Abstract
Most organisms have evolved specific mechanisms to respond to changes in environmental conditions such as light and temperature over the course of day. These periodic changes in the physiology and behaviour of organisms, referred to as circadian rhythms, are a consequence of intricate molecular mechanisms in the form of transcription and translational feedback loops. The plant circadian regulatory network is a complex web of interconnected feedback loops involving various transcription factors such as CCA1, LHY, PRRs, TOC1, LUX, ELF3, ELF4, RVE8, and more. This network enables plants to adapt and thrive in diverse environmental conditions. It responds to entrainment signals, including light, temperature, and nutrient concentrations and interacts with most of the physiological functions such as flowering, growth and stress response. Mathematical modelling of these gene regulatory networks enables a deeper understanding of not only the function but also the perturbations that may affect the plant growth and function with changing climate. Over the years, numerous mathematical models have been developed to understand the diverse aspects of plant circadian regulation. In this review, we have delved into the systematic development of these models, outlining the model components and refinements over time. We have also highlighted strengths and limitations of each of the models developed so far. Finally, we conclude the review by describing the prospects for investigation and advancement of these models for better understanding of plant circadian regulation.
Key message
Plant circadian rhythms are regulated by a complex network of interactions. This review summarizes the mathematical models that have been developed until now to explore plant circadian rhythms.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Circadian rhythms, the self-sustaining biological cycles that follow an approximately 24-h period, play a crucial role in defining plant fitness and several other traits, including the precise control of flowering, crop yield, biomass accumulation and senescence (Fukushima et al. 2009; Graf et al. 2010; Kim et al. 2018). Environmental cues such as light, temperature, nutritional availability entrain these rhythms in plants, guiding responses to their ever-changing surroundings (Paajanen et al. 2021; Quint et al. 2016; Salter et al. 2003). The plant circadian regulation has been studied extensively in Arabidopsis thaliana and involves multiple transcription factors and E3 ubiquitin ligases that form an interconnected network of molecules controlling the phase, period, and amplitude of circadian rhythms (Nohales and Kay 2016). This regulatory network receives input from environmental cues that entrain and align the circadian phase with the external time (Panter et al. 2019). Light, perceived through photoreceptors such as Phytochromes (Phy)- far-red and red-light receptors, Cryptochrome (Cry), ZEITLUPE (ZTL), LOV KELCH PROTEIN 2 (LKP2), FLAVIN-BINDING, KELCH REPEAT, F-BOX 1 (FKF1)- blue light receptors and UV RESISTANCE LOCUS8 (UVR8)- ultraviolet B receptor, regulates the downstream signalling and affects the physiology (Chen et al. 2004).
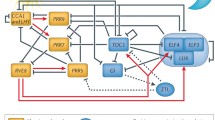
In response to different light conditions, the active form of phytochromes interact with Phytochrome Interacting Factors (PIFs) and regulate multiple genes throughout the day (Martı́nez-Garcı́a et al. 2000). In the morning, with low intensity of red light dominating the light spectrum, Phy C reaches its peak and responds by interacting with Phytochrome Interacting Factor 3 (PIF3), which in turn promotes the expression of downstream genes Circadian Clock Associated 1 (CCA1) and Late Elongated Hypocotyl (LHY) in the central circadian oscillator (Edwards et al. 2015). With changes in the dominant light spectrum from far red to red, Phy D and Phy E become active and their expression peaks. These further interact with PIF4 and PIF5, thus signalling central clock genes and promoting Auxin synthesis (Oakenfull and Davis 2017; Tóth et al. 2001). Further in the noon, under high intensity of sunlight, Phy B and CRY 1 reach their peak expression. Phy B and CRY1 interact with E3 ubiquitin ligase—Constitutive Photomorphogenic 1 (COP1)-Suppressor of Phy A-105 1 (SPA1) complex, thus reducing Elongated Hypocotyl5 (HY5) degradation and promoting photomorphogenesis (Podolec and Ulm 2018; Somers et al. 1998). Towards the evening, as the light intensity decreases, Phy A and CRY 2 become active and assist in the regulation by Phy A interacting with Hypocotyl in Far-Red (HFR1)-PIF3 complex to promote photomorphogenesis even in low light conditions. CRY2 further restricts CONSTANS (CO) degradation through the COP1-SPA1 complex, thus regulating flowering time in plants (Fig. 1) (Podolec and Ulm 2018; Quail et al. 1995; Somers et al. 1998). CCA1 and LHY suppress the expression of transcription factor Timing of CAB expression 1 (TOC1), in the morning (Panter et al. 2019). Further expression of Pseudo Response Regulators (PRR9, PRR7 and PRR5) is sequentially promoted during the day, which interact with B-box 19 (BBX19), a transcription factor with two B-Box motifs, to suppress CCA1 and LHY expression and regulate flowering time (Martı́nez-Garcı́a et al. 2000; Yuan et al. 2021). Towards the evening, expression of Early Flowering 3 (ELF3), ELF4 and LUX is promoted which form a complex known as Evening Complex (EC) and acts as a central hub for crosstalk between circadian and environmental cues by influencing gene expression in photosynthesis, development, and temperature adaptation (Mizuno et al. 2014). At night, the EC suppresses the expression of PRR7, PRR9, and Gigantea (GI). The components of the circadian clock also regulate the expression of EC components, with REVEILLE 8 (RVE8) upregulating ELF4 and LUX expression, TOC1 suppressing ELF4 expression, and CCA1 suppressing ELF3 expression (Fig. 2) (Paajanen et al. 2021).
Light acts as the primary entrainment signal for the plant circadian regulatory network. Phytochrome A & Phytochrome C signals under far-red light, while Phytochrome B, Phytochrome D and Phytochrome E signal under red light conditions. Cryptochrome 1 and Cryptochrome 2 signals under different wavelengths of blue light. These photoreceptors further interact with their downstream components to regulate processes like Flowering, Auxin synthesis and Photomorphogenesis (Oakenfull and Davis 2017)
Plant circadian regulatory network consists of interconnected feedback loops of transcription factors. Different components show rhythmic expression based upon time of day and environmental conditions. CCA1 and LHY form heterodimer (Morning Complex) and show their peak expression near dawn resetting the phase of the rhythm into day/light phase which is further followed by sequential peak expression of PRRs (9, 7 and 5) in the afternoon, towards evening LUX-ELF3-ELF4 form complex (Evening Complex) and reach their peak expression thus initiating resetting of the clock for the night/dark phase. RVE 8,6 and 4 promote expression of PRR7, PRR5, TOC1 and Evening Complex. (Red arrow heads represent transcriptional promotion, green flat heads represent inhibition & black double arrowheads represent protein interactions) (DET1—DE-ETIOLATED 1, TPL—TOPLESS, LWD1/2—LIGHT-REGULATED WD1/2, TCP20/22—Transcription factor TCP20/22, CHE—CCA1 HIKING EXPEDITION) “Created with BioRender.com”
The effect of changes in light intensity on the plant’s circadian clock depends on the timing and duration of the light exposure. A brief light pulse during the subjective night (when the plant would generally be in the dark) can reset the clock to a new phase, while a more prolonged light exposure during the subjective day (when the plant would generally be in the light) has little effect (Ohara et al. 2015a) The timing of the light exposure required to produce a phase shift in the circadian clock, can be studied using the “phase response curve” (PRC) which involves a set of phase shift measurements collected over one cycle of the circadian oscillation (Xu et al. 2021).
