S:PpeDAM6 transgenic European plum than in the control and WT, respectively (Fig. 1a; Supplementary Fig. S1a). There was a significant overlap in the conserved orthogroups (Fisher’s exact test, p < 0.01) in the PmDAM6- and PpeDAM6-overexpressing samples, suggesting the genes regulated by Prunus DAM6 are highly conserved among species. Moreover, Prunus DAM6 is likely a transcription factor that rapidly affects the expression of downstream genes (Fig. 1b; Supplementary Fig. S1b, Tables S1–S2).
Genes significantly up-regulated in DEX-treated 35S:PmDAM6-GR apple and 35S:PpeDAM6 European plum leaves and the associated enriched GO terms (A) Venn diagram of the up-regulated orthogroups in DEX-treated 35S:PmDAM6-GR transgenic apple and 35S:PpeDAM6 transgenic European plum leaves. (B) Visualization of the results of the GO enrichment analysis of commonly up-regulated DEGs by the Revigo platform
Our Gene Ontology (GO) enrichment analysis revealed GO terms related to ‘ethylene biosynthesis and signaling,’ ‘JA biosynthesis and signaling,’ and ‘leaf senescence and abscission’ were enriched among the 1001 differentially expressed genes (DEGs) belonging to the 713 conserved orthogroups up-regulated by the overexpression of Prunus DAM6 (Fig. 1b, Supplementary Table S1). Notably, previous studies did not indicate that DAM6 mediates ethylene biosynthesis and signaling. Thus, we conducted further analyses to address this possibility (described in the following section). The GO enrichment analysis also revealed photosynthesis-associated GO terms, such as ‘photosynthetic electron transport in photosystem I,’ ‘photosystem II assembly,’ ‘photosystem II repair,’ ‘photosynthetic electron transport chain,’ and ‘chlorophyll biosynthetic process,’ were enriched among the 605 DEGs belonging to the 491 conserved orthogroups down-regulated by the overexpression of Prunus DAM6 (Supplementary Fig. S1, Table S2). Additionally, ‘water transport’ and ‘response to water deprivation’ were also enriched GO terms assigned to the genes down-regulated by DAM6 overexpression (Supplementary Figs S1 and S2, Table S2). Of the genes annotated with these water-related GO terms, the expression levels of six AQUAPORIN (AQP) genes (MD01G1148400, MD03G1117900, MD01G1108500, MD01G1062800, MD11G1136200, and MD05G1122600) in 35S:PmDAM6-GR transgenic apple leaves were strongly repressed by the DEX treatment (Supplementary Figs S1 and S2). In hormone-related pathways, the ‘response to auxin’ GO term was enriched among the genes down-regulated by DAM6 overexpression (Supplementary Fig. S1). The expression levels of several genes encoding AUXIN-RESPONSIVE PROTEINS (MD04G1225100, MD12G1241800, MD02G1057200, and MD09G1202300) and AUXIN RESPONSE FACTOR (ARF) (MD00G1103900 and MD10G1192900) were down-regulated in the DEX-treated 35S:PmDAM6-GR transgenic apple leaves (compared with the corresponding control expression levels). However, indole-3-acetic acid (IAA) levels in 35S:PmDAM6-GR transgenic apple leaves treated with DEX did not differ from the IAA levels in the control and WT leaves (Supplementary Fig. S3; Table S2).
Overexpression of Prunus DAM6 promotes ethylene and JA accumulation as well as leaf senescence and abscission in transgenic plants
The DEX treatment of 35S:PmDAM6-GR transgenic apple plants in October promoted leaf abscission and senescence (Fig. 2a), which is in accordance with our GO enrichment analysis (Fig. 1b). The DEX treatment resulted in the following two distinct phenotypes: (1) leaves, which were brown and dehydrated, dropped within approximately 3–5 days; (2) leaves remained attached to branches, but were noticeably yellow (Fig. 2b).
DEX treatment up-regulated the expression of ethylene biosynthesis-related genes and increased ethylene emission from 35S:PmDAM6-GR transgenic apple leaves. (A) DEX treatment induced 35S:PmDAM6-GR transgenic apple leaf senescence and abscission. Scale bar = 1 cm (upper panel) and 30 cm (bottom panel). (B) DEX treatment induced leaf yellowing or browning at 3 weeks post-treatment. Scale bar = 1 cm. (C) Changes in ethylene biosynthesis-related gene expression at 8 and 16 h after 35S:PmDAM6-GR transgenic apple leaves were treated with DEX (as determined by RNA-seq). The heat map presents log2(fold-change) values (expression levels in 35S:PmDAM6-GR transgenic apple/control leaves; n = 2–3). (D) Changes in the expression of ethylene biosynthesis-related genes at 8 and 16 h after the leaves of two 35S:PmDAM6-GR transgenic apple lines (GR21 and GR22) were treated with DEX (as determined by qRT-PCR); S-ADENOSYL-METHIONINE SYNTHETASE (MetK), 1-AMINOCYCLOPROPANE-1-CARBOXYLIC ACID SYNTHASE (ACS), and 1-AMINOCYCLOPROPANE-1-CARBOXYLIC ACID OXIDASE (ACO). (E) DEX treatment induced the accumulation of the ethylene precursor aminocyclopropane-1-carboxylic acid (ACC) in leaves at 24 and 72 h after 35S:PmDAM6-GR transgenic apple leaves were treated with DEX. (F) DEX treatment induced the emission of ethylene from 35S:PmDAM6-GR transgenic apple leaves
