Abstract
The molecular control of bud dormancy establishment and release is still not well understood, although some genes have already been demonstrated to play important roles in this process. The dormancy-associated MADS-box (DAM) genes were first identified in the peach EVERGROWING locus and are considered the main regulators of bud dormancy control. In this work, the apple (Malus × domestica Borkh.), a perennial plant adapted to temperate climates that displays cycles of growth and bud dormancy, was screened for the presence of DAM genes. The candidate genes retrieved were characterized in comparison to DAM genes from other species. Four of them (MdDAM1–4) are structurally very similar to the reported DAM genes. When apple genomic segments containing these candidates were compared to the peach EVERGROWING locus, a highly conserved noncoding region was detected inside their largest intron. Similar sequences were also identified inside introns of apricot and pear DAM genes. Organ expression patterns revealed that MdDAM1–4 are mainly expressed in dormant buds and seeds, with low transcript accumulation in vegetative structures. In addition, the MdDAM genes showed seasonally oscillating patterns of steady-state messenger RNA (mRNA) levels and were downregulated by artificial chilling. Motif analyses in the promoter and in the intronic conserved region of the MdDAM genes disclosed some clues to the regulation of the expression patterns observed. Possible roles for the conserved intronic sequence in dormancy regulation are discussed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Temperate fruit trees developed a cycle of alternating growth and arrest periods as an adaptation to survive winter conditions. Growth cessation usually involves the formation of winter-hard, cold-tolerant specialized buds and the establishment of dormancy, a developmental program defined by the failure to resume growth from meristems under favorable conditions (Rohde and Bhalerao 2007). The timing of the onset and the length of the dormancy period are modulated by both heritable determinants and environmental cues. Dormancy entrance may be elicited by short photoperiods and/or low temperatures, and dormancy release requires a genetically controlled period of exposure to cold temperatures, which is called chilling requirement (CR; Falavigna et al. 2015).
The molecular control of development in plants and most other eukaryotes is largely attributed to the MADS-box family of transcription factors (Kaufmann et al. 2005; Dornelas et al. 2011). The MIKCC-type proteins belong to a plant-specific MADS-box subfamily and play several roles in the formation of reproductive organs specific to seed plants (Kaufmann et al. 2005). Genetic mapping of the EVERGROWING (EVG) locus in peach, whose deletion renders the plant incapable of entering dormancy, revealed six MIKCC-type MADS-box genes (Bielenberg et al. 2008). These genes were named dormancy-associated MADS-box (DAM) genes, and two of them (PpDAM5 and PpDAM6) are progressively more expressed in dormant buds during winter and peak at the winter solstice, at which point their transcript levels start to decrease until budbreak (Li et al. 2009). DAM genes belong to the StMADS11/SVP-clade, represented in Arabidopsis thaliana by SHORT VEGETATIVE PHASE (SVP) and AGAMOUS-LIKE 24 (AGL24). Both genes act antagonistically on the regulation of the flowering time, with SVP expression being modulated by cold exposure (Lee et al. 2007). Homologs of the DAM genes are also found in the rosaceous fruit crops pear (Ubi et al. 2010) and apricot (Sasaki et al. 2011) and also in the less-related weed leafy spurge (Horvath et al. 2010). In the kiwifruit, four SVP-like genes were characterized, and their possible roles in bud dormancy were described (Wu et al. 2012), although the ectopic expression of one of these members (SVP3) in kiwifruit and tobacco did not affect growth and dormancy (Wu et al. 2014). The expression of pear and apricot DAM genes is associated with dormancy progression in a similar way to PpDAM5 and PpDAM6 (Ubi et al. 2010; Sasaki et al. 2011). The overexpression of the apricot PmDAM6 in poplar caused growth retardation and precocious bud set, providing further evidence for the role of DAM genes in bud development and dormancy (Sasaki et al. 2011).
The apple (Malus × domestica Borkh.) is a deciduous tree with well-defined growth and dormancy cycles and with variable CRs across cultivars (Labuschagné et al. 2002). Farmers from mild winter climates must resort to cultivars with a low CR and/or to dormancy-breaking agents to compensate for the insufficient chilling exposure. Despite the importance of dormancy traits for apple production and the availability of many genetic and genomic resources for apple research, no functional genomics approach was reported specifically for the identification of candidate genes for apple dormancy control until very recently (Falavigna et al. 2014; Porto et al. 2015; Wisniewski et al. 2015; Mimida et al. 2015).
With the aim of identifying apple DAM genes, the apple genome was searched for putative PpDAM orthologs. The sequences of six candidate apple DAM genes were thoroughly characterized in silico by comparison to the reported rosaceous DAM genes, and their transcription profiles were determined in several organs and in buds during various dormancy stages and under artificial chilling. In addition, we monitored the transcript accumulation of the candidate apple DAM genes in three genetic backgrounds under field conditions. The results show a strongly conserved dormancy-related expression among the different genotypes for four out of the six candidates. Strikingly, these four genes, as well as all the rosaceous DAM genes analyzed, showed a highly conserved noncoding sequence inside their largest intron containing potential regulatory elements.
Results
Genomic structure and phylogenetic analysis of DAM genes in apple
To find and investigate the DAM candidate genes in apple, the deduced amino acid sequences of all six peach DAM genes (Bielenberg et al. 2008) were used as queries for BlastP analyses in the apple genome (http://www.rosaceae.org, version v1.0), and six predicted proteins repeatedly appeared (E value <10−40). Comparisons with the recent literature yielded matches for five out of these six predicted gene models identified (Table 1, see the “Discussion” section for the proposed nomenclature for the apple DAM genes). One gene, MdDAM1, had been already identified by our group as dormancy-related using suppressive subtraction hybridization (Falavigna et al. 2014). Surprisingly, recent literature revealed that two of the six predicted proteins with high amino acid sequence similarity to peach DAM genes are in fact apple homologs of JOINTLESS, a transcription factor that plays roles in the abscission zone development (Table 1, Nakano et al. 2015).
Because MdDAM2–4 are located in a narrow region in apple chromosome 8 (Table 1), in a similar trend to the localization of DAM genes in peach chromosome 1 (the EVG locus, Bielenberg et al. 2008), we evaluated if the apple genome presents a counterpart to the EVG locus. Syntenic relationships were inferred between peach chromosome 1 and all 17 apple chromosomes by employing a collinearity approach (Wang et al. 2012). The overall collinearity result shows a strong syntenic relation between peach chromosome 1 and apple chromosomes 8, 13, 15, and 16 (Fig. 1, gray lines). Additionally, a pair of collinear blocks containing DAM genes was identified between the apple chromosome 8 and the peach EVG region (Fig. 1, black line). In addition, MdDAM1, located in chromosome 16, is embedded in a small block presenting six genes collinear to the EVG locus. However, the collinearity algorithm only considers a gene as part of a collinear block if it participates in the sequence of pairs of reciprocally similar genes. MdDAM1 was not included as one of these genes, but another DAM-like gene model in the same block, MDP0000255146 (Table 2), paired with PpDAM1. The details about the collinear blocks including DAM genes are shown in the Online Resource 1.
To confirm and check the apple genome annotation, specific primers were designed for the amplification and sequencing of the entire coding regions for all apple DAM candidates, using 5′ and 3′ rapid amplification of complementary DNA (cDNA) ends (RACE; Online Resource 2). We were able to sequence the full coding region of MdDAM1, MdDAM2, MdDAM4, and MdJa (GenBank KT582786 to KT582789, Table 1). Alignments revealed some mispredictions in these four gene models disclosed in the apple genome (Online Resource 3). For MdJb, we were able to obtain ~70 % of the coding sequence (data not shown), while only a small fragment of MdDAM3 could be amplified and sequenced (Online Resource 4).