The circadian oscillator regulates the sensitivity of environmental signalling pathways, resulting in different magnitudes of responses to the same stimulus at different times of the day (referred to as ‘circadian gating’) (Hotta et al. 2007). Changes in light intensity can also affect the amplitude and period of the circadian rhythm in plants. High light intensity can increase the amplitude of the clock, leading to higher expression of clock-associated genes, while low light intensity can decrease the amplitude. Moreover, changes in light intensity can alter the circadian clock’s period, such that more prolonged light exposure can lengthen the period. In contrast, a shorter light exposure can shorten the period (Oakenfull and Davis 2017).
Understanding the intricate relationship between environmental cues and circadian regulation is essential. It enables us to predict how plants will adapt to future climates and may help promote sustainable agriculture. The impact of circadian regulation extends throughout plant physiology, metabolism, and development. Fundamental processes such as stomatal opening and photosynthesis, as well as overall metabolism, are significantly influenced by the rhythmic patterns of the circadian clock (Paajanen et al. 2021). Notably, the circadian system in plants seamlessly interacts with environmental signals across different levels, ranging from individual cells to different tissues across the plant. By unravelling the intricate web of circadian rhythms in plants, we gain valuable insights into their growth mechanisms.
To summarise, the plant circadian clock is a complex system that regulates many physiological processes in response to changes in environmental cues. Modelling this complex system as network and simulating the behaviour of its various components provides an elegant way to understand the physiology of plants in various conditions.
Mathematical modelling is a tool used to integrate mathematics and physiology, in order to understand the living systems. It has proven valuable in understanding how environmental factors regulate the circadian oscillator (Ohara et al. 2015b, 2015a; Pay et al. 2022a). Mathematical modelling to understand the circadian regulation has incorporated methods including boolean formalism and differential equation based modelling (Akman et al. 2012; Dixon et al. 2011; Fogelmark and Troein 2014; Greenwood et al. 2022; Locke et al. 2005a, b; Pokhilko et al. 2010, 2012). These models have tried to capture the interconnected positive–negative feedback loops in varying details (Dixon et al. 2011; Fogelmark and Troein 2014; Greenwood et al. 2019, 2022; Locke et al. 2005a, b; Pay et al. 2022b; Pokhilko et al. 2010, 2012). Boolean models simplify genes into on/off states and have been successfully used to simulate circadian rhythms (Akman et al. 2012). Similarly differential equation based models have successfully reproduced various responses of the Arabidopsis circadian oscillator to light conditions (Olmo et al. 2021). Both methods have their advantages and disadvantages. In this review we have focused primarily on the differential equation-based models that have been developed for the plant circadian regulation.
Over the course of the last two decades, several models have been used to describe plant circadian regulation in detail through various components. In the following, we review the development of these models.
Review of mathematical models of plant circadian regulation
Single loop model
One of the first models for the plant circadian regulation was proposed by Locke et. al in 2005 (Locke et al. 2005a). They modelled the first multigene loop which was found to be responsible for circadian regulation. This negative feedback loop comprised of TOC1 as transcription factor which acts as activator of LHY and CCA1 while they repress the expression of their own activator (Fig. 3). Acute light activation of LHY and CCA1 was simulated using interaction between a hypothetical light sensitive protein P and LHY gene promoter. Their minimal model with a single feedback loop required seven coupled differential equations describing the degradation of proteins and activation of genes in nucleus as well as in cytoplasm. The protein degradation and protein activation were modelled as Michaelis-Menton kinetics and using Hill functions. The model had 29 parameters and were estimated by generating Sobol quasi-random numbers (Locke et al. 2005a). This model, however, had multiple limitations and failed to reproduce the experimental gene expression patterns during night.
Schematic of single loop model inferred from Locke et al. model. This model consisted of single loop between CCA1/LHY and TOC1, along with their concentration in cytoplasm and nucleus (Locke et al. 2005a) “Created with BioRender.com”
Interconnected loop model
The previous model of the CCA1/LHY-TOC1 network, on its own, was insufficient to fully explain certain aspects of circadian behaviour (Locke et al. 2005a). For example, the short period phenotype observed in the CCA1/LHY loss-of-function mutants could not be explained by the model (Alabadı́ D 2001; Salomé and McClung 2004). Also, the light input was included only by activating the LHY expression during dawn and overlooked the sensitivity to light at the end of light period (Más et al. 2003). Furthermore, the network failed to include a component that lowers TOC1 expression, which was crucial for its proper regulation and resulted in irregular TOC1 expression near dusk (Mizoguchi et al. 2002). These findings highlighted the need for additional components and regulatory mechanisms to enhance the accuracy and completeness of circadian clock models. So, this model was expanded with an interconnected loop. Firstly, light stimulated TOC1 transcription was incorporated into the model in order to account for light input at the dusk and decrease TOC1 activation at the end of light period. This allowed for a more comprehensive representation of light-mediated regulation within the network. Additionally, Gene X was also introduced to the network. The nuclear protein X was hypothesized as the initial activator of LHY, circumventing the direct involvement of nuclear TOC1 in this pathway. This addition provided a more accurate depiction of the regulatory cascade leading to LHY activation. Lastly, to account for the efficient degradation of TOC1 protein during the night, the F-box protein ZEITLUPE (ZTL) was incorporated into the model (Locke et al. 2005b). ZTL has been experimentally demonstrated to be crucial in targeting TOC1 for degradation (Más et al. 2003). By including ZTL, the network could better capture the dynamic fluctuations of TOC1 protein levels throughout the circadian cycle. These refinements in the model addressed the identified issues and contributed to a more comprehensive understanding of the circadian clock system.
In this paper a general strategy for comparing model networks with experimental data was provided to estimate parameter values. The authors used two-parameter optimisation methods in this study. First, the authors employed an exhaustive search strategy to explore the parameter space and identify optimal parameter sets thoroughly. They used modified Sobol algorithm to generate 106 quasi-random points by solving their set of equations, each of which represented a vector of parameter values. By evaluating the cost function for each of these 106 points, the authors comprehensively understood the model’s behaviour across various parameter combinations (Locke et al. 2005b).
Second, the authors employed simulated annealing optimisation to refine their search further and identify the most favourable parameter sets. To begin this optimisation process, they chose the sets of 50 parameter that produced the lowest possible cost function scores. Small, random steps were taken in parameter space around each starting point during simulated annealing. Using this approach, they could converge to the optimal parameter set that was close to the original values of the parameters (Bertsimas and Tsitsiklis 1993).
By combining the exhaustive search approach with simulated annealing optimisation, the authors employed a robust methodology to explore the parameter space, identify promising parameter sets, and ultimately converge on the optimal parameter values for their system. This systematic approach ensured a thorough and comprehensive analysis of the system’s behaviour, leading to valuable insights and a more accurate representation of the underlying processes.
Around the same time as these models were being developed the role of PRRs in suppressing the expression of LHY and activation of PRRs by LHY thus forming a negative feedback loop, was revealed, (Farré et al. 2005), making it essential to add PRRs in the model to study the regulation more precisely.
Interlocked feedback loop with Pseudo Response Regulators
In 2006, two pseudo-response regulators, PRR7 and PRR9, were included in a novel computational model of the Arabidopsis circadian clock (Zeilinger et al. 2006). The researchers employed an iterative model identification procedure to add additional elements and methodically verify various structural possibilities. To determine which network topology best matched the results of the experiments, three iteration steps were conducted. The outcomes showed that a detailed explanation of how light stimulus and network components interact had a substantial impact on the model’s functionality. The model structure consisted of 4 negative feedback loops between TOC1-X-LHY, TOC1-LHY-Y, PRR7-LHY & PRR9-LHY (Fig. 4) (Zeilinger et al. 2006).