Our GO enrichment analysis suggested peach and Japanese apricot DAM6 up-regulates the expression of genes associated with ethylene biosynthesis and signaling pathways (Fig. 1b). Ethylene promotes leaf senescence and abscission in model plants (Iqbal et al. 2017). Thus, we hypothesized that Prunus DAM6 may promote leaf senescence and abscission by regulating ethylene metabolism. To test this hypothesis, we examined the expression of genes involved in ethylene metabolic pathways. Consistent with our GO enrichment analysis, the expression levels of most genes involved in the ethylene biosynthesis pathway were significantly up-regulated from 8 h after the DEX treatment (Fig. 2c). Furthermore, our quantitative reverse transcription polymerase chain reaction (qRT-PCR) results confirmed the up-regulated expression of the ethylene biosynthesis-related genes in both 35S:PmDAM6-GR transgenic lines (GR21 and GR22) at 8 h after the DEX treatment (Fig. 2d). Ethylene biosynthesis-related genes, namely S-ADENOSYL-METHIONINE SYNTHETASE (MetK) (MD16G1138300), 1-AMINOCYCLOPROPANE-1-CARBOXYLATE SYNTHASE (ACS) (MD06G1090600), and 1-AMINOCYCLOPROPANE-1-CARBOXYLATE OXIDASE (ACO) (MD17G1106300 and MD09G1114800), had significantly up-regulated expression levels at 16 h after the DEX treatment (Fig. 2d). Similarly, in 35S:PpeDAM6 transgenic European plum lines TG1 and TG2, ethylene biosynthesis-related gene expression levels in leaves were consistently up-regulated (Supplementary Fig. S4a). In addition, there was a significant increase in the abundance of the ethylene biosynthesis precursor aminocyclopropane-1-carboxylate (ACC) in 35S:PmDAM6-GR transgenic apple leaves following the DEX treatment (Fig. 2e). The leaves of both 35S:PmDAM6-GR transgenic lines (GR21 and GR22) accumulated substantially more ethylene than the control and WT leaves at 16 h after the DEX treatment (Fig. 2f). In particular, DEX-treated GR22 leaves had an extremely high ethylene level, which corresponded with their relatively high ethylene biosynthesis-related gene expression levels (compared with the corresponding expression in GR21), suggestive of the positive effect of PmDAM6 on ethylene metabolic pathways (Fig. 2f). Finally, 35S:PmDAM6 transgenic apple lines 35S-2 and 35S-4 underwent earlier leaf abscission and senescence and accumulated more ethylene at 3 and 5 days after sampling than WT in December 2023 (Fig. 3).
Overexpression of PmDAM6 resulted in earlier leaf senescence and abscission and increased ethylene emission from the leaves of transgenic apple plants (A) Leaf senescence and abscission occurred earlier for PmDAM6-overexpressing apple plants than for wild-type plants (photographed on December 13, 2020). Scale bar = 50 cm. (B) Ethylene emitted from the leaves of two apple transgenic lines overexpressing PmDAM6 (35S-2 and 35S-4) and wild-type plants at 3 and 5 days after leaves were collected and incubated in a sealed tube. Leaf samples were collected on December 18, 2023. Data are presented as the mean ± standard error. An asterisk indicates significant differences (t-test, p < 0.05). Scale bar = 50 cm
In addition to the ethylene biosynthesis pathway genes, ethylene signaling pathway genes also had up-regulated expression levels in DEX-treated 35S:PmDAM6-GR transgenic apple and 35S:PpeDAM6 transgenic European plum (Supplementary Figs S4b and S5). In 35S:PmDAM6-GR transgenic apple lines GR21 and GR22, the expression levels of ETHYLENE-INSENSITIVE 3 (EIN3) (MD07G1053800 and MD02G1266200) and ETHYLENE RESPONSE FACTOR (ERF) (MD05G1198700 and MD16G1216900) were significantly up-regulated after the DEX treatment (Supplementary Fig. S5a-b, Table S1). We also assessed the effects of exogenous ethylene on apple leaf senescence and abscission. The application of 500 ppm ethephon significantly induced leaf abscission at 2 weeks post-treatment (Fig. 4).
Interestingly, the seasonal environment appeared to influence the ethylene-mediated leaf senescence and abscission phenotypes of the plants overexpressing PmDAM6. Applying DEX to 35S:PmDAM6-GR transgenic apple leaves in summer resulted in leaf abscission, but not leaf senescence. Moreover, there were no significant differences in the total chlorophyll contents of the DEX-treated leaves and the control in summer (Supplementary Fig. S6a–b). We also investigated the changes in the expression of genes related to ethylene biosynthesis and signaling in leaf samples collected at 24 h after the DEX treatment in summer (relative to the corresponding control and WT gene expression). The expression levels of the ethylene biosynthesis-related genes ACS (MD06G1090600) and ACO (MD17G1106300 and MD09G1114800) were significantly higher in the DEX-treated leaves of 35S:PmDAM6-GR transgenic lines GR21 and GR22 than in the control and WT leaves (Supplementary Fig. S6c). However, the expression of the ethylene signaling-related genes EIN3 (MD07G1053800) and ERF (MD05G1198700 and MD16G1216900) was not up-regulated in GR21 and GR22, which was in contrast to the significantly up-regulated expression of these genes in autumn (Supplementary Fig. S6d). Collectively, these results suggest that the ethylene signaling pathway may need to be activated for PmDAM6-induced leaf senescence in autumn.
Because our comparative transcriptome analysis also revealed the increased expression of genes associated with JA biosynthesis and signaling in transgenic plants overexpressing Prunus DAM6 (Fig. 1b), we examined the expression of JA metabolism-related genes and JA accumulation. In 35S:PmDAM6-GR transgenic apple leaves, the expression levels of genes related to JA biosynthesis and signaling were significantly up-regulated from 8 h after the DEX treatment (Supplementary Fig. S7a–b). Additionally, the accumulation of JA and jasmonic acid-isoleucine (JA-Ile) peaked at 16 h after the DEX treatment (Supplementary Fig. S7c). Furthermore, JA biosynthesis-related gene expression levels were higher and more JA accumulated in 35S:PpeDAM6 transgenic European plum leaves than in WT leaves (Lloret et al. 2021). Similar to the ethephon treatment, the JA treatment induced leaf senescence and abscission, but only when the concentration was extremely high (5 mM), at 3 weeks after apple plants were treated (Supplementary Fig. S8a–b).