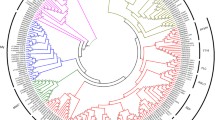
Phylogenetic analysis with DAM- and SVP-like proteins from seven different species (Online Resource 5) indicated that the majority of the proteins are grouped into three major clusters of orthologs and paralogs, with the apple members distributed into two of them (Fig. 2a). All DAM proteins from peach and Japanese apricot grouped in one distinct clade as orthologs. Following the same pattern, MdDAM1–4 formed orthologous pairs with pear DAM proteins in a second clade. Finally, the last clade was composed of MdJa, MdJb, leafy spurge DAM proteins, Arabidopsis SVP, and kiwifruit SVP-like proteins AdeSVP1 and AdeSVP4 (Fig. 2a). Putatively conserved protein domains for all candidate MdDAM genes as well as DAM protein sequences from other species were identified using the software MEME (Bailey et al. 2009). The three characteristic domains of the MIKCC subgroup of MADS box, the MADS (red and purple), I (orange) and K (blue) domains, were found (Fig. 2b). All sequences presented these domains, except EeDAM1 and the out-group Arabidopsis FLOWERING LOCUS C (FLC). Additionally, both sequences presented a short MADS-box domain in relation to the other sequences analyzed (absence of purple box in Fig. 2b). Finally, only the sequences presented in the clade composed of MdJa, MdJb, EeDAM1, EeDAM2, AtSVP, AdeSVP1, and AdeSVP4 do not show a conserved C-terminal domain (green boxes in Fig. 2b).
Phylogenetic analysis and schematic motif structure of DAM proteins and Arabidopsis SVP and AGL24 protein sequences. a Phylogenetic relationships of full-length amino acid sequences. The Arabidopsis FLC was added for tree rooting. Nodal support values are given by the a posteriori probability values shown above the corresponding nodes (when >0.50), and the bar indicates the length for which the number of expected changes per site is equal to 1. Apple DAM candidates (Mdo) are shown inside rectangles. b Schematic view of the conserved domains presented by their deduced protein sequences. Red and purple boxes, MADS domain; orange boxes, I domain; blue boxes, K domain; green boxes, C-terminal domain. Gray lines represent non-conserved sequences. Accession codes used in both analyses are depicted in OR4. Ath Arabidopsis, Ade kiwifruit, Ees leafy spurge, Pmu apricot, Ppe peach, Ppy pear
Characterization of the conserved intronic region of DAM genes
To further investigate the sequence and structural similarities of the DAM candidates, we compared apple genome contigs containing MdDAM genes to the peach EVG locus using graphic alignment (Nix and Eisen 2005). As expected, the regions containing exons displayed the highest level of conservation (Fig. 3a, Online Resource 6). Interestingly, the second exon predicted in the genome for MdDAM1 is not located in a region with high conservation. However, the alignment is in agreement with the MdDAM1 structure inferred from our cDNA sequencing data, which does not predict a protein-coding exon region in this location. On the other hand, a window of highly conserved sequences could be identified between the intronic segments of apple and peach genes (Fig. 3a). This similarity still appears when the pairwise comparisons are shown with a maximum E value <10−15, a stringency cutoff that removes most of the coding region similarity results (Fig. 3b, arrows).
Graphical alignment between the peach EVG locus (upper white bars, reference) and apple genome contigs (lower white bars) containing putative DAM genes. Solid lines indicate a significant similarity of 80-bp sequence windows for a maximum E value of 1e10−5, except for b, where the significance cutoff is 1e10−15. Dashed lines also represent the significant sequence similarity but denote the inverted sequences in relation to the reference. The predicted apple gene models (MDP0000322567 and MDP0000232313) and the sequenced coding regions (MdDAM1 and MdDAM4) are represented below apple contig bars, where boxes and lines indicate exons and introns, respectively. Peach DAM genes (PpDAM1 to PpDAM6) are represented in the same way above the reference bars. The apple genome contigs analyzed were the following: a, b MDC020688.360, c MDC027822.64, and d metacontig composed of the fusion of MDC027822.64 and MDC009761.283. Arrows point to the conserved intronic regions
The genomic prediction for MdDAM4 was only similar to the 3′ end of the peach DAM genes (Fig. 3c). A closer look at the predicted structure suggests that the gene is truncated in the contig MDC027822.64. To determine if the remaining sequences for MdDAM4 are available in another contig, we searched for MdDAM4 segments in the apple genome and found a contig (MDC009761.283) that contains the 5′ end of this gene. A manual concatenation of both contigs created a “metacontig” (see Online Resource 7 for the alignment between the ends of both contigs) that was graphically compared to the peach EVG locus and revealed the missing 5′ portion of MdDAM4, including the conserved intronic sequence (Fig. 3d). The results for MdDAM2 and MdDAM3 resemble those found for MdDAM1, while MdJa and MdJb were less similar to the peach DAM genes (Online Resource 6).
Given that a highly conserved sequence was identified between the intronic segments of apple and peach DAM genes, we examined the occurrence of this sequence in the apple and peach genomes. The conserved intronic sequence was used as a query for BlastN searches in apple contigs and peach scaffolds. Only six significant hits (E value <10−4) were found in peach, all of them inside DAM introns. For apple, nine hits were retrieved and distributed in two small genomic windows in chromosomes 8 and 16. These hits were located inside the introns of MdDAM1–4 as well as in other three uncharacterized gene models. BlastP searches at NCBI using these three genes retrieved DAM, SVP-like, and AGL24-like as best hits (Table 2). The conserved sequence was also found inside introns of the DAM genes of apricot and pear (Table 2), but the strawberry genome did not yield significant hits (data not shown). To evaluate the total extent of sequence conservation in the DAM intronic sequences, we performed multiple alignments using 500 bp up- and downstream of the conserved intronic segments from the apple and peach sequences. The alignment revealed that the segment is located in one of two highly conserved sequence intervals, with approximately 145 and 75 bp, respectively (Online Resource 8, green boxes). They are separated by a less conserved nearly 89-bp-long sequence. The genomic locations of the conserved intronic sequences in apple and peach are shown below the alignment in the Online Resource 8.
MdDAM gene expression patterns
The transcript accumulation of apple MdDAM genes was quantified in several apple organs. The results show that MdDAM1, MdDAM2, MdDAM3, and MdDAM4 were mainly expressed in dormant buds and seeds (Online Resource 9, samples A and S, respectively), with low transcript accumulation in vegetative organs. Interestingly, MdDAM2 expression was 10 times higher in seeds than in dormant buds. Conversely, MdJa and MdJb were expressed in a similar fashion in dormant buds, growing buds, leaves, and seeds, with lower levels in fruits (Online Resource 9).
To evaluate the seasonal oscillation of MdDAM gene expression, messenger RNA (mRNA) levels were quantified in bud samples of three cultivars harvested over a growth-dormancy cycle. These genotypes were selected considering their contrasting CRs, where ‘Fuji Standard’ and ‘Royal Gala’ need higher amounts of cold exposure compared to ‘Castel Gala’ (see the “Methods” section and Denardi and Seccon 2005; Botelho et al. 2006; Anzanello et al. 2014a). MdDAM1, MdDAM3, and MdDAM4 showed peaks of transcript accumulation during winter (June and July) and low transcript levels during spring and summer. Interestingly, ‘Castel Gala’ samples showed an anticipated decrease in their transcript levels in comparison to both high CR cultivars. MdDAM2 transcription peaked in the summer (February), with less expression in the course of the autumn and winter. Finally, MdJa and MdJb presented less variation in their transcript amounts during the year (Fig. 4).