Schematic of a Interconnected feedback loop in Locke Model (Locke et al. 2005b) in which they introduced 2 new components X and Y b Interlocked feedback loop in Zeilinger Model (Zeilinger et al. 2006) which expanded the previous Locke model (Locke et al. 2005b) by introducing PRRs (Red arrow heads represent transcriptional promotion, green flat heads represent inhibition and yellow flashes indicate light activation) “Created with BioRender.com”
The model highlighted that by incorporation of PRR7 and PRR9, its ability to predict gene expression patterns under different environmental conditions can be improved. It also showed that PRR9 plays a role in regulating the phase and amplitude of morning gene expression, contributing to the overall rhythmicity of the circadian clock (Farré et al. 2005).
The parameter optimisation method used in this model was based on minimising the cost function. The researchers also conducted a sensitivity analysis to ensure that their model was robust and not too sensitive to the changes in parameter values. This enabled them to identify the parameters that had the most significant impact on system performance and adjust them accordingly. This approach allowed researchers to develop a highly accurate mathematical model of the Arabidopsis circadian clock that incorporated additional parameters and accurately predicted gene expression under various environmental conditions.
Locke et al. also extended their previous model to incorporate PRR9 and PRR7 as a single component making it a three-loop model (Locke et al. 2006). Using this model, they confirmed GI as major contributor to functions of component Y, but further experimental data showed that it alone could not perform the complete function of component Y (Martin-Tryon et al. 2007).
Further advances made it more evident that additional feedback loops may be required to be included in the model. Especially, dusk regulation was critical for regulating photoperiod in plants, but no model had incorporated it.
An expanded three-loop circuit
To incorporate new experimental discoveries and data, Pokhilko et al. in 2010 introduced an updated three-loop network (Pokhilko et al. 2010). It provided a deeper understanding of how the clock responds to various environmental and genetic disturbances. Interestingly, they separated GI from Y and considered GI’s function in stabilizing ZTL while individually modelling GI and ZTL’s roles.
Additionally, they also added a new component known as the Night Inhibitor (NI), which controls LHY/CCA1 expression. PRR5 was suggested by the researchers as a potential member of the NI, highlighting its important function in regulating the morning gene expression phase. (Farré et al. 2005; Nakamichi et al. 2010).
Furthermore, Pokhilko et al. also removed component X, responsible for activation of CCA1/LHY from the model. A post-translationally modified form of TOC1 (TOC1mod) was introduced instead, for activation of CCA1/LHY. This was assumed to promote LHY/CCA1 transcription in addition to light (Pruneda-Paz et al. 2009). However, PRR9, PRR7, and a “night” inhibitor (NI) that stands for PRR5 hindered the process. Moving on to post-translational modifications, LHY/CCA1 translation was selectively stimulated by light, and later in the day, post-translational changes increased the presence of modified protein (LHYmod). In contrast, LHY/CCA1 mRNA degrades faster in the light than in the dark. The stability of TOC1mod was yet another crucial factor. The delay between TOC1 expression at dusk and the subsequent activation of LHY/CCA1 expression around dawn was assumed to be caused by the fact that it was more stable under darkness.
Regarding protein kinetics and interactions, the ZTL protein facilitated the translation of the TOC1 protein and its targeted destruction (Kim et al. 2007; Más et al. 2003). In the dark, this deterioration was further accelerated. The light was also required to form and dissociate the GI-ZTL complex, with light stabilising the ZTL protein. The acute light-induced transcription of LHY/CCA1, PRR9, GI, and Y after dawn depended on the presence of protein P. Finally, the proteins PRR9, PRR7, and NI degrade more quickly in the dark.
The model successfully described the complex mechanisms involving post-translational modifications, transcription, protein dynamics, and interactions between LHY/CCA1, TOC1, Y, PRR9, PRR7, NI, GI, and ZTL components of the system (Pokhilko et al. 2010).
In this model, the genes involved in the circadian clock system were categorised into three groups: the morning-expressed genes (MEGs), the evening-expressed genes (EEGs), and the central oscillator genes (COGs). The MEGs consisted of LHY/CCA1, PRR9, PRR7, and NI, while the EEGs included TOC1, Y, GI, and ZTL (Fig. 5) (Pokhilko et al. 2010). These genes interacted through a series of regulatory interactions, forming a complex network. The COGs acted as central regulators that integrated the signals from the MEGs and EEGs to generate the overall rhythm of the circadian clock.
Schematic of a Pokhilko et al (2010) Model in which they introduced PRR9, PRR7, NI, GI and ZTL. Also, hypothetical component X from previous model was replaced with TOC1mod. b Pokhilko (2012) Model in which they modified and expanded the previous model by removing hypothetical component Y and TOC1mod while adding Evening Complex (consisting of ELF3, ELF4 & LUX) and COP1 (Red arrow heads represent promotion, green flat heads represent inhibition, yellow flashes indicate light activation and small yellow circles indicate post-translational regulation by light.) (Pokhilko et al. 2010, 2012) “Created with”. BioRender.com”
By considering the transcriptional, translational, and degradation rates, the model provides a quantitative understanding of how the MEGs, EEGs, and COGs interact and contribute to the generation of circadian rhythms.
In 2013, Pokhilko et al. further modified this model by including the negative transcriptional regulations of the core clock genes by TOC1 and the up-regulation of TOC1 expression by ABA signalling. They also added post-translational regulation of TOC1 and the evening complex by GI, ZTL and COP1 (Pokhilko et al. 2013).
Later, a significant breakthrough came in 2011 when a study demonstrated that ELF3, ELF4, and LUX form a complex (referred to as Evening Complex or EC) that binds to the promoters of target genes (Nusinow et al. 2011).
Multi-loop model
As research advancements uncovered new components and provided a better understanding of previously known components, Pokhilko et al. developed an updated version of their previous model, incorporating the newly discovered mechanisms into their framework (Dixon et al. 2011; Helfer et al. 2011; Pokhilko et al. 2012). The model included multiple loops with intricate positive and negative interactions. They modified the evening loop to incorporate the evening complex genes and added post-translational control of the ELF3 protein through COP1 (E3 ubiquitin ligase) (Fig. 5) (Yu et al. 2008).
While GI retained certain functions consistent with the previously studied gene Y (Locke et al. 2005a, b), the researchers found that GI appeared to act more as a modulator than the primary effector. Consequently, they removed the hypothetical gene Y from the network and added the necessary clock elements, namely COP1, LUX, ELF3, and ELF4, to the evening circuit further connecting it to GI (Pokhilko et al. 2012). This modification resulted in evening loop being represented more realistically.
In this new network, the light-dependent degradation of the EC component ELF3 mediated light input, which potentially involved the participation of COP1 and another related E3 ubiquitin ligase (Pokhilko et al. 2012). By incorporating these refinements and mechanisms into the model, the researchers aimed to provide a more comprehensive and accurate representation of the circadian regulatory network, considering both genetic and post-translational regulatory elements and their interactions with light input.
This study looked at how different parameters affected the characteristics of the simulated circadian clock model. These factors affected the clock’s amplitude and period under various light conditions and the period of mutant strains.
Five parameters were restricted using wild-type expression profiles and the time course of a particular mutant strain. In contrast, four parameters were produced from experimental observations and confined based on available data. Multiple data types were fitted to the remaining 16 parameters. Interestingly, while the sensitive factors impacted many different clock model components, only a small percentage of them were connected to the unidentified component Y.