Correlation between PmDAM6 expression and ethylene biosynthesis-related gene expression and ethylene emission in Prunus mume leaves
We selected two P. mume cultivars with contrasting PmDAM6 expression patterns and leaf shedding characteristics. ‘Nanko’ typically exhibits leaf senescence and shedding starting in mid-to-late November, with peak leaf shedding and yellowing in early December, whereas ‘Ellching’ does not exhibit leaf senescence and abscission by early December, but sheds leaves in early January, approximately 1 month later than ‘Nanko’ (Fig. 5a–b). Seasonal PmDAM6 expression patterns in leaves also differed between the two cultivars, with significantly higher expression levels in ‘Nanko’ than in ‘Ellching’ before November 15 (Fig. 5c–d). The PmDAM6 expression level in ‘Nanko’ leaves peaked on November 15, but then subsequently decreased (Fig. 5d). In contrast, in ‘Ellching’ leaves, PmDAM6 expression increased significantly from November 15 to November 30 (peak expression) and then gradually decreased (Fig. 5d). From September 30 to November 15, ACS (LOC103337567) and ACO (LOC103329345) along with PmDAM6 were more highly expressed in ‘Nanko’ leaves than in ‘Ellching’ leaves (Fig. 5c). The expression levels of ethylene biosynthesis-related genes, such as MetK (LOC103323276, LOC103327270, LOC103339824, and LOC103344495), ACS (LOC103337567), and ACO (LOC103329345), were significantly up-regulated following the PmDAM6 expression peak on November 30 in ‘Ellching’ (Fig. 5d). Moreover, ‘Nanko’ emitted significantly more ethylene than ‘Ellching’ in late November (Fig. 5e). We also examined the seasonal changes in ethylene signaling-related gene expression (Supplementary Fig. S9a–d). The results revealed a gradual increase in EIN3 (LOC103331049) expression before leaf senescence and shedding in ‘Nanko’ and ‘Ellching’ (Supplementary Fig. S9b). Similar results were obtained when we compared ‘Nanko’ with ‘SC’, which is another low-chill cultivar. Like ‘Ellching’, ‘SC’ did not exhibit leaf senescence and shedding from late November to December (i.e., leaf senescence and shedding occurred much later for ‘SC’ than for ‘Nanko’) and had lower PmDAM6, MetK, ACS, and ACO expression levels and ethylene emission rates than ‘Nanko’ (Supplementary Fig. S10). Finally, exogenous ethephon (ethylene-releasing compound) induced leaf senescence and abscission in P. mume, similar to its effects on apple (Fig. 6). We detected significant leaf shedding within 2 days of the 500 ppm ethephon treatment; the shed leaves had brown spots and appeared to be dehydrated. Furthermore, from 7 days after ethephon treatments (100 and 500 ppm), the leaves still attached to branches began to turn yellow. Notably, in P. mume, a visible abscission layer formed between the leaf petiole and the branch (Fig. 6d). In addition, the ethephon treatment significantly increased the leaf abscission rate, but JA did not substantially affect leaf abscission (Supplementary Fig. S8c).
Seasonal changes in leaf senescence, abscission, expression of ethylene biosynthesis-related genes, and ethylene emission in two P. mume cultivars with contrasting leaf shedding timing (A, B) Photographs of the whole tree (A) and leaves (B) of two P. mume cultivars with contrasting PmDAM6 expression patterns and leaf abscission characteristics. Scale bar = 2 cm. (C) Heat map of the expression of ethylene biosynthesis-related genes in the P. mume v1.0 genome. The heat map presents log2(fold-change) values [expression levels in Nanko (high-chill)/Ellching (low-chill)]. (D) Seasonal changes in the expression of ethylene biosynthesis-related genes relative to the PmDAM6 expression level in the leaves of Japanese apricot cultivars ‘Nanko’ and ‘Ellching’; S-ADENOSYL-METHIONINE SYNTHETASE (MetK), 1-AMINOCYCLOPROPANE-1-CARBOXYLIC ACID SYNTHASE (ACS), and 1-AMINOCYCLOPROPANE-1-CARBOXYLIC ACID OXIDASE (ACO). Data are presented as the mean ± standard error (n = 3). Left and right Y-axes present the relative expression of ethylene biosynthesis-related genes and PmDAM6, respectively. (E) Ethylene emission from ‘Nanko’ and ‘Ellching’ leaves at 7 days after they were collected on November 23, 2023 and incubated in a sealed tube at room temperature. Data are presented as the mean ± standard error (n = 5). An asterisk indicates significant differences (t-test, p < 0.05)
Effects of an ethephon treatment on leaf senescence and abscission in P. mume. (A) Photographs of shoots treated with 100 or 500 ppm ethephon at 10 days post-treatment. Leaf senescence and abscission were promoted by 100 and 500 ppm ethephon on October 16, 2023. Scale bar = 2 cm. (B) Effects of ethephon on the leaf abscission rate. Data are presented as the mean ± standard error (n = 5). Asterisks indicate significant differences (** or *, p < 0.01 or 0.05; t-test). (C) Photographs of leaves treated with 100 or 500 ppm ethephon at 10 days post-treatment. Scale bar = 2 cm. (D) Ethephon stimulated the formation of a leaf abscission layer. Scale bar = 2 cm
Discussion
The regulatory role of DAM genes in controlling bud dormancy progression has been extensively documented in various Rosaceae species, including peach (P. persica) (Leida et al. 2012; Lloret et al. 2021), Japanese apricot (P. mume) (Yamane et al. 2019; Hsiang et al. 2024), apple (Malus × domestica) (Porto et al. 2016; Wu et al. 2021), and pear (Pyrus pyrifolia white pear group) (Niu et al. 2015; Tuan et al. 2017). However, although DAM mutants [DAM RNAi and DAM-overexpressing transgenic plants (Wang et al. 2002; Bielenberg et al. 2008; Wu et al. 2021)] reportedly show altered phenotypes in bud dormancy as well as other traits, such as leaf shedding, the regulatory effects of DAMs on phenological traits other than bud dormancy remain unclear. In this study, we used DAM-overexpressing transgenic lines to elucidate how Prunus DAM6 regulates the leaf traits of deciduous trees.