Transcript accumulation of MdDAM genes in closed terminal buds of ‘Royal Gala,’ ‘Castel Gala,’ and ‘Fuji Standard’ over one growth-dormancy cycle. Months are represented by their initial letters on the x-axis. Transcript levels are relative to May 2009 samples. Bars are standard errors of the mean (n = 3)
To investigate the effect of the controlled chilling exposure on the transcript accumulation of MdDAM genes and to compare this effect between contrasting CR cultivars, cuttings (brindles with terminal buds) from ‘Royal Gala’ and ‘Castel Gala’ trees were kept in a dark chamber at 6 °C for 1 or 5 weeks. At the time of harvesting, the ‘Royal Gala’ terminal buds were in deep dormancy, while the ‘Castel Gala’ buds were already in ecodormancy, as observed by budbreak rates under forcing conditions (Fig. 5). Extended cold exposure increased the budbreak rate of ‘Royal Gala’ terminal buds but had little effect on the budbreak rate of ‘Castel Gala’ samples. Extended cold exposure decreased the gene expression of all MdDAM genes and MdJa in both cultivars, but not MdJb. The transcript level downregulation was more abrupt for MdDAM2 and MdDAM4, which were barely detectable by real-time PCR after 5 weeks of cold treatment in both cultivars. Additionally, ‘Royal Gala’ and ‘Castel Gala’ buds showed very similar transcript accumulation levels for all genes. Finally, MdJb showed a small variation in the transcript levels during the cold treatment (Fig. 5).
Transcript accumulation of apple DAM candidates in closed terminal buds of two contrasting CR cultivars exposed to 0 week, 1 week (168 h), and 5 weeks (840 h) of chilling (6 °C). Transcript levels are relative to ‘Royal Gala’ initial samples. Budbreak rates of samples analyzed are shown at the bottom. Bars are standard errors of the mean (n = 3)
In silico analyses of cis-elements in the promoter and in the intronic region of MdDAM genes
The promoter analyses of all six candidate MdDAM genes (~1.5 kb upstream of start codon) revealed the presence of both unique and consensus cis-elements (Online Resource 10). Interestingly, MdDAM1–4 showed two CArG-box (binding sites for MADS-box factors) motifs separated by less than 100 bp. In addition, we analyzed the whole largest intron of MdDAM1–4 and, again, unique as well as consensus motifs were found (Online Resource 11). Consensus cis-elements include sites found in the promoters of the homeotic gene AGAMOUS (AG and AGAMOUS), response to dehydration and low temperature (ABRE-like), and CArG-box motifs (CARGCW8GAT). Once again, two CArG-box motifs were found within a window of less than 400 bp in MdDAM1, MdDAM3, and MdDAM4, although outside the conserved intronic region.
Finally, we further characterized the promoter and the conserved intronic region of MdDAM1, which presents high expression in endodormant buds (Falavigna et al. 2014, this work). Four unique cis-elements were found in its promoter region, including a CBF1 motif (specific binding of transcription factor CBF1-C-repeat binding factor/DREB1) and a LTREATLTI78 motif (a putative low-temperature responsive element; Fig. 6). A search for these motifs in the promoter regions of all predicted apple genes available (36,991 predicted genes; see the “Methods” section) revealed that only 154 CBF1 (0.44 % occurrence) and 1050 LTREATLTI78 (2.38 % occurrence) motifs were found (Online Resources 12 and 13). In the same way, the highly conserved region of the largest intron from MdDAM1 was screened (90 base pairs; Online Resource 8), revealing five consensus motifs (Online Resource 11). We searched for cis-elements in the conserved intronic sequences of peach and apricot DAM genes and identified only four out of these five motifs (data not shown). Interestingly, Arabidopsis response regulator 10 (ARR10), a binding site of the GARP family of plant Myb-related transcription factors, is only present in some apple and pear DAM genes (Fig. 6). Moreover, only 641 apple genes (1.73 %) presented ARR10 in their promoter region (Online Resource 14). A gene ontology (GO) search was performed to identify biological processes related for the predicted genes that contain each of these three motifs (Online Resources 12 to 14). As a result, several similar enriched terms were recognized, with the “binding” term being the most general one (Fig. 6).
Gene structure and cis-element analysis of MdDAM1. a The representative image is composed by the promoter region (up to 1.5 kb upstream from the start codon; green box), exons (red boxes), and introns (black lines). The gray box comprehends the conserved intronic region (see OR7). The three cis-elements characterized are shown by a vertical black line, along with the distance from the start codon. b GO analysis of the genes containing CBF1, LTREATLTI78, and ARR10 cis-elements in their promoter regions. The scatterplot shows the cluster representatives (i.e., the terms that remained following redundancy reduction) in a two-dimensional space derived by applying multidimensional scaling to a matrix of the GO term semantic similarities. The bubble color indicates the AgriGO-provided p value (see the legend in the upper right-hand corner); the size indicates the frequency of the GO term in the underlying GO analysis. The bubbles are larger for the more general terms
Discussion
The MADS-box family of transcription factors is known to comprise important regulators of growth and development in plants (Kaufmann et al. 2005; Dornelas et al. 2011). The sequencing of the apple genome revealed a relatively high number of MADS-box genes, possibly due to a recent genome duplication, and many of these are speculated to have roles in the development of the pome fruit, including genes coding for the StMADS11/SVP-clade MADS-box transcription factors (Velasco et al. 2010). Recently, four reports identified genes belonging to this subfamily in apple (Falavigna et al. 2014; Wisniewski et al. 2015; Mimida et al. 2015; Nakano et al. 2015). The first gene, MdDAM1, was identified by our group in a suppression subtractive hybridization assay between endodormant and ecodormant buds (Falavigna et al. 2014). In later studies, two additional apple DAM genes were identified, and their expression patterns were associated with dormancy onset and release (Wisniewski et al. 2015; Mimida et al. 2015). Finally, Nakano et al. (2015) performed a functional analysis of two other members of this subfamily and showed that the ectopic expression of MdJb and, to a lesser extent, MdJa can restore the JOINTLESS-deficient tomato mutant. This finding shows that MdJa and MdJb may be involved in the development of functional pedicel abscission zones rather than with dormancy. In this work, we provide evidence that at least four out of these six apple StMADS11/SVP-clade MADS-box genes (MdDAM1, MdDAM2, MdDAM3, and MdDAM4) participate in dormancy regulation, whereas the data collected for MdJa and MdJb corroborates the hypothesis that these genes may not be acting on dormancy control. Therefore, based on our findings and reports from the literature, we propose a unified nomenclature for DAM genes in apple (Table 1).
To gain knowledge of the possible roles of the six gene models identified, we quantified their transcript accumulation in different apple organs. Interestingly, MdDAM1, MdDAM2, MdDAM3, and MdDAM4 expression was enriched in dormant buds and seeds (Online Resource 9). In peach, the expression of some bud dormancy-related genes is also detected in seeds, including PpDAM1, PpDAM5, and PpDAM6. Additionally, the mRNA levels of these PpDAM genes declined in the seeds after cold stratification (Leida et al. 2012). Our results are in agreement with the ones with peach, suggesting common mechanisms of bud and seed dormancy regulation.