In conclusion, the study clarified the sensitivity of several factors in a simulated circadian clock model. The importance of night regulators, as well as the rates of mRNA and protein synthesis, degradation, and inhibition for different clock features, were emphasised.
Further research in upcoming years identified transcription factor REVEILLE 8 (RVE8) playing a crucial role in regulating these circadian rhythms networks. It was determined that RVE8 regulates the expression of PRR9, PRR5, TOC1, GI, LUX and ELF4 (Rawat et al. 2011).
Extended multi-loop model
The Pokhilko et al. (2010 and 2012) models were extended and revised in the Fogelmark et al. (2014) model of the Arabidopsis circadian clock. This model made significant alterations by eliminating all transcriptional activation and adding two additional elements—the morning-expressed activator RVE8 and the night repressor/activator NOX/BROTHER OF LUX ARRHYTHMO (NOX/BOA) (Fig. 6) (Fogelmark and Troein 2014).
Schematic of extended multi-loop model, as explained in Fogelmark Model (Fogelmark and Troein 2014). They added RVE8 and replaced NI with NOX from the models. (Red arrow heads represent promotion, green flat heads represent inhibition & black double arrowheads represent protein interactions, yellow flashes indicate light activation and small yellow circles indicate post-translational regulation by light.) “Created with BioRender.com”
The transcription factor RVE8 was known to positively regulate the expression of a significant number of clock genes (Dai et al. 2011; Hsu et al. 2013; Rawat et al. 2011). Interestingly, all six targets of RVE8 in the model (PRR9, PRR5, TOC1, GI, LUX, and ELF4) were also binding targets of TOC1, which primarily acted as a repressor in the model. The transcription factor NOX had been discovered to work as a promoter of CCA1 (Helfer et al. 2011). Like LUX, NOX displayed a peak expression in the evening and shared a DNA binding domain. Although its function was only slightly redundant with LUX since LUX showed a more robust clock phenotype, it formed a complex with ELF3 and ELF4 (Nusinow et al. 2011). The authors hypothesised that the evening complex contained transcriptional repressors for NOX and LUX. This model offered insightful information about the regulatory dynamics of the clock system by including RVE8 and NOX as crucial elements and modifying the transcriptional activation mechanisms.
In contrast to the Pokhilko et al. (2012) model, several elements and interactions in this model were eliminated for lack of experimental support or because they were computationally unnecessary. They eliminated direct light-dependent degradation rates for PRR9, PRR5, TOC1 proteins and CCA1 and LHY mRNA. For PRR5, they used ZTL-dependent deterioration to replace the light input. Additionally, they disregarded the possibility that GI’s direct transcriptional response to light existed because COP1’s ability to break down EC was sufficient to account for the morning-related increase in GI transcription.
The LHYmod, hypothetical modified form of LHY/CCA1, which was present in the Pokhilko models, was also eliminated. The elimination of few parameters and clock components balanced the addition of new ones like NOX and RVE8, as well as the separation of CCA1 and LHY. As a result, compared to the Pokhilko model, this model had fewer parameters.
This work successfully showed that an oscillating system model might be fitted utilising a large dataset without the use of a complex cost function based on qualitative elements of the model output. Instead, they used extensive time course data and a direct comparison of simulations to constrain the model’s parameters. This streamlined strategy offered the additional benefit of including time courses that span changes in environmental conditions. The model successfully captured the fundamental dynamics of the circadian clock system and its reaction to environmental changes, demonstrated by faithful recreation the clock’s phase after such transitions. This approach highlights the importance of considering transient behaviour and environmental transitions when studying the circadian clock.
For parameter fitting, the model used all available time course data. However, separately acquired period data from clock period mutants were used to verify the model’s accuracy. The results of the investigation showed a significant agreement between experimental data and the majority of the model’s predictions under constant light (LL) circumstances. With the elf4 mutant, there was one exception, as the predicted period change happened in the opposite direction from what was previously shown in the experiments. Despite a discrepancy in the case of the elf4 mutant, the model overall showed reliable performance and offered insightful information (Fogelmark and Troein 2014).
All these comprehensive models discussed so far have been developed and modified with the aim of incorporating as many components as possible of the plant circadian regulatory network. But none of these models accounted for the differential expression of this network across different tissue with respect to the intensity of light exposure they receive.
Multi-loop model with spatial approach
Bordage et al. aimed to explore the molecular mechanisms underlying the circadian rhythms in mature shoots and roots of Arabidopsis (Bordage et al. 2016). They created an imaging system to track clock gene expression and adapted Pokhilko et al.’s (2012) mathematical model of the Arabidopsis clock to reproduce their experimental results.
The results revealed distinct rhythmic properties and responses to light between shoot and root circadian clocks. Notably, the root clock exhibited entrainment to low-intensity light, even when out of phase with shoot illumination. These variations were more noticeable in the presence of light and remained even in the availability of sucrose. By modifying the light-related parameters in the clock model, the authors were able to recreate the observed discrepancies and overlaps between shoot and root clock. Based on these findings, the study shed light on the field of organ-specific clocks and circadian rhythm regulation in different parts of plants. They estimated the model parameters by varying them both individually and in combination, over a wide range, and then conducted a more detailed exploration within a narrower range (Bordage et al. 2016). By iteratively adjusting the parameters and evaluating the simulated data, the researchers successfully refined their model to closely match the observed characteristics of the circadian clocks in shoots and roots under different lighting conditions.
Other models
Apart from the models discussed above, several other models have been developed to understand the plant circadian regulatory network with diverse objectives.
In 2009, Salazar et al. extended Locke’s single loop and interlocking loop models by adding CONSTANS (CO) and FLOWERING LOCUS T (FT). They demonstrated that FLAVIN, KELCH, F-BOX (FKF1) activates FT independent of known activation of CO by FKF1. Thus, locating a novel and major controller of photoperiodism in Arabidopsis (Salazar et al. 2009).
In 2012, Song et al. further extended this model by adding CYCLING DOF FACTOR 1 (CDF1) mediated promotion of FT to demonstrate that FKF1 induces FT mRNA production through multiple feedforward mechanisms thus regulates flowering time (Song et al. 2012). In 2013, Keily et al. modified Pokhilko et al. 2012 model and added C-REPEAT BINDING FACTOR 3 (CBF3) to demonstrate the crucial role of clock in temperature signal transduction in Arabidopsis (Keily et al. 2013).
In 2012, Akman et al. introduced first Boolean models of plant circadian networks (Akman et al. 2012). They constructed Boolean counterparts of the differential equation-based models and showed Boolean models as being more efficient in reproducing high order properties like photoperiod responses.
In 2015, Ohara et al. modified Locke’s single loop model to understand the functioning of the model under different light conditions (i.e., Red, Blue, and Red & Blue). In this model, instead of using binary value (i.e. either 0 or 1) they kept light input at 0.1 value for constant red light condition and changed its value for duration the stimulus is provided based on wavelength of light stimulus. In a separate model they also tried to analyse phase response curve with varying light intensity and wavelength. They introduced wavelength specific light input by modifying light stimulus from single value to combination of three different stimulus from three different photoreceptors (i.e. Phy A, Phy B and Cry1). The stimulus from different photoreceptors were dependent upon wavelength of light input (i.e. Red and Blue) and its intensity (Ohara et al. 2015a, 2015b).
In 2016, Foo et al. made an ODE based model consisting of 24 ODEs and 40 interactions between components of the model based on publicly available time series data of mRNA and protein expression. Different types of regulations (like post-translational, transcriptional, and light based) considered in model were primarily based on experimentally verified knowledge. The study identified four major negative feedback loops as the essential components referred to as “kernel” that were required for modelling the wildtype molecular rhythms. They also postulated the role of additional components in responding to diverse conditions such as different light conditions (Foo et al. 2016).