A previous study showed that the overexpression of peach PpeDAM6 increases the expression of JA biosynthesis-related genes and JA accumulation in transgenic European plum leaves (Lloret et al. 2021). In the current study, DEX-treated 35S:PmDAM6-GR transgenic apple leaves accumulated significantly more JA and JA-Ile than control leaves, which is consistent with the findings for 35S:PpeDAM6 transgenic European plum (Lloret et al. 2021). A treatment with an extremely high JA concentration promoted leaf senescence. In contrast to the JA treatment, the application of ethephon induced a rapid and significant response, which was characterized by substantial leaf yellowing, dehydration, and abscission within a short period. A comparative transcriptome analysis revealed the conserved regulatory effect of Prunus DAM6 on ethylene biosynthesis. Moreover, 35S:PmDAM6 and 35S:PmDAM6-GR transgenic apple leaves emitted more ethylene than WT and control leaves, respectively. At the transcriptional level, the expression of genes related to ethylene biosynthesis and signaling was significantly up-regulated in the DEX-treated leaves of 35S:PmDAM6-GR transgenic apple and 35S:PpeDAM6 transgenic European plum. In model plants, JA-induced leaf senescence depends on key factors associated with ethylene signaling, including EIN3 (Li et al. 2013). Earlier research indicated the overexpression of EIN3 accelerates leaf senescence, whereas a loss-of-function mutation to this gene (e.g., in the ein3 mutant) leads to delayed JA-induced leaf senescence (Li et al. 2013; Koyama and Sato 2018). On the basis of our results and those of reported studies, we propose that ethylene rather than JA plays a central regulatory role in Prunus DAM6-mediated leaf senescence and abscission. The seasonal environment significantly impacts ethylene-mediated leaf senescence in apples overexpressing PmDAM6. Previous studies show that ethylene signaling genes like EIN2 and EIN3 modulate leaf senescence sensitivity (Yu et al. 2021; Qiu et al. 2015). In our study, DEX treatment in summer did not activate ethylene signaling in 35S:PmDAM6-GR transgenic apple leaves, leading to leaf abscission without senescence. However, in autumn, DEX treatment significantly increased ethylene signaling, resulting in pronounced senescence. This suggests that PmDAM6-induced senescence requires specific environmental cues or developmental stages, particularly in autumn (Fig. 7).
Proposed model of PmDAM6 as a hub gene controlling leaf abscission/senescence and bud dormancy induction in P. mume. During summer, PmDAM6 expression is low in P. mume leaves (Sasaki et al. 2011). In transgenic apple overexpressing PmDAM6 in summer, PmDAM6 promotes ethylene biosynthesis, initiating signals for leaf abscission and dehydration via decreased aquaporins (AQP) expressions. However, ethylene signaling for leaf abscission is reduced or inactive during summer. In contrast, during autumn, with the onset of chilling temperatures and shorter photoperiods, high PmDAM6 expression is observed in P. mume leaves (Sasaki et al. 2011). In autumn, PmDAM6 stimulates not only ethylene biosynthesis but also ethylene signaling in both P. mume and transgenic apple, which led to significant leaf senescence and abscission. PmDAM6 concurrently inducing bud dormancy through changes in hormone metabolism, lipid accumulation, and repression of cell division (Hsiang et al. 2024)
Our transcriptome analysis suggested Prunus DAM6 may have suppressive effects on ‘photosystem I,’ ‘photosystem II assembly,’ ‘chlorophyll biosynthetic process,’ ‘water channel activity,’ and ‘water transport’ in leaves. In several plant species, ethylene inhibits photosynthesis and water loss during the leaf and petal wilting process (Kays and Pallas 1980; Pallas et al. 1982; Mohorović et al. 2023a Qing et al. 2016). Previous studies indicated that a rose AQP protein, Rh-PIP2;1, is a crucial regulator of petal expansion; the inhibited expression of Rh-PIP2;1 due to ethylene suppresses petal expansion (Ma et al. 2008). Another study showed that an ethylene treatment rapidly and significantly down-regulates RhPIP1;1 expression and that silencing RhPIP1;1 expression in rose significantly inhibits petal expansion, resulting in decreases in petal size and fresh weight (Chen et al. 2013). Similar to the growth of other plant organs, leaf growth involves irreversible cell expansion, which is dependent on the water potential. The expression of AQP genes in leaves is critical for maintaining cell growth and expansion because of the associated regulation of the leaf water potential (Uehleln et al. 2003; Volkov et al. 2007; Wei et al. 2007; Heinen et al. 2009; Fricke 2017). In tobacco, overexpressing NtAQP1 leads to increases in leaf area and plant height (Uehleln et al. 2003; Siefritz et al. 2004; Kelly et al. 2014). The overexpression of AtPIP1;2 increases plant growth and leaf transpiration rates as well as stomatal density and photosynthetic efficiency (Aharon et al. 2003). In antisense NtAQP1 transgenic plants, net photosynthesis reportedly decreases, whereas photosynthetic efficiency increases in NtAQP1-overexpressing transgenic plants (Uehleln et al. 2003). Notably, the inhibition of photosynthesis is a necessary part of leaf senescence (Thakur et al. 2016; Krieger-Liszkay et al. 2019; Sakuraba 2021). In addition, water loss and rehydration promote leaf abscission (Agustí et al. 2007; Mahouachi et al. 2007; Dallstream and Piper 2021). For example, water stress enhances ethylene-mediated leaf abscission in cotton (Jordan et al. 1972). After considering these findings, we hypothesize that Prunus DAM6 may induce water stress by suppressing the expression of AQP genes through the activation of ethylene biosynthesis and signaling pathways, while also repressing photosynthesis in leaves, thereby facilitating ethylene-mediated leaf senescence and abscission.