Dormant buds of three distinct cultivars were sampled during a full year and were used to analyze the expression of apple DAM genes (Fig. 4). MdDAM1, MdDAM3, and MdDAM4 showed a seasonal oscillating pattern of transcript accumulation, with a remarkable repression near budbreak (Fig. 4). The same peak of expression for MdDAM1 was also observed in a standard dwarfing apple rootstock (Wisniewski et al. 2015) and in fruit-bearing terminal buds of apple cultivars ‘Jonathan’ and ‘Fuji’ (labeled as MdDAMa in Mimida et al. 2015). MdDAM2 (labeled MdDAMc in Mimida et al. 2015) was more expressed during summer and autumn, as was observed in our data (Fig. 4). Similarly, peach and pear DAM genes exhibit clear seasonal patterns of transcript accumulation, with most genes showing increased mRNA levels during autumn and the beginning of the winter, followed by a decrease as the buds are exposed to low temperatures (Li et al. 2009; Saito et al. 2013; Ubi et al. 2010). Wisniewski et al. (2015) observed that MdJb (labeled by the authors as MdDAM3) presents a low fluctuation over the year.
Our results showed a clear repression of transcript levels during the late winter in field-harvested samples for MdDAM1, MdDAM3, and MdDAM4 (Fig. 4). To investigate if these responses are an effect of chilling exposure, an experiment using controlled temperature treatment was carried out. When chilled under controlled conditions, buds from both contrasting CR cultivars showed a decrease of transcript accumulation for all tested genes, except MdJb (Fig. 5). Interestingly, MdJb also did not present a clear seasonal variation of mRNA levels (Fig. 4). The steady-state mRNA levels of apple DAM genes, as well as the rate of the transcript level repression by cold exposure, did not differ significantly between the cultivars tested, despite the clear differences of budbreak rates at the onset of the experiment (Fig. 5). This result was surprising, given that in field-harvested samples, MdDAM1 and MdDAM3 expression in ‘Castel Gala’ buds declined more rapidly during winter than in ‘Royal Gala’ (Fig. 4). However, this result has to be considered with caution, because, in this experiment, the two apple cultivars were not at the same developmental stage and therefore cannot be strictly compared. In apricot, the expression of three (PmDAM1, PmDAM2, and PmDAM3) out of the six DAM genes under controlled cold exposure was not different between the two contrasting CR cultivars tested, despite the higher budbreak rates of the low CR buds (Sasaki et al. 2011). However, PmDAM1 and PmDAM3 expressions were different between the cultivars in field-harvested buds during winter (Sasaki et al. 2011), as was observed with MdDAM1 and MdDAM3 (Fig. 4; Falavigna et al. 2014). Similarly, peach PpDAM3 was differentially expressed in the field but not under controlled conditions (Jiménez et al. 2010). Genotype-related differences of mRNA level repression rates during winter in some DAM genes may be dependent on other environmental cues in addition to cold exposure.
With the aim of better describing the MdDAM gene structures, we fully characterized them by an in silico analysis approach. We observed that, in addition to the high sequence similarity (Fig. 2), the genomic localization of several MdDAM genes found in this study provides good evidence for their putative role in dormancy. Three of them (MdDAM2, MdDAM3, and MdDAM4) are physically located very close to each other in chromosome 8 (in a 200-kb interval). Two other DAM-like predicted genes were also found inside this interval when the conserved intronic sequence (Online Resource 8) was used as a query in Blast searches against the apple genome (Table 2). These genes were not included in the analyses because they lack one or more typical MIKCC domains (data not shown). Apple chromosome 8 is highly syntenic to the end of peach chromosome 1, where the DAM genes were first mapped (Fig. 1; Bielenberg et al. 2008; Jung et al. 2012). This configuration of the apple DAM genes nearly in tandem resembles the peach EVG locus and may be a vestige remaining from an ancestor chromosome of this particular genomic region. MdDAM1 seems to be an exception to this trend, given that it presents the conserved intron and expression profiles similar to those of MdDAM2, MdDAM3, and MdDAM4, although it is located in apple chromosome 16. MdDAM1 is embedded in a block collinear to the EVG locus but was not included in the sequence of pairs of apple and peach genes that define the collinearity. Instead, a closely located DAM-like gene model (MDP0000255146) participates as a counterpart to PpDAM1. This predicted gene could be a functional apple DAM, although its sequence and structure are not as similar to the peach DAM genes as the ones characterized in this work. The presence of both MdDAM1 and DAM-like genes in a block collinear to the EVG locus indicates a syntenic relationship between them and the peach DAM genes.
Our promoter analysis identified many unique and consensus motifs in the promoter region of all six candidate MdDAM genes (Online Resource 10). Interestingly, MdDAM1–4 promoters displayed at least two CArG-box variants separated by less than 100 bp (Online Resource 10). The distance between the two cis-elements is essential, given that some MADS-box factors exert their transcriptional regulation by forming dimer complexes that bind to two CArG boxes separated by up to 400 bp (Mendes et al. 2013). This finding suggests that apple DAM genes can be directly regulated by themselves or by other MADS-box factors, although further investigation needs to be conducted to unveil their functional roles. We decided to deeply investigate the MdDAM1 promoter, which presents high expression in endodormant buds (Falavigna et al. 2014). We found four unique cis-elements when compared to the other MdDAM members (Online Resource 10), including a low-temperature responsive element (LTRE) and the cis-element for the CBF1 transcription factor. In fact, a peach CBF1 was shown to regulate MdDAM genes in apple (Wisniewski et al. 2015). Additionally, a transient reporter assay in pear showed that the pear CBF2 can interact with the promoter of the pear DAM ortholog PpMADS13-1 (Saito et al. 2015), as has already been suggested for DAM genes in other species (Horvath et al. 2010; Sasaki et al. 2011; Yamane et al. 2011). Taken together, these findings strongly suggest that some DAM genes integrate the cold regulon.
When the genomic regions containing DAM (or DAM-like) genes of apple and peach were compared, a strikingly conserved noncoding region was found inside their intronic regions (Fig. 3, Online Resource 6). As species diverge, the sequences of their orthologous genes accumulate differences. However, regions whose changes will likely compromise protein function, such as exons, remain more conserved (D’haeseleer 2006). Thus, the fact that this intronic region is more conserved than most DAM exonic sequences suggests that an important functional role is involved in its evolutionary maintenance. Interestingly, this sequence is not found in strawberry, a rosaceous species with well-delimited syntenic correlations with apple and peach that does not undergo a typical dormancy cycle (Illa et al. 2011; Jung et al. 2012).
In peach, changes in the CRs of cultivars have been observed in parallel to genomic insertions in the largest intron of DAM genes (Yamane et al. 2011; Zhebentyayeva et al. 2014). Our analysis showed at least two CArG-box variants in the largest introns of MdDAM1, MdDAM3, and MdDAM4, separated within a distance of 350 bp (Online Resource 11). MADS-box proteins could interact with these motifs to regulate their expression. One hypothesis for the changes in the CRs of the cultivars could be the alteration of DAM expression caused by modification of the distance between CArG boxes by genomic insertions (Mendes et al. 2013). However, several pear genotypes with distinct CRs were screened for the presence of insertions in their largest intron, and no relation to dormancy could be drawn (Saito et al. 2013). Furthermore, the insertions in the DAM5 intron described as correlated to the peach CR in the work of Zhebentyayeva et al. (2014) were not located inside the conserved sequence found in this work (data not shown). Thus, the precise effect of these insertions on DAM gene expression regulation remains unknown.