In 2016, DeCaluwe et al. made a reduced model in which they selected eight components of the network (CC1, LHY, PRR9, PRR7, PRR5, TOC1, ELF4 and LUX) and reduced them into four components by combining them sequentially and assuming them as single components (De Caluwé et al. 2016). These assumptions lead to reduced number of equations and parameters. This model has been further extensively explored with different perspectives for spatial modelling by Greenwood et al. They tried to analyse phase difference and delay in phase-resetting over different tissues of plant (i.e. leaf, shoot and root) by introducing light sensitivity of different tissues and local coupling to generate spatial waves (Greenwood et al. 2019, 2022).
In 2018, Joanito et al. used several versions of reduced model to identify incoherent feed-forward switches which help clock network to rapidly switch between different states (Joanito et al. 2018). Same year, Ohara et al. further modified the DeCaluwe reduced model to incorporate sugar responses to the core circadian components. They analysed mechanisms, target genes and effects of sugar signalling in Arabidopsis circadian network (Ohara et al. 2018). In 2022, Huang et al. extended the DeCaluwe 2016 model by adding an extra input of red light and additional regulations such as self-inhibition of CCA1/LHY and replaced activation of PRR9/PRR7 by CCA1/LHY to inhibition in the network. Their aim was to characterise the effects of light quality on plant circadian rhythms (Huang et al. 2022).
In 2022, Pay incorporated DeCaluwe model with COP1 along with 3 photoreceptors (Phy A, Phy B and CRY1) using similar approach to that of Ohara model. They further analysed the model under different light conditions (i.e., Red, Blue, and Red & Blue) in order to identify light condition most favourable for hypocotyl growth (Pay et al. 2022a). Further, they extended their model to incorporate the effect of various light qualities on the flowering time (Pay et al. 2022b).
In 2022, Chew et al. developed Arabidopsis framework model by combining several previous submodels to anticipate the impact of circadian clock gene misregulation on overall plant physiology. The model’s modular structure incorporated detailed submodels of clock genes PRR7, PRR9, and output pathway genes, enabling precise simulations of flowering time and various phenotypic outcomes in Arabidopsis. They integrated starch metabolism and light signalling pathways to explain complex molecular interactions in plants. By integrating interactions among the clock, photoperiod, phenology, and functional-structural submodels, they illustrated the interconnected regulatory processes governing plant development. Through simulations, the model accurately reproduced the altered dynamics of clock gene expression in mutant plants, offering valuable insights into the molecular mechanisms of circadian regulation and its influence on plant growth (Chew et al. 2022).
In a recently published model by Chan et al. 2024, they constructed an ODE based model analysing effects of various light properties (photoperiod, intensity, light–dark order) over Arabidopsis thaliana growth and development using days-to-flower (DTF) and hypocotyl growth as measures. It consisted of 27 ODEs and light as input, while growth-related circadian gene expression as output. The expression of these genes was used to compute hypocotyl length and DTF. They demonstrated that not only the photoperiod but ratio of combination of lights as well as their order can affect hypocotyl growth and DTF of plants. Also, the same combination of these properties can have different effects on hypocotyl growth and DTF. Thus, concluding optimum combinations of these properties separately for hypocotyl growth and DTF (Chan et al.2024).
Future prospects in plant circadian modelling
While remarkable progress has been made in recent years in the complex world of plant circadian biology, many of the knowledge gaps of this complex network continue to defy our understanding.
With changing climate, it is pertinent to understand the cross talk between circadian signalling and effect of temperature. Although, there have been some studies that have tried to model the effect of change in temperature on the circadian regulation (Avello et al. 2019), change in day length along with change in temperature (at 22 °C and 27 °C) on plant development (Seaton et al. 2015) and change in various aspects of light signal (photoperiod, phase of dawn, light offset, light intensity, duration of twilight and period) with constant temperature (22 °C) over hypocotyl length and flowering time (Pereira et al. 2021), a comprehensive mathematical model that captures the effects of intra-day variation in light and temperature is still missing. Such a model can account for seasonal changes in environmental cues for plants and may pave way for better management strategies in the face of climate change (Panter et al. 2019). The future modelling studies may also focus on connecting the circadian and seasonal rhythms that has huge implications in crop yield. A recent study by Nagano et al. showed that significant change in day length along with temperature is observed throughout the year. Although, there is a lag between change in day length and temperature changes still annual temperature changes have greater influence over seasonal oscillations of gene expressions as compared to change in day length (Nagano et al. 2019). Furthermore, most of the mathematical models for plant circadian regulation have focused on model plant, Arabidopsis. Though crucial for fundamental understanding of the physiological process, with more data being generated for various crops, it becomes important to develop models for other more economically and socially relevant plants. This is particularly important as it has already been established that circadian rhythms affect several agricultural traits (Bendix et al. 2015).
Recently several new interactions between the components plant circadian network have been identified. Some of these include- the interaction of BBX19 and TPL with PRRs to assist in their transcriptional activities (Yuan et al. 2021), multiple interactions of CRY2 with PHYB, COP1-SPA1 complex and CIB1 (Lopez et al. 2021), the interaction of LNKs with RVEs which helps in targeting promoters (Ma et al. 2018), RVEs interact with TOC1, PRR5 and PRR7 and provide DNA binding specificity (Ma et al. 2018).
Incorporation of these new interactions in the mathematical models may provide more accurate description of the circadian rhythms in diverse conditions.
Although blue light regulates and aids in several critical functions in plant development, such as flowering time, auxin synthesis and photomorphogenesis, models to date have only incorporated CRY1 and no other factors involved in blue light signalling such as CRY2 and ZTL (Kim et al. 2007; Ohara et al. 2015a; Pay et al. 2022a). Several models have been successful in accounting for different wavelengths of light but not intensity with respect to the time of the day. These models have separated white light into blue and red light, thus allowing Phytochrome A, Phytochrome B and CRY1 (Ohara et al. 2015a; Pay et al. 2022a). The role of other photoreceptors such as PHYC, PHYD, PHYE and CRY2 have not been included yet in any model and needs to be explored in the context of changing light intensity and wavelengths.
Limitations
Model complexity
The ODE-based models have been able to predict the behaviour of the network well, but there are some limitations as we increase the number of components and interactions. One such limitation is the complexity of the model and the dependency of the model on computationally derived parameters. Parameters such as rate of transcription, rate of inhibition, rate of translation and so on are challenging to derive experimentally. Thus, their values need to be estimated through parameter estimation methods using the available experimental data. To overcome this limitation recently, S-formalism has been introduced to the ODE-based models (Foo et al. 2020). This technique was crafted with intricate biochemical networks in mind and offered a comparatively more seamless means of incorporating supplementary regulators into the model.
Data availability
Mathematical modelling is dependent upon the experimental data available. These data are required from parameter optimisation to validation of the model under different conditions. In case of mathematical modelling of plant circadian regulation, primarily expression profiles of various network components are utilized. With advances in sequencing technologies, although there is considerable amount of expression data now available in the public databases, not all of that is helpful for modelling purposes. Often, there is a need to integrate the data from multiple experiments that poses its own challenges, such as accounting for different experimental conditions, sample collection time, experimental errors etc. Higher data availability improves the accuracy of models in precisely predicting the observable under different conditions. More experiments are thus required with varying photoperiods, temperatures, and varying combinations of them for more complex and realistic modelling of the plant circadian regulation.