To date, many studies have been conducted to clarify the molecular basis of the regulatory roles of Rosaceae DAMs in dormant buds. These studies revealed the regulatory functions of DAMs affecting phytohormone metabolism (Tuan et al. 2017; Busov 2019; Falavigna et al. 2019, 2021; Yamane et al. 2019; Yang et al. 2020; Lloret et al. 2021; Wu et al. 2021; Hsiang et al. 2024), cell division (Yang et al. 2020; Falavigna et al. 2021; Wu et al. 2021; Hsiang et al. 2024), and lipid catabolism (Hsiang et al. 2024), thereby control bud dormancy. However, the tree dormancy period is divided into several distinct phases, including dormancy induction (leaf abscission, terminal bud set, and bud development), dormancy establishment (complete developmental arrest), and dormancy release (release from bud break repression). It remains unclear which process is primarily affected by DAMs. In the present study, we determined that PmDAM6 regulates ethylene metabolism and promotes leaf senescence and abscission in autumn, which precedes the onset of bud dormancy (Fig. 7). This may imply that in Rosaceae, DAM may be involved in multiple metabolic pathways during the tree dormancy induction phase in autumn. Furthermore, DAM may be a hub gene linking leaf abscission with bud dormancy onset during the regulation of the tree growth/dormancy cycle.
Data availability
All pertinent information is included within the manuscript and its accompanying materials. The RNA-seq data for the 35S:PmDAM6-GR apple transformants and control produced in this study were archived in the Gene Expression Omnibus database of the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/geo) under the accession number PRJNA1072146.
References
Agustí J, Conesa A, Cercós M, Talón M, Tadeo FR (2007) Calcium signaling in water stress-induced leaf abscission in citrus plants. Adv Plant Ethyl Res. https://doi.org/10.1007/978-1-4020-6014-4_66
Aharon R, Shahak Y, Wininger S, Bendov R, Kapulnik Y, Galili G (2003) Overexpression of a plasma membrane aquaporin in transgenic tobacco improves plant vigor under favorable growth conditions but not under drought or salt stress. Plant Cell 15:439. https://doi.org/10.1105/TPC.009225
Beil I, Kreyling J, Meyer C, Lemcke N, Malyshev AV (2021) Late to bed, late to rise—warmer autumn temperatures delay spring phenology by delaying dormancy. Glob Chang Biol 27:5806–5817. https://doi.org/10.1111/GCB.15858
Bielenberg DG, Wang Y, Li Z, Zhebentyayeva T, Fan S, Reighard GL, Scorza R, Abbott AG (2008) Sequencing and annotation of the evergrowing locus in peach [Prunus persica (L.) Batsch] reveals a cluster of six MADS-box transcription factors as candidate genes for regulation of terminal bud formation. Tree Genet Genomes 4:495–507. https://doi.org/10.1007/s11295-007-0126-9
Busov VB (2019) Plant development: dual roles of poplar SVL in vegetative bud dormancy. Curr Biol 29:68–70. https://doi.org/10.1016/j.cub.2018.11.061
Callahan AM, Zhebentyayeva TN, Humann JL, Saski CA, Galimba KD, Georgi LL, Scorza R, Main D, Dardick CD (2021) Defining the ‘HoneySweet’ insertion event utilizing NextGen sequencing and a de novo genome assembly of plum (Prunus domestica). Hortic Res. https://doi.org/10.1038/S41438-020-00438-2
Chen W, Yin X, Wang L, Tian J, Yang R, Liu D, Yu Z, Ma N, Gao J (2013) Involvement of rose aquaporin RhPIP1;1 in ethylene-regulated petal expansion through interaction with RhPIP2;1. Plant Mol Biol 83:219–233. https://doi.org/10.1007/S11103-013-0084-6/METRICS
Chen S, Zhou Y, Chen Y, Gu J (2018) fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34:i884–i890. https://doi.org/10.1093/BIOINFORMATICS/BTY560
da Falavigna V, S, Guitton B, Costes E, Andrés F, (2019) I want to (bud) break free: the potential role of DAM and SVP-Like genes in regulating dormancy cycle in temperate fruit trees. Front Plant Sci 9:1–17. https://doi.org/10.3389/fpls.2018.01990
da Falavigna V, S, Severing E, Lai X, Estevan J, Farrera I, Hugouvieux V, Revers LF, Zubieta C, Coupland G, Costes E, Andrés F, Falavigna V da S, Severing E, Lai X, Estevan J, Farrera I, Hugouvieux V, Revers LF, Zubieta C, Coupland G, Costes E, Andrés F, (2021) Unraveling the role of MADS transcription factor complexes in apple tree dormancy. New Phytol 232:2071–2088. https://doi.org/10.1111/nph.17710
Daccord N, Celton JM, Linsmith G, Becker C, Choisne N, Schijlen E, Van De Geest H, Bianco L, Micheletti D, Velasco R, Di Pierro EA, Gouzy J, Rees DJG, Guérif P, Muranty H, Durel CE, Laurens F, Lespinasse Y, Gaillard S, Aubourg S, Quesneville H, Weigel D, Van De Weg E, Troggio M, Bucher E (2017) High-quality de novo assembly of the apple genome and methylome dynamics of early fruit development. Nat Genet 49:1099–1106. https://doi.org/10.1038/ng.3886
Dallstream C, Piper FI (2021) Drought promotes early leaf abscission regardless of leaf habit but increases litter phosphorus losses only in evergreens. Aust J Bot 69:121–130. https://doi.org/10.1071/BT20052
Dobin A, Gingeras TR (2015) Mapping RNA-seq reads with STAR. Curr Protoc Bioinform. https://doi.org/10.1002/0471250953.BI1114S51