Finally, we analyzed the conserved intronic region presented by rosaceous DAM genes and identified several cis-elements related to light regulation (Online Resource 11). It is intriguing that some apple and pear DAM genes present a unique cis-element called ARR10 (Fig. 6), which does not appear in the conserved intron of Prunus species (Online Resource 8). This motif is the binding site of at least four type-B ARRs (ARR1, AAR2, AAR10, and ARR12), which have significant roles in modulating cytokinin signaling and the control of meristem cell division (Hosoda et al. 2002; Hill et al. 2013). Interestingly, although the ARR10 motif rarely appears (1.73 %) in the apple gene promoters analyzed, it is commonly presented in the promoter and in the whole intronic sequences of MdDAM genes (Online Resources 10 and 11). This finding suggests that cytokinin levels can indirectly repress apple DAM genes, given that this hormone is related to cell division and growth.
The role of intronic regions in the control of transcript accumulation is well documented in the Arabidopsis AGAMOUS (AtAG) gene. Reporter gene expression under the sole control of the AtAG promoter did not match the expected transcriptional pattern. However, this pattern is recovered when the reporter is controlled by the promoter and the largest intron (Sieburth and Meyerowitz 1997). Indeed, the presence of cis-elements acting as docking sites for other transcription factors in this intron is essential for the spatial and temporal control of AtAG expression (Hong et al. 2003). Functional analyses are needed to evaluate possible roles of conserved elements in introns of DAM genes over their expression, particularly the sequences found in this work for rosaceous homologs.
We identified and characterized four DAM genes in apple through extensive in silico analyses of their genomic structure. In addition, MdDAM1, MdDAM2, MdDAM3, and MdDAM4 presented expression profiles with close resemblance to the ones reported for DAM genes of peach (Li et al. 2009), pear (Ubi et al. 2010; Saito et al. 2013), and apricot (Sasaki et al. 2011). These four genes also shared a highly conserved sequence in their largest intron with all the reported rosaceous DAM genes. This conserved sequence is a promising new feature of DAM genes. Combined, our results provide additional support to help unveil the conserved role of DAM genes in apple regarding endodormancy establishment and maintenance.
Methods
Plant material
Field-grown ‘Royal Gala’ and ‘Castel Gala’ plants were located in a commercial orchard (Papanduva, SC, Brazil; −26.434870, −50.106103, and 788 m altitude). Six-year-old ‘Royal Gala’ trees were cultivated on Marubakaido rootstock with M9 as an interstock. ‘Castel Gala’ samples were from 3-year-old trees grafted on M9 rootstock. ‘Fuji Standard’ trees were from an experimental field at Empresa de Pesquisa Agropecuária e Extensão Rural de Santa Catarina (Caçador, SC, Brazil; −26.836971, −50.975246, and 935 m altitude). The plants studied were 7-year-old trees grafted on M7.
Both ‘Royal Gala,’ ‘Gala Baigent,’ and ‘Castel Gala’ are phenologically stable cultivars derived from spontaneous bud sport mutations from trees of the original cv. Gala (‘Gala Standard’). ‘Royal Gala’ and ‘Gala Baigent’ trees have the same CR of ‘Gala Standard’ (around 600 CH), while their fruit skins show more concentration of red pigments. ‘Castel Gala’ fruits are identical in appearance to the ‘Gala Standard’ fruits, but the CR is approximately half the CH needed to break the ‘Gala Standard’ bud dormancy (Anzanello et al. 2014a). ‘Fuji Standard’ buds require over 1000 CH to break bud dormancy in Southern Brazil (Petri and Stuker 1988).
All bud samples were terminal buds in phenological stage A (closed) according to the Fleckinger scale (EPPO 1984). Sampling was always performed at 9:00 am for ‘Royal Gala’ and ‘Castel Gala’ and at 11:00 a.m. for ‘Fuji Standard.’ Field-harvested buds were immediately frozen in liquid N2 and stored at −80 °C until analysis. Cold treatments were performed as previously described (Anzanello et al. 2014a, b). Briefly, 20–30 cm stem cuttings with terminal buds were cut and kept moistened until processing. Cuttings were surface-sterilized, air-dried, wrapped in plastic bags, and maintained at 6 °C in a BOD-type chamber in a vertical position in the dark. After 1 and 5 weeks, a batch of samples was either forced to budbreak (40 cuttings) or harvested for bud sampling (20 cuttings) as above. Forcing was done for 56 days in a growth chamber at 25 °C, with a 12-h photoperiod and 70 % relative humidity. The percent maximum budbreak was calculated by dividing the number of apical buds showing green tissue by the total number of viable apical buds.
The plant material for tissue/organ analysis was harvested from ‘Gala Baigent’ trees located at an experimental station of Empresa Brasileira de Pesquisa Agropecuária in Vacaria, RS, Brazil (−28.513777, −50.881465, and 972 m altitude). The trees were 3 years old and grafted on Marubakaido rootstocks with M9 as an interstock. Sample stages were selected following the Fleckinger scale (EPPO 1984): closed terminal buds (A stage), buds with green tips (C stage), pre-anthesis flowers and young leaves (E2 stage), whole-set fruits ~10 mm in diameter and leaves (I stage), and unripe fruits ~40 mm in diameter that were divided into pulp and skin (J stage). We also sampled mature fruits ~70 mm in diameter, partitioning them into pulp, seeds, and skin (named M).
RNA extraction, cDNA synthesis, sequencing, and real-time PCR
The total RNA was extracted from samples composed of 10–20 buds by LiCl precipitation, as described by Zeng and Yang (2002), following the modifications of Reid et al. (2006). The integrity of the RNA samples was checked on 1 % agarose/ethidium bromide gels. RNAse inhibitor (RNAseOUT, Invitrogen, Waltham, MA, USA) was added to samples (1–2 μl) before storing at −80 °C. Contaminating DNAs were removed with the Turbo DNA-free Kit (Applied Biosystems). Synthesis of cDNA was done from 1 μg DNA-free total RNA using the GeneAmp RNA PCR Core Kit (Applied Biosystems) with oligo dT16 (included in the kit), according to the manufacturer’s instructions. Assessment of genomic DNA contamination was done by PCR amplification of cDNAs with a primer pair for a WD40 domain containing gene (Perini et al. 2014) spanning an intron.
Real-time PCR analysis was performed as in Falavigna et al. (2014). The only difference is that the annealing temperature for MdDAM3 was set to 55 °C for all experiments except the tissue/organ assays, performed as described and with a different set of primers. Amplification efficiencies were determined using LinRegPCR v. 2012.0 (Ruijter et al. 2009). Relative expression levels were calculated from amplification data using Pfaffl mathematics (Pfaffl 2001). The reference genes used were chosen among the most stable for apple buds (Perini et al. 2014). The reference gene adopted for samples exposed to cold under controlled conditions was ACCUMULATION AND REPLICATION OF CHLOROPLAST (ARC5), and all other analyses employed MALATE DEHYDROGENASE (MDH) as the reference gene.
For the sequencing of apple DAM candidates, the oligonucleotide sequences designed for real-time PCR analyses were used as gene-specific primers for RACE using the SMARTer RACE cDNA Amplification Kit (Clontech, Mountain View, USA), according to the manufacturer’s instructions. RACE products were sequenced by Sanger’s method using an ABI-PRISM 3100 Genetic Analyzer and associated chemistry (Applied Biosystems).