Overfitting
The problem of overfitting looms big in any mathematical modelling approach especially when dealing with sparse data. When a model is overfitted to the sparse data set, it captures noise and anomalies rather than the underlying patterns. As a result, a model may perform exceptionally well on the already available data but may fail to explain new, unobserved data. Overfitting becomes a real worry in plant circadian regulation networks since the underlying mechanisms are intricate and complex, possibly compromising the model’s predictive accuracy and limiting its capacity to offer insightful information (Locke et al. 2005a). Therefore, achieving an accurate and trustworthy mathematical description of plant circadian regulatory networks requires careful balance between capturing fundamental dynamics and reliance on sparse data.
Conclusion
There have been continuous advancements in the mathematical modelling of plant circadian regulatory networks, parallel to the experimental exploration of the network components and increased availability of expression data. The models have increased in complexity from a single-loop model in 2005 to interconnected model with multiple feedback loops in 2014 by incorporating new experimental findings and data (Table 1).
With single cell models reaching enough explanatory power with respect to the available experimental data, the focus, in recent years has shifted towards spatial models which try to capture tissue specific heterogeneity in plant circadian responses. One possible reason for that can be an increase in the complexity of the network and computational dependency for parameter optimisation. New approaches and methods must be designed to overcome these limitations, as exploring this network in more detail might provide solutions for contemporary problems of climate change and food security by sustainable farming.
A deeper understanding of how plants perceive and respond to light cues can improve agricultural efforts and crop yields. Deciphering the complex pathways governing plant physiology is the prerequisite for targeted strategies for crop growth. Examining the system’s behaviour under different light conditions, seasons, and locations can provide valuable insights into vegetation dynamics and identify specific agricultural behaviour. Scientists armed with a comprehensive understanding of plant circadian communication can strengthen the sustainability of agriculture, opening new pathways and constantly consuming the growing demands of our global population.
References
Akman OE, Watterson S, Parton A, Binns N, Millar AJ, Ghazal P (2012) Digital clocks: simple boolean models can quantitatively describe circadian systems. J R Soc Interface 9(74):2365–2382. https://doi.org/10.1098/rsif.2012.0080
Alabadı́ D, Oyama T, Yanovsky MJ, Harmon FG, Más P, Kay SA, (2001) Reciprocal regulation between TOC1 and LHY / CCA1 within the Arabidopsis circadian clock. Science 293(5531):880–883. https://doi.org/10.1126/science.1061320
Avello PA, Davis SJ, Ronald J, Pitchford JW (2019) Heat the clock: entrainment and compensation in Arabidopsis circadian rhythms. J Circadian Rhythms 17(1):5. https://doi.org/10.5334/jcr.179
Bendix C, Marshall CM, Harmon FG (2015) Circadian clock genes universally control key agricultural traits. Mol Plant 8(8):1135–1152. https://doi.org/10.1016/j.molp.2015.03.003
Bertsimas D, Tsitsiklis J (1993) Simulated annealing. Stat Sci 8(1):10–15. https://doi.org/10.1214/ss/1177011077
Bordage S, Sullivan S, Laird J, Millar AJ, Nimmo HG (2016) Organ specificity in the plant circadian system is explained by different light inputs to the shoot and root clocks. New Phytol 212(1):136–149. https://doi.org/10.1111/nph.14024
Chan AMH, Pay ML, Christensen J, He F, Roden LC, Ahmed H, Foo M (2024) Red, blue or mix: choice of optimal light qualities for enhanced plant growth and development through in silico analysis. In Silico Plants 6(1):008. https://doi.org/10.1093/insilicoplants/diae008
Chen M, Chory J, Fankhauser C (2004) Light signal transduction in higher plants. Annu Rev Genet 38(1):87–117. https://doi.org/10.1146/annurev.genet.38.072902.092259
Chew YH, Seaton DD, Mengin V, Flis A, Mugford ST, George GM, Moulin M, Hume A, Zeeman SC, Fitzpatrick TB, Smith AM, Stitt M, Millar AJ (2022) The Arabidopsis framework model version 2 predicts the organism-level effects of circadian clock gene mis-regulation. In Silico Plants 4(2):010. https://doi.org/10.1093/insilicoplants/diac010
Dai S, Wei X, Pei L, Thompson RL, Liu Y, Heard JE, Ruff TG, Beachy RN (2011) BROTHER OF LUX ARRHYTHMO is a component of the Arabidopsis circadian clock. Plant Cell 23(3):961–972. https://doi.org/10.1105/tpc.111.084293
De Caluwé J, Xiao Q, Hermans C, Verbruggen N, Leloup JC, Gonze D (2016) A compact model for the complex plant circadian clock. Front Plant Sci 7:74. https://doi.org/10.3389/fpls.2016.00074
Del Olmo M, Grabe S, Herzel H (2021) Mathematical modeling in circadian rhythmicity. Methods in molecular biology. Springer, Berlin, pp 55–80
Dixon LE, Knox K, Kozma-Bognar L, Southern MM, Pokhilko A, Millar AJ (2011) Temporal repression of core circadian genes is mediated through EARLY FLOWERING 3 in Arabidopsis. Curr Biol 21(2):120–125. https://doi.org/10.1016/j.cub.2010.12.013
Edwards KD, Guerineau F, Devlin PF, Millar AJ, Edwards KD, Guerineau F, Devlin PF, Millar AJ (2015) Low-temperature-specific effects of PHYTOCHROME C on the circadian clock in Arabidopsis suggest that PHYC underlies natural variation in biological timing. bioRxiv. https://doi.org/10.1101/030577
Farré EM, Harmer SL, Harmon FG, Yanovsky MJ, Kay SA (2005) Overlapping and distinct roles of PRR7 and PRR9 in the Arabidopsis circadian clock. Curr Biol 15(1):47–54. https://doi.org/10.1016/j.cub.2004.12.067
Fogelmark K, Troein C (2014) Rethinking transcriptional activation in the Arabidopsis circadian clock. PLoS Comput Biol 10(7):e1003705. https://doi.org/10.1371/journal.pcbi.1003705
Foo M, Somers DE, Kim PJ (2016) Kernel architecture of the genetic circuitry of the Arabidopsis circadian system. PLoS Comput Biol 12(2):e1004748. https://doi.org/10.1371/journal.pcbi.1004748
Foo M, Bates DG, Akman OE (2020) A simplified modelling framework facilitates more complex representations of plant circadian clocks. PLoS Comput Biol 16(3):e1007671. https://doi.org/10.1371/journal.pcbi.1007671
Fukushima A, Kusano M, Nakamichi N, Kobayashi M, Hayashi N, Sakakibara H, Mizuno T, Saito K (2009) Impact of clock-associated Arabidopsis pseudo-response regulators in metabolic coordination. Proc Natl Acad Sci 106(17):7251–7256. https://doi.org/10.1073/pnas.0900952106