Emms DM, Kelly S (2019) OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol 20:1–14. https://doi.org/10.1186/S13059-019-1832-Y/FIGURES/5
Faust M, Erez A, Rowland LJ, Wang SY, Norman HA (1997) Bud dormancy in perennial fruit trees. HortScience 32:623–629
Ferrante A, Francini A (2006) Ethylene and leaf senescence. Ethyl Action Plant. https://doi.org/10.1007/978-3-540-32846-9_3/COVER
Fricke W (2017) Water transport and energy. Plant Cell Environ 40:977–994. https://doi.org/10.1111/PCE.12848
Guak S, Fuchigami LH (2001) Effects of applied ABA on growth cessation, bud dormancy, cold acclimation, leaf senescence and N mobilization in apple nursery plants. J Hortic Sci Biotechnol 76(4):459–464. https://doi.org/10.1080/14620316.2001.11511394
He Y, Fukushige H, Hildebrand DF, Gan S (2002) Evidence supporting a role of jasmonic acid in Arabidopsis leaf senescence. Plant Physiol 128:876–884. https://doi.org/10.1104/PP.010843
Heinen RB, Ye Q, Chaumont F (2009) Role of aquaporins in leaf physiology. J Exp Bot 60:2971–2985. https://doi.org/10.1093/JXB/ERP171
Hsiang TF, Yamane H, Gao-Takai M, Tao R (2024) Regulatory role of Prunus mume DAM6 on lipid body accumulation and phytohormone metabolism in the dormant vegetative meristem. Hortic Res. https://doi.org/10.1093/hr/uhae102
Hu Y, Jiang Y, Han X, Wang H, Pan J, Yu D (2017) Jasmonate regulates leaf senescence and tolerance to cold stress: crosstalk with other phytohormones. J Exp Bot 68:1361–1369. https://doi.org/10.1093/JXB/ERX004
Iqbal N, Khan NA, Ferrante A, Trivellini A, Francini A, Khan MIR (2017) Ethylene role in plant growth, development and senescence: interaction with other phytohormones. Front Plant Sci 8:235913. https://doi.org/10.3389/FPLS.2017.00475/BIBTEX
Jiménez S, Lawton-Rauh AL, Reighard GL, Abbott AG, Bielenberg DG (2009) Phylogenetic analysis and molecular evolution of the dormancy associated mads-box genes from peach. BMC Plant Biol 9:1–12. https://doi.org/10.1186/1471-2229-9-81
Jordan WR, Morgan PW, Davenport TL (1972) Water stress enhances ethylene-mediated leaf abscission in cotton. Plant Physiol 50:756–758. https://doi.org/10.1104/PP.50.6.756
Kays SJ, Pallas JE (1980) Inhibition of Photosynthesis by Ethylene Nat 285:51–52. https://doi.org/10.1038/285051a0
Kelly G, Sade N, Attia Z, Secchi F, Zwieniecki M, Holbrook NM, Levi A, Alchanatis V, Moshelion M, Granot D (2014) Relationship between hexokinase and the aquaporin PIP1 in the regulation of photosynthesis and plant growth. PLoS ONE 9:e87888. https://doi.org/10.1371/JOURNAL.PONE.0087888
Kitamura Y, Habu T, Yamane H, Nishiyama S, Kajita K, Sobue T, Kawai T, Numaguchi K, Nakazaki T, Kitajima A, Tao R (2018) Identification of QTLs controlling chilling and heat requirements for dormancy release and bud break in Japanese apricot (Prunus mume). Tree Genet Gen 14:33. https://doi.org/10.1007/s11295-018-1243-3
Koyama T (2014) The roles of ethylene and transcription factors in the regulation of onset of leaf senescence. Front Plant Sci 5:116514. https://doi.org/10.3389/FPLS.2014.00650/BIBTEX
Koyama T, Sato F (2018) The function of ETHYLENE RESPONSE FACTOR genes in the light-induced anthocyanin production of Arabidopsis thaliana leaves. Plant Biotechnol 35:87–91. https://doi.org/10.5511/PLANTBIOTECHNOLOGY.18.0122B
Krieger-Liszkay A, Krupinska K, Shimakawa G (2019) The impact of photosynthesis on initiation of leaf senescence. Physiol Plant 166:148–164. https://doi.org/10.1111/PPL.12921
Leida C, Conesa A, Llácer G, Badenes ML, Ríos G (2012) Histone modifications and expression of DAM6 gene in peach are modulated during bud dormancy release in a cultivar-dependent manner. New Phytol 193:67–80. https://doi.org/10.1111/j.1469-8137.2011.03863.x
Li W, Godzik A (2006) Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22:1658–1659. https://doi.org/10.1093/BIOINFORMATICS/BTL158
Li Z, Reighard GL, Abbott AG, Bielenberg DG (2009) DORMANCY-ASSOCIATED MADS genes from the EVG locus of peach [Prunus persica (L.) Batsch] have distinct seasonal and photoperiodic expression patterns. J Exp Bot 60:3521–3530. https://doi.org/10.1093/jxb/erp195
Li Z, Peng J, Wen X, Guo H (2013) ETHYLENE-INSENSITIVE 3 is a senescence-associated gene that accelerates age-dependent leaf senescence by directly repressing miR164 transcription in Arabidopsis. Plant Cell 25:3311–3328. https://doi.org/10.1105/TPC.113.113340
Li S, Yang Y, Zhang Q, Liu N, Xu Q, Hu L (2018) Differential physiological and metabolic response to low temperature in two zoysiagrass genotypes native to high and low latitude. PLoS ONE 13:1–20. https://doi.org/10.1371/journal.pone.0198885
Liao Y, Smyth GK, Shi W (2014) featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30:923–930. https://doi.org/10.1093/BIOINFORMATICS/BTT656
Lloret A, Quesada-Traver C, Conejero A, Arbona V, Gómez-Mena C, Petri C, Sánchez-Navarro JA, Zuriaga E, Leida C, Badenes ML, Ríos G (2021) Regulatory circuits involving bud dormancy factor PpeDAM6. Hortic Res. https://doi.org/10.1038/s41438-021-00706-9
Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:1–21. https://doi.org/10.1186/s13059-014-0550-8