Phylogenetic analysis and conserved protein motif analyses
Complete amino acid sequences of DAM proteins from apple, peach, apricot, pear, leafy spurge, kiwi, and StMADS11/SVP-clade MIKCC proteins from A. thaliana, with A. thaliana FLC as the out-group, were aligned using Muscle (Edgar 2004). The accession numbers of all sequences used are presented in Online Resource 5. The phylogenetic tree was inferred using MrBayes version 3.2.1, employing the mixed amino acid substitution model in default settings (Ronquist et al. 2012). We performed a 1,000,000 generation run, sampled every 100 generations and the first 25 % of the trees were discarded as burn-in. The remaining ones were summarized in a consensus tree that was visualized and edited using FigTree v.1.4 (http://tree.bio.ed.ac.uk/software/figtree/).
Protein sequences from the translations of resequenced coding regions for MdDAM1, MdDAM2, MdDAM4, and MdJa, as well as translations from apple genome versions of MdDAM3 and MdJb (genome codes MDP0000527190 and MDP0000209705, respectively), were searched for conserved protein motif discovery using the MEME software version 4.10.1 (Bailey et al. 2009). Along with the apple DAM-like translations, DAM protein sequences from the peach, apricot, pear, kiwi, and leafy spurge, as well as the StMADS11/SVP-clade proteins SVP and AGL24 from Arabidopsis, were used as inputs. The software parameters were set to the defaults, except for the following: motif distribution parameter set to any number of repetitions, maximum number of motifs equal to 5, presented in at least 19 sequences, and motif width between 12 and 100 amino acids.
Sequence alignment
Contigs (codes MDC020948.189, MDC020688.360, MDC008471.150, MDC017035.448, MDC016474.222, and MDC027822.64, available at http://www.rosaceae.org) bearing candidate MdDAM genes were aligned to the EVG locus using the GATAligner software (Nix and Eisen 2005). The parameters used were the defaults, except for the adoption of an 80-bp sequence window. Approximately 1000 bp of each apple and peach intron sequence that contains the conserved regions identified were aligned using ClustalW (Larkin et al. 2007), and the resulting alignment was plotted using JalView (Waterhouse et al. 2009). For the collinearity analyses, all the proteins mapped to peach chromosome 1 were used as queries for BlastP against the predicted apple proteins. Only the top five matches from the results of each query were used for a maximum E value <1e −10. These results and the gene positions were used as inputs for the MCScanX algorithm (Wang et al. 2012). The “family circle plotter” utility from the same software was used for the plot construction. The resulting image was converted to grayscale and improved for chromosome drawing and labeling in a vector graphics editor.
Cis-element discovery and gene ontology analyses
All cis-element searches were performed using the Web tool PlantPAN version 1.0 (Plant Promoter Analysis Navigator; http://plantpan.mbc.nctu.edu.tw). For the MdDAM promoter region analysis, ~1.5 kb upstream of the start codon was used. Additionally, the sequence of the largest MdDAM intron or only 90 bp of the highly conserved intronic region of MdDAM1 was used in the cis-element motif analysis. Only the given strand was considered, and only cis-elements larger than six nucleotides were used in further analyses. CBF1, LRTEATLTI78, and ARR10 motifs were used in a genome-wide search in the promoter regions of all apple genes predicted in the 1.0 version of the genome. Promoter sequences (1 kb upstream of predicted start codons) were extracted from genome data using BEDTools (Quinlan and Hall 2010) and in-house Perl scripts. The sequences extracted (36,991) were screened for conserved cis-elements using FIMO (Grant et al. 2011), available at http://meme.nbcr.net/meme/tools/fimo, with a match p value of less than e −4 and the given strand only. Genes with these motifs were scanned for enrichment of GO terms using agriGO (Du et al. 2010), available at http://bioinfo.cau.edu.cn/agriGO/, and REVIGO (Supek et al. 2011), available at http://revigo.irb.hr/. Only terms with a p value lower than log 10−1 were analyzed. Consensus and unique motifs were analyzed using online software available at http://bioinformatics.psb.ugent.be/webtools/Venn/.
References
Anzanello R, Fialho FB, Santos HP, Bergamaschi H, Marodin GAB (2014a) Bud dormancy in apple trees after thermal fluctuations. Pesq Agrop Brasileira 49:457–464
Anzanello R, Fialho FB, dos Santos HP, Bergamaschi H, Marodin GAB (2014b) Biological methods for assessment of budbreak in apple trees for modeling dormancy. Semina: Ciênc Agrár 35:1163–1176
Bailey TL, Bodén M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS (2009) MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 37:W202–W208
Bielenberg DG, Wang Y (E), Li Z, Zhebentyayeva T, Fan S, Reighard GL, Scorza R, Abbott AG (2008) Sequencing and annotation of the evergrowing locus in peach [Prunus persica (L.) Batsch] reveals a cluster of six MADS-box transcription factors as candidate genes for regulation of terminal bud formation. Tree Genet Genomes 4:495–507
Botelho RV, Ayub RA, Muller MML (2006) Sommatory of chilling hours and chilling units at different regions of Paraná state. Sci Agric 7:89–96
D’haeseleer P (2006) How does DNA sequence motif discovery work? Nat Biotechnol 24:959–961
Denardi F, Seccon JJ (2005) ‘Castel Gala’—mutação da macieira ‘Gala’ com baixa necessidade de frio e maturação precoce. Agropec Catar 18:78–82
Dornelas MC, Patreze CM, Angenent GC, Immink RGH (2011) MADS: the missing link between identity and growth? Trends Plant Sci 16:89–97
Du Z, Zhou X, Ling Y, Zhang Z, Su Z (2010) AgriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res 38:64–70
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797
EPPO (1984) EPPO crop growth stage keys—apple and pear. EPPO Bull 14:291–294
Falavigna VS, Porto DD, Buffon V, Margis-Pinheiro M, Pasquali G, Revers LF (2014) Differential transcriptional profiles of dormancy-related genes in apple buds. Plant Mol Biol Rep 32:796–813
Falavigna VS, Porto DD, Silveira CP, Revers LF (2015) Recent advances in genetics and molecular control of bud dormancy in pipfruits. In: Anderson JV (ed) Advances in plant dormancy. Springer, Switzerland, pp 107–122
Grant CE, Bailey TL, Noble WS (2011) FIMO: scanning for occurrences of a given motif. Bioinformatics 27:1017–1018
Hill K, Mathews DE, Kim HJ, Street IH, Wildes SL, Chiang YH, Mason MG, Alonso JM, Ecker JR, Kieber JJ, Schaller GE (2013) Functional characterization of type-B response regulators in the Arabidopsis cytokinin response. Plant Physiol 162:212–224
Hong RL, Hamaguchia L, Busch MA, Weigel D (2003) Regulatory elements of the floral homeotic gene AGAMOUS identified by phylogenetic footprinting and shadowing. Plant Cell 15:1296–1309
Horvath DP, Sung S, Kim D, Chao W, Anderson J (2010) Characterization, expression and function of DORMANCY ASSOCIATED MADS-BOX genes from leafy spurge. Plant Mol Biol 73:169–179