Graf A, Schlereth A, Stitt M, Smith AM (2010) Circadian control of carbohydrate availability for growth in Arabidopsis plants at night. Proc Natl Acad Sci 107(20):9458–9463. https://doi.org/10.1073/pnas.0914299107
Greenwood M, Domijan M, Gould PD, Hall AJW, Locke JCW (2019) Coordinated circadian timing through the integration of local inputs in Arabidopsis thaliana. PLoS Biol 17(8):e3000407. https://doi.org/10.1371/journal.pbio.3000407
Greenwood M, Tokuda IT, Locke JCW (2022) A spatial model of the plant circadian clock reveals design principles for coordinated timing. Mol Syst Biol 18(3):e10140. https://doi.org/10.15252/msb.202010140
Helfer A, Nusinow DA, Chow BY, Gehrke AR, Bulyk ML, Kay SA (2011) LUX ARRHYTHMO encodes a nighttime repressor of circadian gene expression in the Arabidopsis core clock. Curr Biol 21(2):126–133. https://doi.org/10.1016/j.cub.2010.12.021
Hotta CT, Gardner MJ, Hubbard KE, Baek SJ, Dalchau N, Suhita D, Dodd AN, Webb AAR (2007) Modulation of environmental responses of plants by circadian clocks. Plant Cell Environ 30(3):333–349. https://doi.org/10.1111/j.1365-3040.2006.01627.x
Hsu PY, Devisetty UK, Harmer SL (2013) Accurate timekeeping is controlled by a cycling activator in Arabidopsis. Elife 2:e00473. https://doi.org/10.7554/eLife.00473
Huang T, Shui Y, Wu Y, Hou X, You X (2022) Red light resets the expression pattern, phase, and period of the circadian clock in plants: a computational approach. Biology 11(10):1479. https://doi.org/10.3390/biology11101479
JF Martı́nez-Garcı́a, E Huq, PH Quail, (2000) Direct targeting of light signals to a promoter element-bound transcription factor. Science 288(5467):859–863. https://doi.org/10.1126/science.288.5467.859
Joanito I, Chu J, Wu S, Hsu C (2018) An incoherent feed-forward loop switches the Arabidopsis clock rapidly between two hysteretic states. Sci Rep 8(1):13944. https://doi.org/10.1038/s41598-018-32030-z
Keily J, MacGregor DR, Smith RW, Millar AJ, Halliday KJ, Penfield S (2013) Model selection reveals control of cold signalling by evening-phased components of the plant circadian clock. Plant J 76(2):247–257. https://doi.org/10.1111/tpj.12303
Kim JY, Song HR, Taylor BL, Carré IA (2003) Light-regulated translation mediates gated induction of the Arabidopsis clock protein LHY. EMBO J 22(4):935–944. https://doi.org/10.1093/emboj/cdg075
Kim WY, Fujiwara S, Suh SS, Kim J, Kim Y, Han L, David K, Putterill J, Nam HG, Somers DE (2007) ZEITLUPE is a circadian photoreceptor stabilized by GIGANTEA in blue light. Nature 449(7160):356–360. https://doi.org/10.1038/nature06132
Kim H, Kim HJ, Vu QT, Jung S, McClung CR, Hong S, Nam HG (2018) Circadian control of ORE1 by PRR9 positively regulates leaf senescence in Arabidopsis. Proc Natl Acad Sci 115(33):8448–8453. https://doi.org/10.1073/pnas.1722407115
Locke JCW, Millar AJ, Turner MS (2005a) Modelling genetic networks with noisy and varied experimental data: the circadian clock in Arabidopsis thaliana. J Theor Biol 234(3):383–393. https://doi.org/10.1016/j.jtbi.2004.11.038
Locke JCW, Southern MM, Kozma-Bognár L, Hibberd V, Brown PE, Turner MS, Millar AJ (2005b) Extension of a genetic network model by iterative experimentation and mathematical analysis. Mol Syst Biol 1(2005):0013. https://doi.org/10.1038/msb4100018
Locke JCW, Kozma-Bognár L, Gould PD, Fehér B, Kevei É, Nagy F, Turner MS, Hall A, Millar AJ (2006) Experimental validation of a predicted feedback loop in the multi-oscillator clock of Arabidopsis thaliana. Mol Syst Biol 2(1):59. https://doi.org/10.1038/msb4100102
Lopez L, Fasano C, Perrella G, Facella P (2021) Cryptochromes and the circadian clock: the story of a very complex relationship in a spinning world. Genes 12(5):672. https://doi.org/10.3390/genes12050672
Ma Y, Gil S, Grasser KD, Mas P (2018) Targeted recruitment of the basal transcriptional machinery by LNK clock components controls the circadian chythms of nascent RNAs in Arabidopsis. Plant Cell 30(4):907–924. https://doi.org/10.1105/tpc.18.00052
Makino S, Kiba T, Imamura A, Hanaki N, Nakamura A, Suzuki T, Taniguchi M, Ueguchi C, Sugiyama T, Mizuno T (2000) Genes encoding pseudo-response regulators: insight into His-to-Asp phosphorelay and circadian rhythm in Arabidopsis thaliana. Plant Cell Physiol 41(6):791–803. https://doi.org/10.1093/pcp/41.6.791
Martin-Tryon EL, Kreps JA, Harmer SL (2007) GIGANTEA acts in blue light signaling and has biochemically separable roles in circadian clock and flowering time regulation. Plant Physiol 143(1):473–486. https://doi.org/10.1104/pp.106.088757
Más P, Kim WY, Somers DE, Kay SA (2003) Targeted degradation of TOC1 by ZTL modulates circadian function in Arabidopsis thaliana. Nature 426(6966):567–570. https://doi.org/10.1038/nature02163
Mizoguchi T, Wheatley K, Hanzawa Y, Wright L, Mizoguchi M, Song HR, Carré IA, Coupland G (2002) LHY and CCA1 are partially redundant genes required to maintain circadian rhythms in Arabidopsis. Dev Cell 2(5):629–641. https://doi.org/10.1016/S1534-5807(02)00170-3
Mizuno T, Nomoto Y, Oka H, Kitayama M, Takeuchi A, Tsubouchi M, Yamashino T (2014) Ambient temperature signal feeds into the circadian clock transcriptional circuitry through the EC night-time repressor in Arabidopsis thaliana. Plant Cell Physiol 55(5):958–976. https://doi.org/10.1093/pcp/pcu030
Nagano AJ, Kawagoe T, Sugisaka J, Honjo MN, Iwayama K, Kudoh H (2019) Annual transcriptome dynamics in natural environments reveals plant seasonal adaptation. Nature Plants 5(1):74–83. https://doi.org/10.1038/s41477-018-0338-z
Nakamichi N, Kiba T, Henriques R, Mizuno T, Chua NH, Sakakibara H (2010) PSEUDO-RESPONSE REGULATORS 9, 7, and 5 are transcriptional repressors in the Arabidopsis circadian clock. Plant Cell 22(3):594–605. https://doi.org/10.1105/tpc.109.072892
Nohales MA, Kay SA (2016) Molecular mechanisms at the core of the plant circadian oscillator. Nat Struct Mol Biol 23(12):1061–1069. https://doi.org/10.1038/nsmb.3327
Nusinow DA, Helfer A, Hamilton EE, King JJ, Imaizumi T, Schultz TF, Farré EM, Kay SA (2011) The ELF4–ELF3–LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature 475(7356):398–402. https://doi.org/10.1038/nature10182
Oakenfull RJ, Davis SJ (2017) Shining a light on the Arabidopsis circadian clock. Plant Cell Environ 40(11):2571–2585. https://doi.org/10.1111/pce.13033
Ohara T, Fukuda H, Tokuda IT (2015a) An extended mathematical model for reproducing the phase response of Arabidopsis thaliana under various light conditions. J Theor Biol 382:337–344. https://doi.org/10.1016/j.jtbi.2015.07.016
Ohara T, Fukuda H, Tokuda IT (2015b) Phase response of the Arabidopsis thaliana circadian clock to light pulses of different wavelengths. J Biol Rhythms 30(2):95–103. https://doi.org/10.1177/0748730415576426