Ma N, Xue J, Li Y, Liu X, Dai F, Jia W, Luo Y, Gao J (2008) Rh-PIP2;1, a rose aquaporin gene, is involved in ethylene-regulated petal expansion. Plant Physiol 148:894–907. https://doi.org/10.1104/PP.108.120154
Mahouachi J, Arbona V, Gómez-Cadenas A (2007) Hormonal changes in papaya seedlings subjected to progressive water stress and re-watering. Plant Growth Regul 53:43–51. https://doi.org/10.1007/S10725-007-9202-2/METRICS
Malyshev AV, Beil I, Zohner CM, Garrigues R, Campioli M (2024) The clockwork of spring: bud dormancy timing as a driver of spring leaf-out in temperate deciduous trees. Agric Meteorol 349:109957. https://doi.org/10.1016/J.AGRFORMET.2024.109957
Mohorović P, Geldhof B, Holsteens K, Rinia M, Daems S, Reijnders T, Ceusters J, Van den Ende W, Van de Poel B (2023a) Ethylene inhibits photosynthesis via temporally distinct responses in tomato plants. Plant Physiol. https://doi.org/10.1093/PLPHYS/KIAD685
Mohorović P, Vaughan-Hirsch J, Ceusters J, Van de Poel B (2023b) The role of ethylene in photosynthate partitioning and source-sink modulation in crops. Plant Horm Ethyl Stress Acclim Agric Appl. https://doi.org/10.1016/B978-0-323-85846-5.00010-2
Moser M, Asquini E, Miolli GV, Weigl K, Hanke MV, Flachowsky H, Si-Ammour A (2020) The MADS-box gene MdDAM1 controls growth cessation and bud dormancy in apple. Front Plant Sci 11:1–13. https://doi.org/10.3389/fpls.2020.01003
Nakano R, Ogura E, Kubo Y, Inaba A (2003) Ethylene biosynthesis in detached young persimmon fruit is initiated in calyx and modulated by water loss from the fruit. Plant Physiol 131:276–286. https://doi.org/10.1104/PP.010462
Niu Q, Li J, Cai D, Qian M, Jia H, Bai S, Hussain S, Liu G, Teng Y, Zheng X, Li J, Hussain S, Niu Q, Zheng X, Liu G, Qian M, Bai S (2015) DORMANCY-ASSOCIATED MADS-box genes and microRNAs jointly control dormancy transition in pear ( Pyrus pyrifolia white pear group) flower bud. J Exp Bot 67:239–257. https://doi.org/10.1093/jxb/erv454
Oikawa A, Fujita N, Horie R, Saito K, Tawaraya K (2011) Solid-phase extraction for metabolomic analysis of high-salinity samples by capillary electrophoresis–mass spectrometry. J Sep Sci 34:1063–1068. https://doi.org/10.1002/JSSC.201000890
Pallas JE, JR, Kays SJ, (1982) Inhibition of photosynthesis by ethylene—a stomatal effect. Plant Physiol 70:598–601. https://doi.org/10.1104/PP.70.2.598
Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C (2017) Salmon provides fast and bias-aware quantification of transcript expression. Nat Method 14:417–419. https://doi.org/10.1038/nmeth.4197
Porto DD, da Falavigna V, S, Arenhart RA, Perini P, Buffon V, Anzanello R, dos Santos HP, Fialho FB, de Oliveira PRD, Revers LF, (2016) Structural genomics and transcriptional characterization of the DORMANCY-ASSOCIATED MADS-box genes during bud dormancy progression in apple. Tree Genet Genome. https://doi.org/10.1007/s11295-016-1001-3
Qiu K, Li Z, Yang Z, Chen J, Wu S, Zhu X, Gao S, Gao J, Ren G, Kuai B, Zhou X (2015) EIN3 and ORE1 accelerate degreening during ethylene-mediated leaf senescence by directly activating chlorophyll catabolic genes in arabidopsis. PLoS Genet 11:e1005399. https://doi.org/10.1371/journal.pgen.1005399
Sakuraba Y (2021) Light-mediated regulation of leaf senescence. Int J Mol Sci 22:3291. https://doi.org/10.3390/IJMS22073291
Sasaki R, Yamane H, Ooka T, Jotatsu H, Kitamura Y, Akagi T, Tao R (2011) Functional and expressional analyses of PmDAM genes associated with endodormancy in Japanese apricot. Plant Physiol 157:485–497. https://doi.org/10.1104/pp.111.181982
Saure MC (1985) Dormancy release in deciduous fruit trees. Hortic Rev 7:239–300. https://doi.org/10.1002/9781118060735.ch6
Siefritz F, Otto B, Bienert GP, Van Der Krol A, Kaldenhoff R (2004) The plasma membrane aquaporin NtAQP1 is a key component of the leaf unfolding mechanism in tobacco. Plant J 37:147–155. https://doi.org/10.1046/J.1365-313X.2003.01947.X
Thakur N, Sharma V, Kishore K (2016) Leaf senescence: an overview. Indian J Plant Physiol 21:225–238. https://doi.org/10.1007/S40502-016-0234-3
Tuan PA, Bai S, Saito T, Ito A, Moriguchi T (2017) Dormancy-associated MADS-box (DAM) and the abscisic acid pathway regulate pear endodormancy through a feedback mechanism. Plant Cell Physiol 58:1378–1390. https://doi.org/10.1093/pcp/pcx074
Uehleln N, Lovisolo C, Siefritz F, Kaldenhoff R (2003) The tobacco aquaporin NtAQP1 is a membrane CO2 pore with physiological functions. Nat 425:734–737. https://doi.org/10.1038/nature02027
Velasco R, Zharkikh A, Affourtit J, Dhingra A, Cestaro A, Kalyanaraman A, Fontana P, Bhatnagar SK, Troggio M, Pruss D, Salvi S, Pindo M, Baldi P, Castelletti S, Cavaiuolo M, Coppola G, Costa F, Cova V, Dal Ri A, Goremykin V, Komjanc M, Longhi S, Magnago P, Malacarne G, Malnoy M, Micheletti D, Moretto M, Perazzolli M, Si-Ammour A, Vezzulli S, Zini E, Eldredge G, Fitzgerald LM, Gutin N, Lanchbury J, MacAlma T, Mitchell JT, Reid J, Wardell B, Kodira C, Chen Z, Desany B, Niazi F, Palmer M, Koepke T, Jiwan D, Schaeffer S, Krishnan V, Wu C, Chu VT, King ST, Vick J, Tao Q, Mraz A, Stormo A, Stormo K, Bogden R, Ederle D, Stella A, Vecchietti A, Kater MM, Masiero S, Lasserre P, Lespinasse Y, Allan AC, Bus V, Chagné D, Crowhurst RN, Gleave AP, Lavezzo E, Fawcett JA, Proost S, Rouzé P, Sterck L, Toppo S, Lazzari B, Hellens RP, Durel CE, Gutin A, Bumgarner RE, Gardiner SE, Skolnick M, Egholm M, Van De Peer Y, Salamini F, Viola R (2010) The genome of the domesticated apple (Malus × domestica Borkh.). Nat Genet 42:833–839. https://doi.org/10.1038/ng.654