Hosoda K, Imamura A, Katoh E, Hatta T, Tachiki M, Yamada H, Mizuno T, Yamazaki T (2002) Molecular structure of the GARP family of plant Myb-related DNA binding motifs of the Arabidopsis response regulators. Plant Cell 14:2015–2029
Illa E, Sargent DJ, Girona EL, Bushakra J, Cestaro A, Crowhurst R, Pindo M, Cabrera A, van der Knaap E, Iezzoni A, Gardiner S, Velasco R, Arús P, Chagné D, Troggio M (2011) Comparative analysis of rosaceous genomes and the reconstruction of a putative ancestral genome for the family. BMC Evol Biol 11:9
Jiménez S, Reighard GL, Bielenberg DG (2010) Gene expression of DAM5 and DAM6 is suppressed by chilling temperatures and inversely correlated with bud break rate. Plant Mol Biol 73:157–167
Jung S, Cestaro A, Troggio M, Main D, Zheng P, Cho I, Folta KM, Sosinski B, Abbott A, Celton J-M, Arús P, Shulaev V, Verde I, Morgante M, Rokhsar D, Velasco R, Sargent DJ (2012) Whole genome comparisons of Fragaria, Prunus and Malus reveal different modes of evolution between Rosaceous subfamilies. BMC Genomics 13:129
Kaufmann K, Melzer R, Theissen G (2005) MIKC-type MADS-domain proteins: structural modularity, protein interactions and network evolution in land plants. Gene 347:183–198
Labuschagné I, Louw JH, Schmidt K, Sadie A (2002) Genetic variation in chilling requirement in apple progeny. J Am Soc Hortic Sci 127:663–672
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948
Lee JH, Yoo SJ, Park SH, Hwang I, Lee JS, Ahn JH (2007) Role of SVP in the control of flowering time by ambient temperature in Arabidopsis. Genes Dev 21:397–402
Leida C, Conejero A, Arbona V, Gómez-Cadenas A, Llácer G, Badenes ML, Ríos G (2012) Chilling-dependent release of seed and bud dormancy in peach associates to common changes in gene expression. PLoS One 7:e35777
Li Z, Reighard GL, Abbott AG, Bielenberg DG (2009) Dormancy-associated MADS genes from the EVG locus of peach [Prunus persica (L.) Batsch] have distinct seasonal and photoperiodic expression patterns. J Exp Bot 60:3521–3530
Mendes MA, Guerra RF, Berns MC, Manzo C, Masiero S, Finzi L, Kater MM, Colombo L (2013) MADS domain transcription factors mediate short-range DNA looping that is essential for target gene expression in Arabidopsis. Plant Cell 25:2560–2572
Mimida N, Saito T, Moriguchi T, Suzuki A, Komori S, Wada M (2015) Expression of DORMANCY-ASSOCIATED MADS-BOX (DAM)-like genes in apple. Biol Plantarum 59:237–244
Nakano T, Kato H, Shima Y, Ito Y (2015) Apple SVP family MADS-box proteins and the tomato pedicel abscission zone regulator JOINTLESS have similar molecular activities. Plant Cell Physiol. doi:10.1093/pcp/pcv034
Nix DA, Eisen MB (2005) GATA: a graphic alignment tool for comparative sequence analysis. BMC Bioinform 6:9
Perini P, Pasquali G, Margis-Pinheiro M, Oliveira PRD, Revers LF (2014) Reference genes for transcriptional analysis of flowering and fruit ripening stages in apple (Malus × domestica Borkh.). Mol Breed 34:829–842
Petri JL, Stuker H (1988) Effect of temperature and length of cold storage of nursery plants on the subsequent development of young apple trees. Acta Hortic 232:117–126
Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29:e45
Porto DD, Bruneau M, Perini P, Anzanello R, Renou J, dos Santos HP, Fialho HP, Revers LF (2015) Transcription profiling of the chilling requirement for budbreak in apples: a putative role for FLC-like genes. J Exp Bot 66:2659–2672
Quinlan AR, Hall IM (2010) BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26:841–842
Reid K, Olsson N, Schlosser J, Peng F, Lund S (2006) An optimized grapevine RNA isolation procedure and statistical determination of reference genes for real-time RT-PCR during berry development. BMC Plant Biol 6:27–37
Rohde A, Bhalerao RP (2007) Plant dormancy in the perennial context. Trends Plant Sci 12:217–223
Ronquist F, Teslenko M, Mark PVD, Ayres DL, Darling A, Hohna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol 61:539–542
Ruijter JM, Ramakers C, Hoogaars WM, Karlen Y, Bakker O, van den Hoff MJ, Moorman AF (2009) Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res 37:e45
Saito T, Bai S, Ito A, Ubi BE, Imai T, Moriguchi T (2013) Expression and genomic structure of the dormancy-associated MADS box genes MADS13 in Japanese pears (Pyrus pyrifolia Nakai) that differ in their chilling requirement for endodormancy release. Tree Physiol 33:654–667
Saito T, Bai S, Imai T, Ito A, Nakajima I, Moriguchi T (2015) Histone modification and signalling cascade of the dormancy-associated MADS-box gene, PpMADS13-1, in Japanese pear (Pyrus pyrifolia) during endodormancy. Plant Cell Environ 38:1157–1166
Sasaki R, Yamane H, Ooka T, Jotatsu H, Kitamura Y, Akagi T, Tao R (2011) Functional and expressional analyses of PmDAM genes associated with endodormancy in Japanese apricot. Plant Physiol 157:485–497
Sieburth L, Meyerowitz E (1997) Molecular dissection of the AGAMOUS control region shows that cis elements for spatial regulation are located intragenically. Plant Cell 9:355–365
Supek F, Bosnjak M, Skunca N, Smuc T (2011) REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One 6:e21800
Ubi B, Sakamoto D, Ban Y, Shimada Y, Ito A, Nakajima I, Takemura Y, Tamura F, Saito T, Moriguchi T (2010) Molecular cloning of dormancy-associated MADS-box gene homologs and their characterization during seasonal endodormancy transitional phases of Japanese pear. J Am Soc Hortic Sci 135:174–182
Velasco R, Zharkikh A, Affourtit J, Dhingra A, Cestaro A, Kalyanaraman A, Fontana P, Bhatnagar SK, Troggio M, Pruss D, Salvil S, Pindo M, Baldi P, Castellettil S, Cavaiuolo M, Coppola G, Costa F, Cova V, Dal Ri A, Goremykin V, Komjanc M, Longhi S, Magnago P, Malacarne G, Malnoyl M, Micheletti D, Moretto M, Perazzolli M, Si-Ammour A, Vezzulli S, Zini E, Eldredge G, Fitzgerald LM, Gutin N, Lanchbury J, Macalma T, Mitchell JT, Reid J, Wardell B, Kodira C, Chen Z, Desany B, Niazi F, Palmer M, Koepke T, Jiwan D, Schaeffer S, Krishnan V, Wu C, Chu VT, King ST, Vick J, Tao Q, Mraz A, Stormo A, Stormo K, Bogden R, Ederle D, Stella A, Vecchietti A, Kater MM, Masiero S, Lasserre P, Lespinasse Y, Allan AC, Bus V, Chagné D, Crowhurst RN, Gleave AP, Lavezzo E, Fawcett JA, Proost S, Rouzé P, Sterck L, Toppo S, Lazzari B, Hellens RP, Durel CE, Gutin A, Bumgarner RE, Gardiner SE, Skolnick M, Egholm M, Peer YV, Salamini F, Viola R (2010) The genome of the domesticated apple (Malus × domestica Borkh.). Nat Genet 42:833–839
Wang Y, Tang H, Debarry JD, Tan X, Li J, Wang X, Lee TH, Jin H, Marler B, Guo H, Kissinger JC, Paterson AH (2012) MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res 40:e49
Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ (2009) Jalview version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25:1189–1191
Wisniewski M, Artlip T, Norelli J (2015) Overexpression of a peach CBF gene in apple: a model for understanding the integration of growth, dormancy, and cold hardiness in woody plants. Front Plant Sci 6:85
Wu R, Walton EF, Richardson AC, Wood M, Hellens RP, Varkonyi-Gasic E (2012) Conservation and divergence of four kiwifruit SVP-like MADS-box genes suggest distinct roles in kiwifruit bud dormancy and flowering. J Exp Bot 63:797–807