Ohara T, Hearn TJ, Webb AAR, Satake A (2018) Gene regulatory network models in response to sugars in the plant circadian system. J Theor Biol 1(457):137–151. https://doi.org/10.1016/j.jtbi.2018.08.020
Paajanen P, de Barros L, Dantas L, Dodd AN (2021) Layers of crosstalk between circadian regulation and environmental signalling in plants. Curr Biol 31(8):R399–R413. https://doi.org/10.1016/j.cub.2021.03.046
Panter PE, Muranaka T, Cuitun-Coronado D, Graham CA, Yochikawa A, Kudoh H, Dodd AN (2019) Circadian regulation of the plant transcriptome under natural conditions. Front Genet 10:1239. https://doi.org/10.3389/fgene.2019.01239
Pay ML, Christensen J, He F, Roden L, Ahmed H, Foo M (2022a) An extended plant circadian clock model for characterising flowering time under different light quality conditions. 22nd international conference on control, automation and systems (ICCAS), 1848–1853. https://doi.org/10.23919/ICCAS55662.2022.10003836
Pay ML, Kim DW, Somers DE, Kim JK, Foo M (2022b) Modelling of plant circadian clock for characterizing hypocotyl growth under different light quality conditions. In Silico Plants 4(1):001. https://doi.org/10.1093/insilicoplants/diac001
Pereira J, Mouazen AM, Foo M, Ahmed H (2021) A framework of artificial light management for optimal plant development for smart greenhouse application. PLoS ONE 16(12):e0261281. https://doi.org/10.1371/journal.pone.0261281
Podolec R, Ulm R (2018) Photoreceptor-mediated regulation of the COP1/SPA E3 ubiquitin ligase. Curr Opin Plant Biol 45:18–25. https://doi.org/10.1016/j.pbi.2018.04.018
Pokhilko A, Hodge SK, Stratford K, Knox K, Edwards KD, Thomson AW, Mizuno T, Millar AJ (2010) Data assimilation constrains new connections and components in a complex, eukaryotic circadian clock model. Mol Syst Biol 6:416. https://doi.org/10.1038/msb.2010.69
Pokhilko A, Fernández AP, Edwards KD, Southern MM, Halliday KJ, Millar AJ (2012) The clock gene circuit in Arabidopsis includes a repressilator with additional feedback loops. Mol Syst Biol 8:574. https://doi.org/10.1038/msb.2012.6
Pokhilko A, Mas P, Millar AJ (2013) Modelling the widespread effects of TOC1 signalling on the plant circadian clock and its outputs. BMC Syst Biol 7:23. https://doi.org/10.1186/1752-0509-7-23
Pruneda-Paz JL, Breton G, Para A, Kay SA (2009) A functional genomics approach reveals CHE as a component of the Arabidopsis Circadian Clock. Science 323(5920):1481–1485. https://doi.org/10.1126/science.1167206
Quail PH, Boylan MT, Parks BM, Short TW, Xu Y, Wagner D (1995) Phytochromes: photosensory perception and signal transduction. Science 268(5211):675–680. https://doi.org/10.1126/science.7732376
Quint M, Delker C, Franklin KA, Wigge PA, Halliday KJ, van Zanten M (2016) Molecular and genetic control of plant thermomorphogenesis. Nature Plants 2(1):15190. https://doi.org/10.1038/nplants.2015.190
Rawat R, Takahashi N, Hsu PY, Jones MA, Schwartz J, Salemi MR, Phinney BS, Harmer SL (2011) REVEILLE8 and PSEUDO-REPONSE REGULATOR5 form a negative feedback loop within the Arabidopsis circadian clock. PLoS Genet 7(3):e1001350. https://doi.org/10.1371/journal.pgen.1001350
Salazar JD, Saithong T, Brown PE, Foreman J, Locke JCW, Halliday KJ et al (2009) Prediction of photoperiodic regulators from quantitative gene circuit models. Cell 139(6):1170–1179. https://doi.org/10.1016/j.cell.2009.11.029
Salomé PA, McClung CR (2004) The Arabidopsis thaliana Clock. J Biol Rhythms 19(5):425–435. https://doi.org/10.1177/0748730404268112
Salter MG, Franklin KA, Whitelam GC (2003) Gating of the rapid shade-avoidance response by the circadian clock in plants. Nature 426(6967):680–683. https://doi.org/10.1038/nature02174
Seaton DD, Smith RW, Song YH, MacGregor DR, Stewart K, Steel G, Foreman J, Penfield S, Imaizumi T, Millar AJ, Halliday KJ (2015) Linked circadian outputs control elongation growth and flowering in response to photoperiod and temperature. Mol Syst Biol 11(1):776. https://doi.org/10.15252/msb.20145766
Somers DE, Webb AAR, Pearson M, Kay SA (1998) The short-period mutant, toc1-1, alters circadian clock regulation of multiple outputs throughout development in Arabidopsis thaliana. Development 125(3):485–494. https://doi.org/10.1242/dev.125.3.485
Song YH, Smith RW, To BJ, Millar AJ, Imaizumi T (2012) FKF1 conveys timing information for CONSTANS stabilization in photoperiodic flowering. Science 336(6084):1045–1049. https://doi.org/10.1126/science.1219644
Tóth R, Kevei É, Hall A, Millar AJ, Nagy F, Kozma-Bognár L (2001) Circadian clock-regulated expression of phytochrome and cryptochrome genes in Arabidopsis. Plant Physiol 127(4):1607–1616. https://doi.org/10.1104/pp.010467
Xu X, Yuan L, Xie Q (2021) Circadian rhythm: phase response curve and light entrainment. In: Staiger D, Davis S, Davis AM (eds) Plant circadian networks methods in molecular biology. Springer, Berlin, pp 1–13
Yu JW, Rubio V, Lee NY, Bai S, Lee SY, Kim SS, Liu L, Zhang Y, Irigoyen ML, Sullivan JA, Zhang Y, Lee I, Xie Q, Paek NC, Deng XW (2008) COP1 and ELF3 control circadian function and photoperiodic flowering by regulating GI stability. Mol Cell 32(5):617–630. https://doi.org/10.1016/j.molcel.2008.09.026
Yuan L, Yu Y, Liu M, Song Y, Li H, Sun J, Wang Q, Xie Q, Wang L, Xu X (2021) BBX19 fine-tunes the circadian rhythm by interacting with PSEUDO-RESPONSE REGULATOR proteins to facilitate their repressive effect on morning-phased clock genes. Plant Cell 33(8):2602–2617. https://doi.org/10.1093/plcell/koab133
Zeilinger MN, Farré EM, Taylor SR, Kay SA, Doyle FJ (2006) A novel computational model of the circadian clock in Arabidopsis that incorporates PRR7 and PRR9. Mol Syst Biol 2:58. https://doi.org/10.1038/msb4100101
Acknowledgements
SS acknowledges financial support by Indian Institute of Technology Gandhinagar and Ministry of Education, India. AS acknowledges support from DBT Ramalingaswami Re-entry Fellowship, SERB Start up Research Grant and Indian Institute of Technology Gandhinagar.
Funding
This study was funded by DBT Ramalingaswami Re–entry Fellowship, SERB Start up Research Grant.
Author information
Authors and Affiliations
Contributions
SS and AS conceptualized the idea for the article. SS performed the literature search and data analysis. SS wrote the first draft. SS and AS both drafted and critically revised the work.
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Singh, S.K., Srivastava, A. Decoding the plant clock: a review of mathematical models for the circadian regulatory network. Plant Mol Biol 114, 93 (2024). https://doi.org/10.1007/s11103-024-01493-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11103-024-01493-2