Volkov V, Hachez C, Moshelion M, Draye X, Chaumont F, Fricke W (2007) Water permeability differs between growing and non-growing barley leaf tissues. J Exp Bot 58:377–390. https://doi.org/10.1093/JXB/ERL203
Wada M, Nishitani C, Komori S (2020) Stable and efficient transformation of apple. Plant Biotechnol 37:163–170. https://doi.org/10.5511/PLANTBIOTECHNOLOGY.20.0602A
Walde MG, Wenden B, Chuine I, Gessler A, Saurer M, Vitasse Y (2024) Stable water isotopes reveal the onset of bud dormancy in temperate trees, whereas water content is a better proxy for dormancy release. Tree Physiol. https://doi.org/10.1093/TREEPHYS/TPAE028
Wang J, Gao Z, Li H, Jiu S, Qu Y, Wang L, Ma C, Xu W, Wang S, Zhang C (2020) DORMANCY-ASSOCIATED MADS-box (DAM) genes influence chilling requirement of sweet cherries and co-regulate flower development with SOC1 gene. Int J Mol Sci 21(3):921. https://doi.org/10.3390/ijms21030921
Wang F, Zhang R, Lin J, Zheng J, Hänninen H, Wu J (2022) High autumn temperatures increase the depth of bud dormancy in the subtropical Torreya grandis and Carya illinoinensis and delay leaf senescence in the deciduous Carya. Trees - Struct Funct 36:1053–1065. https://doi.org/10.1007/S00468-022-02272-6/METRICS
Wei W, Alexandersson E, Golldack D, Miller AJ, Kjellbom PO, Fricke W (2007) HvPIP1;6, a barley (Hordeum vulgare L.) plasma membrane water channel particularly expressed in growing compared with non-growing leaf tissues. Plant Cell Physiol 48:1132–1147. https://doi.org/10.1093/PCP/PCM083
Wu R, Cooney J, Tomes S, Rebstock R, Karunairetnam S, Allan AC, Macknight RC, Varkonyi-Gasic E (2021) RNAi-mediated repression of dormancy-related genes results in evergrowing apple trees. Tree Physiol 41:1510–1523. https://doi.org/10.1093/treephys/tpab007
Yamane H, Kashiwa Y, Ooka T, Tao R, Yonemori K (2008) Suppression subtractive hybridization and differential screening reveals endodormancy-associated expression of an SVP/AGL24-type MADS-box gene in lateral vegetative buds of Japanese apricot. J Am Soc Hortic Sci 133:708–716. https://doi.org/10.21273/jashs.133.5.708
Yamane H, Wada M, Honda C, Matsuura T, Ikeda Y, Hirayama T, Osako Y, Gao-Takai M, Kojima M, Sakakibara H, Tao R (2019) Overexpression of Prunus DAM6 inhibits growth, represses bud break competency of dormant buds and delays bud outgrowth in apple plants. PLoS ONE 14:1–24. https://doi.org/10.1371/journal.pone.0214788
Yang Q, Yang B, Li J, Wang Y, Tao R, Yang F, Wu X, Yan X, Ahmad M, Shen J, Bai S, Teng Y (2020) ABA-responsive ABRE-BINDING FACTOR3 activates DAM3 expression to promote bud dormancy in Asian pear. Plant Cell Environ 43:1360–1375. https://doi.org/10.1111/pce.13744
Yu G, Wang LG, Han Y, He QY (2012) ClusterProfiler: an R package for comparing biological themes among gene clusters. Omi A J Integr Biol 16:284–287. https://doi.org/10.1089/omi.2011.0118
Yu Y, Qi Y, Xu J, Dai X, Chen J, Dong CH, Xiang F (2021) Arabidopsis WRKY71 regulates ethylene-mediated leaf senescence by directly activating EIN2, ORE1 and ACS2 genes. Plant J 107:1819–1836. https://doi.org/10.1111/tpj.15433
Zhang Y, Wang HL, Li Z, Guo H (2020) Genetic network between leaf senescence and plant immunity: crucial regulatory nodes and new insights. Plants 9:495. https://doi.org/10.3390/PLANTS9040495
Zhang Y, Tan S, Gao Y, Kan C, Wang HL, Yang Q, Xia X, Ishida T, Sawa S, Guo H, Li Z (2022) CLE42 delays leaf senescence by antagonizing ethylene pathway in Arabidopsis. New Phytol 235:550–562. https://doi.org/10.1111/NPH.18154
Zhu Y, Li Y, Xin D, Chen W, Shao X, Wang Y, Guo W (2015) RNA-Seq-based transcriptome analysis of dormant flower buds of Chinese cherry (Prunus pseudocerasus). Gene 555:362–376. https://doi.org/10.1016/j.gene.2014.11.032
Funding
This research was supported by a Grant-in-Aid for Scientific Research (KAKENHI) from the Japan Society for the Promotion of Science, Japan (Grant-in-Aid KAKENHI Nos 26252005, 18H02198, and 21H02186) to HY.
Author information
Authors and Affiliations
Contributions
HY: conceptualization; TFH, YYC, RN, AO, TM, and YI: analysis and investigation; HY: resources; TFH and HY: writing–original draft; TFH and YYC: visualization; HY and YI: writing–review & editing. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
No conflict of interest declared.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Hsiang, TF., Chen, YY., Nakano, R. et al. Dormancy regulator Prunus mume DAM6 promotes ethylene-mediated leaf senescence and abscission. Plant Mol Biol 114, 99 (2024). https://doi.org/10.1007/s11103-024-01497-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11103-024-01497-y