Wu R, Wang T, McGie T, Voogd C, Allan AC, Hellens RP, Varkonyi-Gasic E (2014) Overexpression of the kiwifruit SVP3 gene affects reproductive development and suppresses anthocyanin biosynthesis in petals, but has no effect on vegetative growth, dormancy, or flowering time. J Exp Bot 65:4985–4995
Yamane H, Tao R, Ooka T, Jotatsu H, Sasaki R, Yonemori K (2011) Comparative analyses of dormancy-associated MADS-box genes, PpDAM5 and PpDAM6, in low- and high-chill peaches (Prunus persica L.). J Jpn Soc Hortic Sci 80:276–283
Zeng Y, Yang T (2002) RNA isolation from highly viscous samples rich in polyphenols and polysaccharides. Plant Mol Biol Rep 20:417a–417e
Zhebentyayeva T, Fan S, Chandra A, Bielenberg DG, Reighard GL, Okie WR, Abbott AG (2014) Dissection of chilling requirement and bloom date QTLs in peach using a whole genome sequencing of sibling trees from an F2 mapping population. Tree Genet Genomes 10:35–51
Acknowledgments
We would like to thank the “Empresa de Pesquisa Agropecuária do Estado de Santa Catarina” (EPAGRI, Caçador, SC, Brazil, especially to M.Sc. Frederico Denardi and Dr. Marcus V. Kvitschal), Mr. Décio Amorim (Papanduva, SC, Brazil), and Dr. João Caetano Fioravanço (Embrapa Uva e Vinho, Vacaria, RS, Brazil) for providing us access to apple orchards. We also thank Carolina Pereira Silveira for the very helpful discussions and technical assistance. This work was supported by the Empresa Brasileira de Pesquisa Agropecuária (Embrapa), Conselho Nacional de Desenvolvimento Tecnológico (CNPq), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), Financiadora de Estudos e Projetos (FINEP), and Fundação de Amparo à Pesquisa do Estado do Rio Grande do Sul (FAPERGS).
Authors’ contributions
DDP, VSF, PP, RA, FBF, HPS, and LFR performed the samplings in the field. PP and RA harvested the samples under controlled conditions. DDP, VSF, PP, and VB isolated the mRNA and performed the real-time PCR. VSF and PP searched the apple genome for DAM orthologs. DDP generated and sequenced the RACE products and performed the graphic alignments and collinearity analyses. VSF constructed the phylogenetic trees. RAA performed the cis-element and gene ontology analyses. DDP, VSF, RAA, and LFR wrote the manuscript. HPS, FBF, PRDO, and LFR conceived of and designed the experiments. All authors read and approved the final manuscript.
Data archiving statement
The nucleotide sequences generated in this work were submitted to the GenBank database under accession numbers KT582786, KT582787, KT582788, and KT582789 (Online Resource 5).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by V. Decroocq
Electronic supplementary material
Below is the link to the electronic supplementary material.
Online Resource 1
Gene composition of the apple DAM-containing genome blocks collinear to the peach EVG locus. The gene pairs were established by the MCScanX program. (PDF 99 kb)
Online Resource 2
Primers used for real-time PCR and generation/sequencing of RACE products. (PDF 29 kb)
Online Resource 3
Alignments of the coding sequences predicted in the genome with those obtained from RACE. (PDF 63 kb)
Online Resource 4
Sequence alignment between genomic and RACE sequences of MdDAM3. (PDF 678 kb)
Online Resource 5
Accession codes of DAM predicted proteins used in the phylogenetic and motif analyses. (PDF 43 kb)
Online Resource 6
Graphical alignments between the peach EVG locus (upper bars, reference) and MdDAM2 (MDP0000259294), MdDAM3 (MDP0000527190), MdJa (MDP0000233948) and MdJb (MDP0000209705). Each line linking the two bars indicates the significant reciprocal similarity of an 80 bp sequence window for the E value cutoff indicated inside the image. Dashed red lines have the same meaning but denote inverted sequences in relation to the reference. The locations of peach and apple DAM genes are indicated by blue arrows. Alongside the arrows, the protein-coding exons and introns are represented as red boxes and lines, respectively. Light blue lines indicate 10 kb intervals. (PDF 357 kb)
Online Resource 7
Construction of the “metacontig” containing the whole coding region for MdDAM4. Two contigs containing the MdDAM4 gene were assembled into one, and the fusion region is shown in red. The last 910 bp of the MDC009761.283 contig aligned with the first 910 bp of the complementary sequence from the MDC027822.64 contig, except for some nucleotides (blue amidst the red region) that are recorded in the apple genome in the form of single nucleotide polymorphisms. (PDF 461 kb)
Online Resource 8
Multiple alignment of intronic regions containing the conserved sequence for peach DAM genes, MdDAM1, MdDAM2 and MdDAM3, and three other apple DAM-like genes. The conserved noncoding sequence found in this work and used for the searches in rosaceous genomes is indicated in the red rectangle. Sequences considered highly conserved are highlighted inside green boxes. (PDF 589 kb)
Online Resource 9
Relative levels of transcript accumulation for MdDAM genes in different tissues and organs. The codes A (closed buds), C (buds with green tips), E2 (flower buds), I (10 mm diameter fruits), and J (40 mm diameter fruits) designate phenological stages according to the Fleckinger scale (EPPO 1984). ‘M’ stands for mature fruits. LV, leaves; IN, inflorescences; FR, fruit; PU, fruit pulp; SK, fruit skin; S, seeds of mature fruits. Relative transcript levels in closed terminal buds (A) were set to 1. Bars are standard errors of the mean (n = 3). (PDF 579 kb)
Online Resource 10
List of all cis-elements found in the promoter region of MdDAM genes (up to 1.5 kb upstream from start codon). Cis-element names have links to web pages presenting their description. (PDF 317 kb)
Online Resource 11
List of all cis-elements found in the largest introns of MdDAM1–4 and in the conserved intronic region of MdDAM1. Cis-element names have links to web pages presenting their description. (PDF 644 kb)
Online Resource 12
List and GO analysis of the genes containing the CBF1 cis-element in the promoter region. GO codes in REVIGO results have links to web pages presenting their description. (PDF 122 kb)
Online Resource 13
List and GO analysis of the genes containing the LTREATLI78 cis-element in the promoter region. GO codes in REVIGO results have links to web pages presenting their description. (PDF 323 kb)
Online Resource 14
List and GO analysis of the genes containing the ARR10 cis-element in the promoter region. GO codes in REVIGO results have links to web pages presenting their description. (PDF 213 kb)
Rights and permissions
About this article
Cite this article
Porto, D.D., da Silveira Falavigna, V., Arenhart, R.A. et al. Structural genomics and transcriptional characterization of the Dormancy-Associated MADS-box genes during bud dormancy progression in apple. Tree Genetics & Genomes 12, 46 (2016). https://doi.org/10.1007/s11295-016-1001-3
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11295-016-1001-3